Abstract
Objective(s):
In the current research, the prevalence of Staphylococcus aureus clones and genes encoding antimicrobial resistance and toxins were examined among 120 S. aureus strains from nosocomial infections in tehran, Iran.
Materials and Methods:
Antimicrobial susceptibility was examined, based on disk diffusion and PCR method to identify resistance and toxin-encoding genes. Based on the polymorphisms in SCCmec, agr, spa, and MLST, the isolates were typed.
Results:
Among 120 S. aureus isolates, 85 (70.8%) were methicilin resistant S. aureus (MRSA), and 35 (29.2%) were methicilin sensetive S. aureus (MSSA). The tested isolates contained resistance genes, including ant(4΄)-Ia (90%), aac(6΄)-Ie/aph(2˝) (80%), aph(3΄)-IIIa (30%), erm(A) (26.7%), erm(B) (10.8%), erm(C) (11.7%), msr(A) (40.8%), msr(B) (14.2%), tet(M) (45.8%), and mupA (8.3%). The MRSA strains were clustered into six different clones. The most common genotypes included ST239-SCCmec III/t037 (23.3%), ST239-SCCmec III/t388 (22.5%), ST22-SCCmec IV/t790 (8.3%), ST15-SCCmec IV/t084 (7.5%), ST585-SCCmec III/t713 (5%), and ST239-SCCmec III/t924 (4.2%), respectively. ST182/t196 (8.3%) and ST123/t171 (5%) belonged exclusively to MSSA strains. Overall, 10 (66.7%) and 5 (33.3%) out of 15 isolates with pvl genes were attributed to clones ST22-SCCmec IV/t790 and ST15-SCCmec IV/t084, respectively. ST22-SCCmec IV/t790, ST239-SCCmec III/t037, and ST15-SCCmec IV/t084, were related to high-level mupirocin-resistant phenotypes.
Conclusion:
The genetic diversity of S. aureus was confirmed in our hospitals, and ST239-SCCmec III/t037 showed a relatively high prevalence in our study. It seems that assessment of resistance and virulence genes in different S. aureus molecular types is necessary for proper antibiotic consumption.
Key Words: Agr, MLST, MRSA, SCCmec, Spa, Staphylococcus aureus
Introduction
Staphylococcus aureus, which is described as a common nosocomial pathogen, is responsible for various diseases, such as food poisoning, osteomyelitis, wound infections, and even fatal conditions, such as endocarditis (1). Over the past few decades, it has been well-documented that the pathogen’s resistance potential to antimicrobial agents, especially methicillin, may lead to its persistence in the hospital and community (2). In 1961, the first case of methicillin-resistant S. aureus (MRSA) occurred in the UK (3). The prevalence of this infection has steadily increased since then, as confirmed in several studies, raising major concerns about the global increase in its prevalence, as well as its associated mortality and morbidity in the healthcare setting, especially intensive care units (ICUs) due to MRSA infections (1-3).
Resistance to methicillin is attributed to β-lactamase expression or changes in the structure of mecA gene-encoded penicillin-binding protein-2. Generally, mecA gene (21-67 kbp) is recognized as a staphylococcal cassette chromosome mec (SCCmec). Generally, SCCmec is categorized into 11 types with respect to mec genes and ccr gene complexes (4). Identification of SCCmec type among S. aureus clinical isolates can be useful in molecular typing of MRSA strains (5).
Based on previous findings, SCCmec I-III and IV-V are respectively responsible for the most common hospital-acquired and community-acquired MRSA (HA-MRSA and CA-MRSA, respectively) infections. HA- and CA-MRSA strains can be distinguished with respect to some genotypic, phenotypic, and epidemiological characteristics, as well as virulence factors (4, 5).
The emergence and prevalence of MRSA infections containing multidrug-resistant (MDR) genes have significantly limited the availability of antibiotics over the past decades. In addition, the growing emergence of MDR-MRSA strains poses a major global health concern (1). Wide resistance to β-lactams, besides other antibiotics, including aminoglycosides, lincosamides, and macrolides, has been shown in MRSA strains (6).
