Abstract
Objective(s):
The rs712 polymorphism in a let-7 microRNA-binding site at KRAS gene has been associated with cancer. To examine its association with rs712 polymorphism, we analyzed Mexican individuals with colorectal cancer (CRC) and healthy subjects.
Materials and Methods:
Genotyping of the rs712 polymorphism was performed by polymerase chain reaction in 281 controls and 336 CRC patients.
Results:
The observed frequencies of rs712 polymorphism indicated an associated protective factor for CRC (P=0.032). An association between genotype and the disease was evident in: colon localization (allele T, odds ratio (OR) 3.82, 95% confidence Intervals (CI) 2.77-5.28, P=0.0001), node metastasis (genotype TT, OR 2.49, 95% CI 1.45-4.28, P=0.0009), poor differentiation (genotype GT, OR 2.35, 95% CI 1.35-4.1, P=0.0033), and poor chemotherapy response (genotype GT, OR 2.6, 95% CI 1.7-4.24, P=0.0001).
Conclusion:
Comparison of the data from patients with control group showed that polymorphism of rs712 in KRAS gene was protective factor, which was associated with susceptibility for CRC. However, the genotypes TT and GT of rs712 polymorphism in KRAS could contribute significantly to colon localization, node metastasis, poor differentiation and poor chemotherapy response in CRC patients in this sample population.
Key Words: Colorectal cancer, KRAS, let-7, Mexican population, Polymorphism
Introduction
Colorectal cancer (CRC) is a serious public health problem in Mexico and the world (1-3), and its incidence varies between different ethnic groups (2-5). In Mexico, CRC is associated with 4% of cancer dead (6). CRC is thought to develop through a gradual accumulation of genetic changes that could modify the intestinal cells (1, 3). In this sense, many studies have shown the relationship of KRAS gene with CRC, so that the K-Ras protein (first identified in Kirsten rat sarcoma virus) is a part of RAS/MAPK signaling pathway, and their function is through GTPase that acts like switch by converting the active GTP molecule to inactive GDP, which is essential in the cellular signal process that control the growth, maturation and cellular death (7). Two copies of the KRAS gene exist, the pseudogene KRAS1 and KRAS2 gene, which are localized at 12q12.1 chromosome, and contain 6 exons of which the exons 1, 5 and 6 are non-coding, and 4 exon join by alternative splicing to make 2 mRNA transcripts known as isoforms 4A (active) and 4B (inactive) (7, 8). The KRAS protein is a proto-oncogene, which is regulated by proteins that bind to the promoter regions of the gene in the initiation of transcription by microRNAs (miRNAs) molecules that act on the elongation phase of transcription. The miRNAs regulate gene expression in the 3’UTR region of mRNA. These interactions between microRNA and mRNA destabilize the mRNA and repress protein synthesis. MicroRNAs such as let-7, lin-4 and bentam, regulate proliferation and differentiation of the cells, and are altered in cancer. These are divided into two groups: oncomirs (on-regulated) that act as oncogenes and anti-oncomirs, which act as tumor suppressors, targeting oncogenes, repressing the cell cycle and cell division in cancer cells. The microRNA let-7 is an oncogene-anti-oncomir, which regulates the levels of KRAS protein and decreases the rate of cell proliferation (7-9). The rs712 polymorphism that is located in 3′UTR of KRAS mRNA is product of one base change of G to T, and it has been hypothesized that this polymorphism modifies the function of the gene and consequently promotes the cell proliferation and migration in the intestinal mucosa, contributing to CRC carcinogenesis (10). The allele T (rs712) showed a frequency of 16% - 36% among controls Chinese population (11). Also, a significant association has been demonstrated between the rs712 polymorphism and different cancers in some studies (12). However, in the Mexican population, these associations remain unknown. Thus, the objective of this investigation was to evaluate the association of the rs712 polymorphism in a let-7 microRNA-binding KRAS gene in Mexicans with CRC.