Aminoglycosides play a key role in serious anti-staphylococcal therapies. According to the previous researches, resistance to aminoglycosides is attributed to aminoglycoside-modifying enzymes (AMEs) including aminoglycoside nucleotidyltransferases, aminoglycoside phosphotransferases and aminoglycoside acetyltransferases (7). mupA and mupB genes are responsible for resistance to mupirocin which is used to treat various types of skin diseases caused by S. aureus (8). Resistance to macrolides as protein synthesis inhibitors is mediated by msr genes activating efflux pumps and erm genes modifying the ribosomal binding site (9). Thus, in spite of new antibiotics introduction, concerning the emergence and dissemination of antibiotic resistance genes, MRSA infections treatment is still a great dilemma worldwide.
This study was conducted to identify antibiotic resistance patterns and the carriage of resistance and virulence genes as well as major MRSA clones by MLST, spa, SCCmec and agr techniques in clinical samples taken from patients in Tehran, Iran.
Materials and Methods
Sampling, MRSA isolation and antibacterial susceptibility testing
This cross-sectional study included 368 clinical samples from wound, blood, and urine specimens during April-December 2016. Ethics Committee of Shahid Beheshti University of Medical Sciences approved the implementation of this study (IR.SBMU.SM.REC.1395.157). All patients signed written informed consent forms.
After the rapid transfer of the specimens to the laboratory, S. aureus identification was performed, based on the conventional biochemical tests. S. aureus identification was confirmed based on the PCR assay for nucA gene (10). According to the Clinical and Laboratory Standards Institute (CLSI) standards, resistance to methicillin was examined with oxacillin and cefoxitin (1 and 30 µg, respectively) disks in Mueller-Hinton agar plates (Merck; Germany) containing 4% sodium chloride (11). For further molecular analysis, confirmed isolates were stored in Tryptic Soy Broth with 15% glycerol (Merck; Germany) at a temperature of -70 °C. Afterwards, based on the Kirby-Bauer method, the susceptibility profiles to 12 antibiotics including tetracycline (T 30 µg), clindamycin (CD 2 µg), ciprofloxacin (CIP 5 µg), trimethoprim- sulfamethoxazole (TS 2.5 µg), kanamycin (K 30 µg), ceftriaxon (CRO 30 µg), quinupristin-dalfopristin (SYN 15 µg), erythromycin (E 15 µg), amikacin (AK 30 µg), gentamicin (GM 10 µg), tobramycin (TN 10 µg), teicoplanin (TEC 30 µg), penicillin (PG 10 µg), and linezolid (LZD 30 µg) (Mast, UK) were determined, based on the CLSI criteria (11).
Using E-test strips (bioMe´rieux), the minimum inhibitory concentrations (MICs) were measured for mupirocin and vancomycin. Resistance to three antibiotic groups or more, besides beta-lactams, was defined as MDR. High mupirocin resistance was defined as antibiotic use ≥ 256 mg/l. Growth in a well containing clindamycin and erythromycin (0.5 and 4 μg/ml, respectively) indicated inducible macrolide-lincosamide-streptogramin B and/or clindamycin resistance phenotypes; otherwise, constitutive MLSB and/or clindamycin resistance phenotype was confirmed (11). ATCC29213 and ATCC25923 (S. aureus) were considered as the reference strains for the quality control purposes.
Extraction of genomic DNA
For extracting genomic DNA, pure overnight S. aureus cultures were used on 5% sheep blood agar (BA; Merck, Germany), based on the protocols of InstaGene Matrix kit (BioRad, USA).
Resistance and toxin genes profiling
To identify toxin (etb, tst, pvl, eta) and resistance (tet(M), aac (6΄)-Ie/aph (2˝), mupA, erm(A), msr(A), msr(B), erm(B), erm(C), ant (4΄)-Ia, aph (3΄)-IIIa) genes, PCR assay was carried out. The details of the degenerated primers in this study are described in Table 1.
Table 1.
Primer and oligo sequences used in present research
| Gene | Primer | Primer sequence (5´ 3´) | Length (bp) | Reference |
|---|---|---|---|---|
| mecA | F | AGA AGA TGG TAT GTG GAA GTT AG | 583 | (10) |
| R | ATG TAT GTG CGA TTG TAT TGC | |||
| tst-1 | F | TTA TCG TAA GCC CTT TGT TG | 398 | (10) |
| R | TAA AGG TAG TTC TAT TGG AGT AGG | |||
| nucA | F | GCG ATT GAT GGT GAT ACG GTT | 270 | (12) |
| R | AGC CAA GCC TTG ACG AAC TAA AGC | |||
| luk-PV | F | TTC ACT ATT TGT AAA AGT GTC AGA CCC ACT | 180 | (12) |
| R | TAC TAA TGA ATT TTT TTA TCG TAA GCC CTT | |||
| etb | F | ACA AGC AAA AGA ATA CAG CG | 226 | (13) |
| R | GTT TTT GGC TGC TTC TCT TG | |||
| eta | F | GCA GGT GTT GAT TTA GCA TT | 93 | (13) |
| R | AGA TGT CCC TAT TTT TGC TG | |||
| tet(M) | F | AGT GGA GCG ATT ACA GAA | 158 | (13) |
| R | CAT ATG TCC TGG CGT GTC TA | |||
| aph(3΄)-IIIa | F | CTT GAT CGA AAA ATA CCG CTG C | 269 | (14) |
| R | TCA TAC TCT TCC GAG CAA A | |||
| ant(4΄)-Ia | F | AAT CGG TAG AAG CCC AA | 135 | (14) |
| R | GCA CCT GCC ATT GCT A | |||
| aac(6΄)-Ie/aph(2˝) | F | CCA AGA GCA ATA AGG GCA TAC C | 222 | (14) |
| R | CAC ACT ATC ATA ACC ACT | |||
| erm(B) | F | CTA TCT GAT TGT TGA AGA AGC ATT | 141 | (15) |
| R | GTT TAC TCT TGG TTT AGG ATC AAA | |||
| erm(A) | F | TAT CTT ATC GTT GAG AAG GGA TT | 139 | (15) |
| R | CTA CAC TTG GCT GAT GAA A | |||
| erm(C) | F | AAT CGT CAA TTC CTG CAT GT | 299 | (16) |
| R | TAA TCG TGG AAT ACG GGT TTG | |||
| msr(B) | F | TAT GAT ATC CAT AAT AAT TAT CCA ATC | 595 | (16) |
| R | AAG TTA TAT CAT GAA TAG ATT GTC CTG TT | |||
| msr(A) | F | GGC ACA ATA AGA GTG TTT AAA GG | 940 | (16) |
| R | AAG TTA TAT CAT GAA TAG ATT GTC CTG TT | |||
| mupA | F | CCC ATG GCT TAC CAG TTG A | 1158 | (17) |
| R | CCA TGG AGC ACT ATC CGA |
Multiplex PCR for SCCmec typing
According to a study by Boy and colleagues, for SCCmec typing, multiplex PCR amplification was performed with specific primers (4). The controls comprised of the MRSA strains, i.e., ATCC 10442, N315, 85/2082, MW2, and WIS (attributed to types I, II, III, IV, and V, respectively).
Multiplex PCR amplification for agr typing
In addition, for agr typing, multiplex PCR amplification was carried out, using forward (Pan) and reverse (agr1 to agr4) primers for the agr groups as previously recommended by Gilot et al (18). The specific oligonucleotide primers are listed in Table 1.
spa typing
On the other hand, spa gene was detected according to a study by Harmsen and colleagues (19). After the positive spa PCR products were purified, DNA sequencing was carried out in both strands (Macrogen; South Korea). Chromas 1.45 (Australia) was used to edit the sequences. To assign the sequences to specific spa types, the Ridom SpaServer database was searched.
MLST technique
Via amplification and sequencing, MLST was carried out on S. aureus isolates. The internal fragments of housekeeping genes were used to identify the allelic profiles; these genes included gmk, arcC, aroE, glpF, pta, yqiL, and tpi. The isolate was assigned a sequence type (ST) after comparing the sequences with the S. aureus MLST database.