Materials and Methods
Study population
In this study, DNA samples from 281 healthy volunteer donor and 336 CRC patients with confirmed diagnosis not matched by age and sex, were collected. DNA samples were collected since 2013 to 2017 as part of genetic library of our laboratory, which has been analyzed for other genetic markers polymorphisms (3). DNA samples from parental familial were excluded. Procedures were in accordance with 1964 Helsinki declaration, and the participant in the study signed written informed consent approved by 1305 ethical committee of Centro de Investigación Biomédica de Occidente, Instituto Mexicano del Seguro Social.
Methods
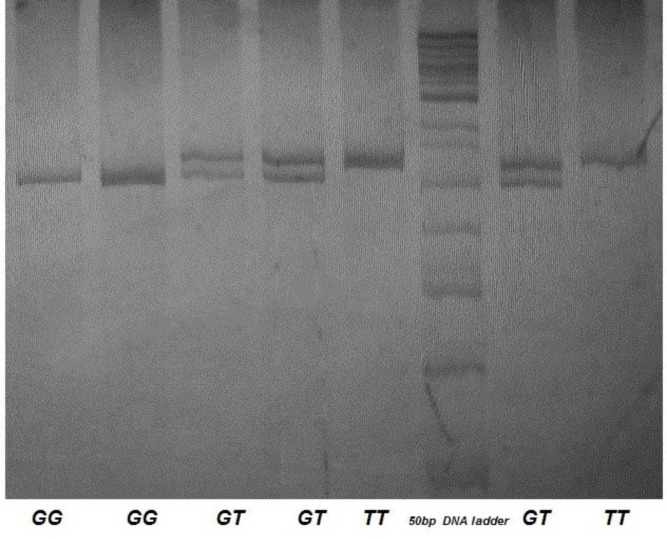
DNA sample was obtained by salt precipitation method (13), for the amplification by polymerase chain reaction (PCR). To evaluate the rs712 polymorphism, the following reagents and primers were utilized. The primers: 5′-ATGACAGTGGAAGTTTTTTTTTCCTC-3′ and 5′-GAATCATCATCAGGAAGCCCAT-3′ (14), with 50 ng of genomic DNA, 7.5 pmol of primers, 2.5 mM MgCl2, 2.5 U of Taq polymerase (Invitrogen, Carlsbad, CA USA), 0.2 mM dNTPs and 0.1% BSA (Bovine Serum Albumin, New England BioLabs Inc; Beverly, MA, USA) in a 15 ml of total volume. Annealing temperature was 56.5 °C. The allele was identified on 6% polyacrylamide gel by previous enzymatic digestion (Taq I; New England BioLabs Inc; Beverly, MA, USA) stained by silver nitrate (15). GG genotype (wild type) was identified as the digested fragment and TT genotype (polymorphic type) as the undigested fragment (Figure 1).
Figure 1.
Genotypes representation of the rs712 polymorphism of KRAS gene and detection of GG [wild genotype, 299 base pairs (bp) and 25 bp bands, digested with Taq I restriction enzyme], TT (polymorphic genotype, 324 pb band, undigested) and GT (heterozygous genotype, 324 bp, 299 bp and 25 bp bands)
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was tested in the control group. The association analysis was performed by OR using the PASW Statistic Base 18 software, 2009 (Chicago, IL, USA).
Results
Epidemiological data
Comparative epidemiological data of the studied groups are shown in the Table 1. In CRC patients, the average age was 58.53 (range 30 to 92 years old). Fifty-four percent (180/336) of these patients were male and 77% were ≥50 years old.
Table 1.
Demographic data for the study group
| CRC (n=336) | Controls (n=281) | P -value | |||
|---|---|---|---|---|---|
| Age (years) | |||||
| Mean (SD)* | 58.53 | (12.49) | 40.49 | (10.61) | <0.0001 |
| < 50 years [(n), %] | (76) | 23.0 | (215) | 77.0 | |
| ≥ 50 years [(n), %] | (260) | 77.0 | (66) | 23.0 | <0.0001 |
| Gender | |||||
| Male [(n), %] | (180) | 54.0 | (144) | 51.0 | 0.572 |
| Female [(n), %] | (156) | 46.0 | (137) | 49.0 | |
Colorectal cancer (CRC), SD (standard deviation),
Student's t-test.