Results
Sampling and antibiotic susceptibility
In this study, out of 368 samples obtained from various clinical specimens, 120 isolates (83 (69.2%) obtained from men and 37 (30.8%) from women) were identified as S. aureus. These isolates originated from wound (60%), blood (20.8%) and urinary tract infections (19.2%). Of the 120 S. aureus clinical isolates obtained from the hospitalized patients, 85 (70.8%) were MRSA and 35 (29.2%) were methicillin susceptible S. aureus (MSSA).
None of the isolates were susceptible to all of the antimicrobial agents tested regarding in vitro antimicrobial susceptibility tests. All isolates were susceptible to vancomycin, among which 55 (45.8%), 48 (40%) and 17 (14.2%) isolates had a MIC of 0.5, 1 and 2 µg/ml, respectively.
Among the 35 MSSA isolates, no mupirocin resistance was detected, whereas 30 MRSA isolates (35.3%) were mupirocin resistant. Of these mupirocin resistant isolates, 14 (46.7%) and 16 (53.3%) had high and low resistance levels, respectively. All the high-level mupirocin-resistant (HLMUPR) isolates were collected from wound samples. Patients diagnosed with mupirocin-susceptible and -resistant MRSA infections were not significantly different in terms of age and gender (P= 0.145 and 0.128, respectively). The frequency of resistance for MRSA and MSSA isolates to different antibacterial agents are presented in Table 2.
Table 2.
The frequency of antimicrobial resistance in 120 S. aureus isolates obtained from clinical specimens
| Antibiotics | MSSA (n=35) n (%) |
MRSA (n=85) n (%) |
All (n=120) n (%) |
|||
|---|---|---|---|---|---|---|
| S | R | S | R | S | R | |
| penicillin | 2(5.7) | 33(94.3) | 6(7.1) | 79(92.9) | 8(6.7) | 112(93.3) |
| kanamycin | 17(48.6) | 18(51.4) | 10(11.8) | 75(88.2) | 27(22.5) | 93(77.5) |
| gentamicin | 21(60) | 14(40) | 9(10.6) | 76(89.4) | 30(25) | 90(75) |
| ciprofloxacin | 23(65.7) | 12(34.3) | 12(14.1) | 73(85.9) | 35(29.2) | 85(70.8) |
| tetracycline | 21(60) | 14(40) | 15(17.6) | 70(82.4) | 36(30) | 84(70) |
| amikacin | 27(77.1) | 8(22.9) | 11(12.9) | 74(87.1) | 38(31.7) | 82(68.3) |
| tobramycin | 29(82.9) | 6(17.1) | 20(23.5) | 65(76.5) | 49(40.8) | 71(59.2) |
| erythromycin | 29(82.9) | 6(17.1) | 21(24.7) | 64(75.3) | 50(41.7) | 70(58.3) |
| clindamycin | 30(85.7) | 5(14.3) | 32(37.6) | 53(62.4) | 62(51.7) | 58(48.3) |
| ceftriaxone | 30(85.7) | 5(14.3) | 35(41.2) | 50(58.8) | 65(54.2) | 55(45.8) |
| trimetoprim-sulfamethoxazole | 33(94.3) | 2(5.7) | 53(62.4) | 32(37.6) | 86(71.7) | 34(28.3) |
| mupirocin | 35(100) | 0(0) | 55(64.7) | 30(35.3) | 90(75) | 30(25) |
| quinupristin-dalfopristin | 31(88.6) | 4(11.4) | 74(87.1) | 11(12.9) | 105(87.5) | 15(12.5) |
| linzolid | 35(100) | 0(0) | 85(100) | 0(0) | 120(100) | 0(0) |
| teicoplanin | 35(100) | 0(0) | 85(100) | 0(0) | 120(100) | 0(0) |
| vancomycin | 35(100) | 0(0) | 85(100) | 0(0) | 120(100) | 0(0) |
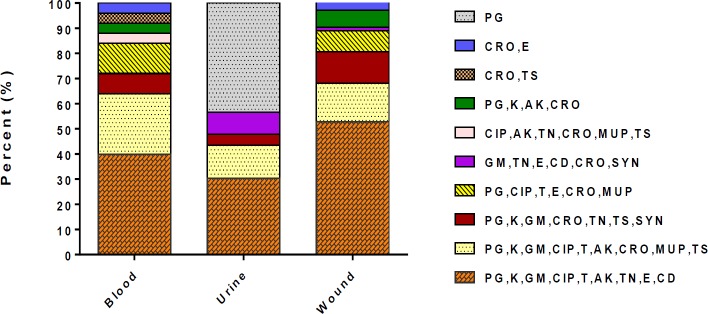
Of the 120 S. aureus isolates, 87.5% (105/120) were defined as MDR. The predominant multiple drug resistance profile among the MDR isolates were resistance to 9 and 7 antibiotics found in 75 (62.5%) and 12 (10%) isolates, respectively. Distribution of resistance profile and different clinical sample in S. aureus isolated from nosocomial infections are presented in Figure 1.
Figure 1.
Resistance pattern of Staphylococcus aureus obtained from clinical samples
The distribution of resistance genes
In addition, the frequency of antibiotic resistance genes was measured. The genes included ant(4΄)-Ia (90%), aac(6΄)-Ie/aph(2˝) (80%), aph(3΄)-IIIa (30%), erm(A) (26.7%), erm(B) (10.8%), erm(C) (11.7%), msr(A) (40.8%), msr(B) (14.2%), tet(M) (45.8%), and mupA (8.3%) (Figure 2). The MRSA strains contained ant(4΄)-Ia, aac(6΄)-Ie/aph(2˝), and mecA genes. Other detected antibiotic resistance genes included tet(M) (64.7%), msr(A) (57.6%), aph(3΄)-IIIa (42.3%), erm(A) (37.6%), msr(B) (20%), erm(C) (16.5%), erm(B) (15.3%), and mupA (11.8%), while 31.4% and 56.7% of MSSA strains were found to respectively carry aac(6΄)-Ie/aph(2˝) and ant(4΄)-Ia. Particularly, the MRSA strains contained more resistance genes, compared to the MSSA isolates.
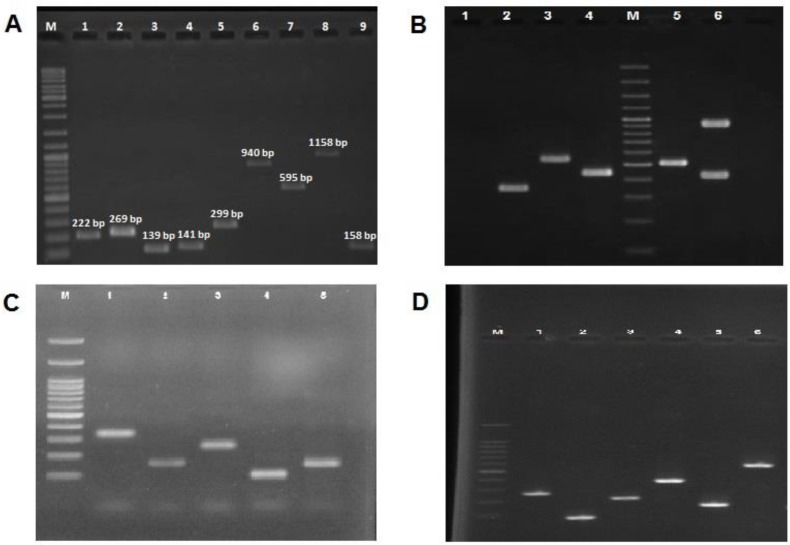
Figure 2.