Genotype frequency
Table 2 shows the genotype distribution of the rs712 polymorphism between CRC patients and controls. The genotype TT was observed as protective factor in 18% (59/336) of the patients with CRC and 25% (70/281) of the controls (OR 0.64, 95% CI = 0.43-0.94, P=0.0326). The genotype distribution of rs712 polymorphism was in HWE in the control group. Additional differences were observed when the genotypes were analyzed by recessive model (OR 1.55, 95%CI = 1.03-2.34, P=0.0326), as a risk factor.
Table 2.
Genotype and allelic distribution of the rs712 polymorphism of KRAS in studied groups
| rs712 polymorphism | CRC | Controls * | OR | Confidence intervals (95%) | P -value | |||
|---|---|---|---|---|---|---|---|---|
| Model | Genotype | (n=336) | % | (n=281) | % | |||
| GG | (98) | 29 | (69) | 24.5 | ||||
| GT | (179) | 53 | (142) | 50.5 | 1.13 | (0.82-1.55) | 0.4993 | |
| TT | (59) | 18 | (70) | 25 | 0.64 | (0.43-0.94) | 0.0326 | |
| Dominant | GG | (98) | 29 | (69) | 24.5 | |||
| GT/TT | (238) | 71 | (212) | 75.5 | 0.79 | (0.55-1.13) | 0.1991 | |
| Recessive | TT | (59) | 17 | (70) | 25 | |||
| GG/GT | (277) | 83 | (211) | 75 | 1.55 | (1.03-2.34) | 0.0326 | |
| Allele (2n=672) | (2n=562) | |||||||
| G | (456) | 0.6785 | (353) | 0.6281 | 1.09 | (0.89-1.33) | 0.3894 | |
| T | (417) | 0.3215 | (354) | 0.3719 | 0.91 | (0.74-1.11) | 0.3894 | |
Controls genotype, Hardy-Weinberg equilibrium in controls group (chi-square test=0.032; P=0.8577), Colorectal cancer (CRC).
Comparative analysis with genotypes and clinical characteristics of CRC patients
Significant differences were found with regards to clinical characteristics of the CRC group and genotypes, alleles and genetics model of the rs712 polymorphism. Colon cancer localization and allele T (OR 3.82, 95% CI 2.77-5.28, P=0.0001), positive tumor node metastasis with TT genotype (OR 2.49, 95% CI 1.45-4.28, P=0.0009), poor differentiation (OR 2.35, 95% CI 1.35-4.1, P=0.0033) and poor chemotherapy response (OR 2.6, 95% CI 1.7-4.24, P=0.0001) with heterozygous (GT) genotype, were a risk factor for CRC (Table 3).
Table 3.
Clinical variables of the colorectal cancer and their association with the rs712 polymorphism of KRAS gene
| Clinical variable * | Genotype | Model | Allele | OR | Confidence intervals (95%) | P -value |
|---|---|---|---|---|---|---|
| Location: colon versus recto | T | 3.82 | (2.77-5.28) | 0.0001 | ||
| G | 0.26 | (0.18-0.36) | 0.0001 | |||
| Node metastasis: positive | TT | 2.49 | (1.45-4.28) | 0.0009 | ||
| recessive | 0.40 | (0.23-0.68) | 0.0011 | |||
| Differentiation: poor | GT | 2.35 | (1.35-4.1) | 0.0033 | ||
| **poor chemotherapy response | GT | 2.6 | (1.7-4.24) | 0.0001 |
Non-significance clinical variables in analysis included: gender (male, female), age (<50, ≥ 50 years), tobacco and alcohol consumption, stage (I-II, III-IV), metastasis .
non-response to treatment with pro-drug 5-floururacil (5-FU) and capecitabine was evaluated according to the pathological Ryan's classification described as follows: 1. moderate response (single cells or small groups of cancerous cells), 2. minimum response (residual cancer surrounded by fibrosis), and 3. poor response (minimal or no tumor destruction, extensive residual cancer) (26).