A) Lane M, 100-bp DNA Ladder (Fermentas, UK); Lane 1 PCR product of aac(6΄)-Ie/aph(2˝) encoding gene, Lane 2 PCR product of PCR product of aph(3΄)-IIIa encoding gene, Lane 3 PCR product of erm(A) encoding gene, Lane 4 PCR product of erm(B) encoding gene, Lane 5 PCR product of erm(C) encoding gene, Lane 6 PCR product of msr(A) encoding gene, Lane 7 PCR product of msr(B) encoding gene, Lane 8 PCR product of mupA encoding gene, and Lane 9 PCR product of tet(M) encoding gene. B) Lane M, DNA Ladder; lane 1 negative control, Lane 2 the 323 bp PCR product of agr type III, lane 3 the 575 bp PCR product of agr type II, lane 4 the 441 bp PCR product of agr type I, lane 5 the 518 bp PCR product of SCCmec Type III, lane 6 the 937 and 415 bp PCR products of SCCmec Type IV. C) Lane M, DNA ladder; lane 1-5, variable PCR product of spa. D) Lane M, DNA Ladder; Lane 1the 270 bp PCR product of nucA gene, Lane 2the 93 bp PCR product of eta gene, Lane 3the 226 bp PCR product of etb gene, Lane 4the 398 bp PCR product oftst-1 gene, Lane 5the 180 bp PCR product of luk-PV gene, Lane 6the 583 bp PCR product of mecA gene
Fourteen (11.7%) out of 30 mupirocin-resistance S. aureus isolates were identified as HLMUPR-MRSA, while mupA gene was confirmed in 10 isolates (71.4%). In addition, ant(4΄)-Ia (108, 90%), followed by aac(6΄)-Ie/aph(2˝) (96, 80%), was recognized as the most common aminoglycoside resistance gene. A total of 75 (62.5%) isolates contained ant(4΄)-Ia, as well as aac(6΄)-Ie/aph(2˝). On the other hand, 15 (12.5%) isolates harbored aph(3΄)-IIIa, besides ant(4΄)-Ia. Also, eleven (9.2%) isolates harbored ant(4΄)-Ia, aph(3΄)-IIIa, and aac(6΄)-Ie/aph(2˝) genes, while 10 (8.3%) contained aac(6΄)-Ie/aph(2˝), as well as aph(3΄)-IIIa; however, ant(4΄)-Ia alone was confirmed in 7 (5.8%) strains. Resistance to tetracycline was observed among 84 (70%) S. aureus isolates, 55 (45.8%) of which harbored tet(M) gene. Antibiotic resistance genes showed the highest prevalence among MRSA strains from wound infections.
Virulence gene profiling
Among toxin-encoding genes, the highest and lowest frequencies were attributed to tst (58; 48.3%) and etb (3; 2.5%) genes, respectively (Figure 2). In present work, 12.5% of the isolates were positive for pvl gene and eta gene was identified in 7.5% of the isolates. The only toxin encoding gene among the MSSA isolates was tst gene (10 out of 120 isolates, 8.3%). S. aureus isolates harbouring tst gene were isolated from wound (32; 55.2%), blood (17; 29.3%), and urinary tract infections (9; 15.5%) while pvl positive isolates were detected in wound (12; 80%) and blood (3; 20%) samples. Eight isolates carrying pvl and tst genes, simultaneously, had mupA gene.
Distribution of SCCmec types
According to SCCmec typing, 66 (77.6%) and 19 (22.4%) MRSA isolates contained SCCmec types III and IV, respectively. No isolate harbored SCCmec type V, II, or I. Based on the multiplex PCR, isolates positive for PVL were attributed to SCCmec type IV, while tst gene was found in MRSA isolates from SCCmec III and IV. The HA-MRSA origin was emphasized by the presence of SCCmec type III.
Frequency of agr types
agr typing indicated that type I was the predominant agr type present in 91 isolates (75.8%), followed by type III which was present in 20 isolates (16.7%). agr type II was detected in 9 isolates (7.5%). Among 35 MSSA isolates, 20 and 15 harboured agr types I and III respectively (Figure 2).