Discussion
In Mexico, the CRC is a growing morbidity-mortality problem (1-4). Our results do agree with previous reports in the literature, which described an average age of 50 years in individuals with CRC (1-5). Probably, lifestyle changes in terms of diet and changes in longevity have influenced the increased frequency of this disease in the Mexican population (3).
Knowledge on the molecular mechanisms of colorectal carcinogenesis is major goal; many relevant studies have demonstrated association between CRC and different polymorphisms on KRAS gene, such as rs712 polymorphism that has been shown to alter the let-7 binding site and regulate KRAS activity affecting gene expression and promoting the cell proliferation in the intestinal mucosa (12, 16).
Studies have demonstrated an association between polymorphism and an increased severity and susceptibility to various diseases including CRC (11, 12). However, the association of this polymorphism with the cancer depends probably on environmental factors that can promote tumoral epigenetic changes (17). The result observed in this study show that the presence of TT variant was a protective factor for CRC. These results were different from a meta-analysis in digestive system cancer of China’s population (11). However, other studies have demonstrated the association with decreased risk for the T allele, and dominant model GT/TT of rs712 polymorphism in breast cancer of Iranian population (18). It has been suggested that let-7 acts as tumor suppressor that can regulate the expression of distinct pathways required in the tumoral behavior (19).
Nevertheless, the T allele and genotype TT were observed as risk factors in patients with colon localization, and nodule metastasis, respectively. As we mentioned, let-7 has an important participation in the development of metastasis, and the polymorphisms in Let-7 modifies binding site that regulates KRAS activity by affecting gene expression and promoting cell proliferation (12).
Other data shows risk association between genotype GT of rs712 let-7 polymorphism in CRC patients with poor differentiation; similar data were found by Jiang et al (10) that suggested the possible participation of rs712 in the progression of CRC.
The association of GT genotypes with non-response at adjuvant chemotherapy treatment in patients with CRC [Capecitabine (Xeloda, Roche, oral drug) and pro-drug 5-floururacil (5-FU) are chemotherapeutic drugs commonly used in CRC individuals] was evident. Regarding the rs712 polymorphism in let-7 region on KRAS gene, it could alter the expression mechanism by affecting let-7 binding region that regulates the constitutive expression of KRAS (12). It has been observed that approximately 30% of CRC tumors have mutations in KRAS that are predictive of high grade, and aggressive tumors with poor response to chemotherapy, which lead to poor prognosis (20). These mutations alter the GTPase activity of KRAS and could activate proliferative signaling pathways (12. 21). However, the precise mechanism to understanding the therapeutic efficacy of miRNAs is not well-defined. Also, it has been observed that the presence of metastasis is associated with adverse clinical outcome and might alter the expression of different molecular factors included miRNAs such miR-21 and let-7, which participate in the regulation of cellular processes (12, 22-24). It has also been observed that the response to drugs not only is related to the monogenic inheritance of a protein variant but also depends on several genes encoding proteins involved in multiple metabolic pathways, posttranslational modifications, gene interactions, and epigenetics (25).
Conclusion
Our results showed an association of polymorphism as protective factor with CRC when compared with controls. However, this polymorphism could be a good marker in patients for 1) colon localization, 2) node metastasis, 3) poor differentiation, and 4) poor response to chemotherapy factors that could contribute to CRC susceptibility in Mexican population. Similar analyses are necessary to confirm these observed evidences.
Acknowledgment
The authors would like to thank to nurses for assisting in the extracting blood samples. This research was financially supported in part through Fondo de Investigación en Salud FIS/IMSS/PROT/G15/1463 grants.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Alberts SR, Citrin D, Schwartz D, Rodriguez M. Colon, Rectal, and Anal Cancers. Cancer Management. 2016. http://www.cancernetwork.com/cancer-management/colon-rectal-and-anal-cancers.