spa typing
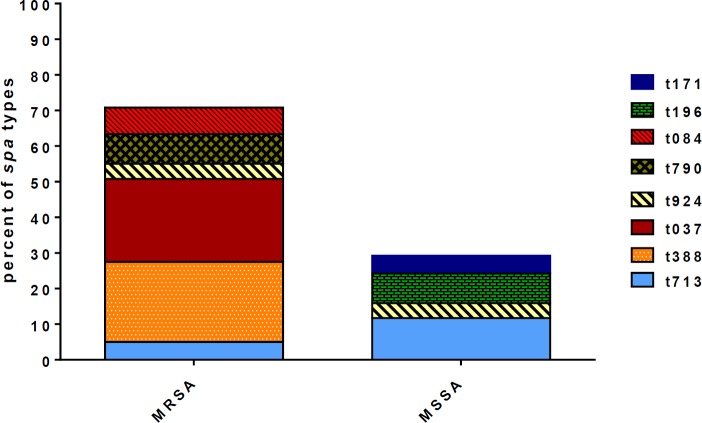
spa typing was performed for all S. aureus isolates. spa typing discriminated eight different types: t037 (23.3%), t388 (22.5%), t713 (16.8%), t924 (8.3%), t790 (8.3%), t196 (8.3%) t084 (7.5%) and t171 (5%) (Figure 2). All the spa types except t196 and t171 were found in MRSA strains. Distribution of spa types among methicillin resistance and methicillin sensitive strains are presented in Figure 3. The most prevalent spa type among MSSA strains was t713 (11.7%, 14/120), followed by t196 (8.3, 10/120), t171 (5%, 6/120) and t924 (4.2%, 5/120), respectively.
Figure 3.
Distribution of spa types in methicilin resistant S. aureus (MRSA) and methicilin sensetive S. aureus (MSSA) strains isolated from nosocomial infections
MLST
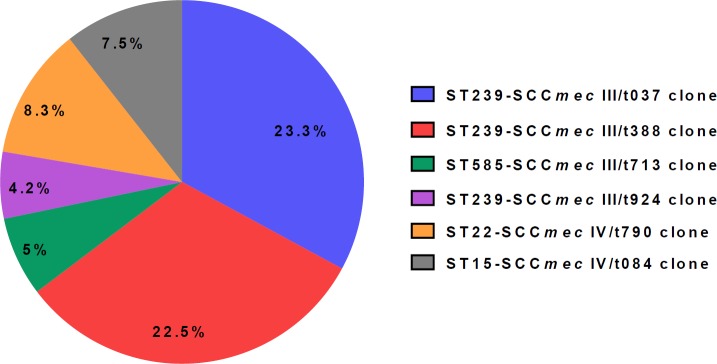
Apparently, 120 isolates belonged to six different STs including ST239 (65 strains), ST22 (10 strains), ST182 (10 strains), ST15 (9 strains), ST585 (20 strains) and ST123 (6 strains). ST239, ST585, ST182 and ST123 were found in MSSA strains. It should be noted that of these STs, ST182 and ST123 belonged exclusively to MSSA strains. In conclusion, MRSA strains are clustered into six different groups. ST239-SCCmec III/t037 was found to be the most prominent MRSA clone identified in this study. Distribution of MRSA clones isolated from nosocomial infections are presented in Figure 4.
Figure 4.
Distribution of molecular types in 85 methicilin resistant S. aureus (MRSA) strains isolated from nosocomial infections
Fifteen PVL-carrying strains in our study belonged to ST22-SCCmec IV/t790 (10 isolates, 66.7%) and ST15-SCCmec IV/t084 (5 isolates, 33.3%) clones. Among the isolates under study, 58 (48.3%) isolates harboring tst-1 were distributed in ST239-SCCmec III/t037 (15 isolates, 25.9%), ST239-SCCmec III/t388 (11 isolates, 19%), ST15-SCCmec IV/t084 (8 isolates, 13.8%), ST585-SCCmec III/t713 (6 isolates, 10.3%), ST239-SCCmec III/t924 (5 isolates, 8.6%), and ST22-SCCmec IV/t790 (3 isolates, 5.2%) clones. Among examined isolates, nine isolates (7.53%) were found to carry the eta gene. The eta positive isolates were distributed in ST15-SCCmec IV/t084 (4 isolates, 44.4%), ST22-SCCmec IV/t790 (3 isolates, 33.3%) and ST239-SCCmec III/t037 (2 isolates, 22.2%) clones. The etb gene was detected in ST585-SCCmec III/t713 (66.7%, 2/3) and ST22-SCCmec IV/t790 (33.3%, 1/3) clones. MRSA clones resistance profile varied. Resistance to mupirocin was detected in all the MRSA clones with the exception of ST585-SCCmec III/t713 clone. Interestingly, mupirocin resistant MSSA isolates belonged to ST182/spa type t196. HLMUPR-MRSA strains were detected in ST22-SCCmec IV/t790 (42.8%, 6/14), ST15-SCCmec IV/t084 (28.6%, 4/14) and ST239-SCCmec III/t037 (28.6%, 4/14) clones.