- 2.Pourhoseingholi MA. Epidemiology and burden of colorectal cancer in asia-pacific region: what shall we do now? Transl Gastrointest Cancer . 2014;3:169–173. [Google Scholar]
- 3.Gutiérrez IA, Puebla AM, Delgado JI, Figuera LE, Zúñiga GM, Gómez K, et al. Association between TNF-α-308G>A and -238G>A gene polymorphisms and TNF-α serum levels in mexican colorectal cancer patients. Genet Mol Res . 2016;15:1–11. doi: 10.4238/gmr.15028199. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136:359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.García S, Téllez FI, Méndez N, Uribe M. Results of the first program of colorectal cancer screening in mexico. Endoscopia. 2015;27:59–63. [Google Scholar]
- 7.OMIM. 190070. V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog; KRAS. https://www.omim.org/entry/190070. [DOI] [PubMed]
- 8.Jancík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol . 2010;2010:150960–150973. doi: 10.1155/2010/150960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Jakubowski M, Hunt JL. KRAS gene mutation in colorectal cancer is correlated with increased proliferation and spontaneous apoptosis. Am J Clin Pathol. 2011;135:245–252. doi: 10.1309/AJCP7FO2VAXIVSTP. [DOI] [PubMed] [Google Scholar]
- 10.Jiang QH, Peng HX, Zhang Y, Tian P, Xi ZL, Chen H. rs712 polymorphism within let-7 microRNA-binding site might be involved in the initiation and progression of colorectal cancer in chinese population. Onco Targets Ther . 2015;8:3041–3045. doi: 10.2147/OTT.S89746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du XY, Hu YY, Xie C, Deng CY, Liu CY, Luo ZG, et al. Significant association between Let-7-KRAS rs712 G > T polymorphism and cancer risk in the chinese population: a meta-analysis. Oncotarget . 2017;8:13863–13871. doi: 10.18632/oncotarget.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci . 2015;22:9–21. doi: 10.1186/s12929-015-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215–1216. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li ZH, Pan XM, Han BW, Guo XM, Zhang Z, Jia J, et al. A let-7 binding site polymorphism rs712 in the KRAS 3’ UTR is associated with an increased risk of gastric cancer. Tumour Biol . 2013;34:3159–3163. doi: 10.1007/s13277-013-0885-x. [DOI] [PubMed] [Google Scholar]
- 15.Sanguinetti CJ, Dias E, Simpson AJ. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 1994;17:914–921. [PubMed] [Google Scholar]
- 16.Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, et al. A let-7 microRNA-binding site polymorphism in 3’-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol . 2011;22:104–109. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulte PA, Whittaker C, Curran CP. Considerations for using genetic and epigenetic information in occupational health risk assessment and standard setting. J Occup Environ Hyg . 2015;12:69–81. doi: 10.1080/15459624.2015.1060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanaei S, Hashemi M, Eskandari E, Hashemi SM, Bahari G. KRAS gene polymorphisms and their impact on breast cancer risk in an iranian population. Asian Pac J Cancer Prev . 2017;18:1301–1305. doi: 10.22034/APJCP.2017.18.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 20.Cong T, Xiang D. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol . 2012;18:5171–5180. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deschoolmeester V, Baay M, Specenier P, Lardon F, Vermorken JB. A review of the most promising biomarkers in colorectal cancer: one step closer to targeted therapy. Oncologist . 2010;15:699–731. doi: 10.1634/theoncologist.2010-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull . 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, Yu J, Ng SS. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag Res. 2014;6:405–422. doi: 10.2147/CMAR.S35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, Mo YY. Role of microRNAs in breast cancer. Cancer Biol Ther . 2013;14:201–212. doi: 10.4161/cbt.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinedo HM, Peters GF. Fluorouracil: biochemistry and pharmacology. J Clin Oncol . 1988;6:1653–1664. doi: 10.1200/JCO.1988.6.10.1653. [DOI] [PubMed] [Google Scholar]
- 26.Ryan R, Gibbons D, Hyland JM, Treanor D, White A, Mulcahy HE, et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology. 2005;47:141–146. doi: 10.1111/j.1365-2559.2005.02176.x. [DOI] [PubMed] [Google Scholar]