cMLSB phenotype was detected in ST239-SCCmec III/t037 (43.5%, 20/46), ST239-SCCmec III/t388 (43.5%, 20/46), ST585-SCCmec III/t713 (4.3%, 2/46), ST15-SCCmec IV/t084 (4.3%, 2/46), ST239-SCCmec III/t924 (2.2%, 1/46) and ST22-SCCmec IV/t790 (2.2%, 1/46) while iMLSB phenotype distributed among 3 major clones ST239-SCCmec III/t388 (55.6%, 5/9), ST22-SCCmec IV/t790 (33.3%, 3/9) and ST15-SCCmec IV/t084 (11.1%, 1/9). Of 12 MSSA isolates with cMLSB phenotype, 5 isolates belonged to ST182/t196, 4 isolates belonged to ST239/t924, 2 isolates belonged to ST585/t713 and 1 isolate belonged to ST123/t171. All the iMLSB phenotype among MSSA strains belonged to ST585/t713. Characteristics of MRSA clones are presented in Table 3.
Table 3.
Distribution of MRSA molecular types isolated from nosocomial infections
| Molecular types | agr class | Type of toxin (No;%) | Genotypic resistance patterns (No;%) | Phenotypic resistance patterns (No;%) | No (%) |
|---|---|---|---|---|---|
| ST239- SCCmecIII/t037 | I | tst (15;53.6), eta (2;7.1) | mecA (28;100), aph(3΄)-IIIa (20;71.4), aac(6΄)Ie/aph (2˝)(28;100),erm(A) (9;32.1),erm(B) (4;14.3), erm(C) (8;28.6), msr(A) (20;71.4), msr(B) (4;14.3), ant(4΄)-Ia (28;100), tet(M) (28;100) | PG, GM, K, CIP, AK, T, TN, E, CD (18;64.3) | 28 (23.3) |
| PG, GM, K, CIP, AK, T, CRO, MUP, TS (8;28.6) | |||||
| GM, TN, E, CD, CRO, SYN (2;7.1) | |||||
| ST239- SCCmecIII/t388 | I | tst (11;40.7) | mecA (27;100), aph(3΄)-IIIa (10;37), erm(A) (12; 44.4),erm(B) (8;29.6), msr(A) (12;44.4), aac(6΄)-Ie/aph(2˝) (27;100),msr(B) (8;29.6), ant(4΄)-Ia (27;100),tet(M) (12;44.4) | PG, K, CIP, GM, AK, T, CD, TN, E (20;74.1) | 27 (22.5) |
| PG, E, CIP, CRO, T, MUP (5;18.5) | |||||
| PG, K, AK, CRO (2;7.4) | |||||
| ST585- SCCmecIII/t713 | I | tst (6;100), etb (2;7.7) | mecA (6;100), aph(3΄)-IIIa (1;16.7), erm(A) (3;50), ant(4΄)-Ia (6;100), erm(C) (5;83.3), msr(A) (5;83.3), aac(6΄)-Ie/aph(2˝) (6;100), | PG, K, CIP, GM, AK, T, CD, TN, E (2;17.9) | 6 (5) |
| PG, TS, K, TN, GM, CRO, SYN (4;35.7) | |||||
| ST239- SCCmecIII/t924 | III | tst (5;100) | aph(3΄)-IIIa(5;100), mecA (5;100), aac(6΄)-Ie/aph(2˝) (5;100), erm(A) (5; 100), erm(B) (1;20), ant(4΄)-Ia (5;100), | PG, CD, K, CIP, GM, AK,T, TN, E (1;20) | 5 (4.2) |
| PG, TS, K, GM, TN, CRO, SYN (3;60) | |||||
| CIP, AK, TN, CRO, MUP, TS (1,20) | |||||
| ST22-SCCmecIV/t790 | I | pvl (10;100), tst (3;30), eta (3;30), etb (1;10) | mecA (10;100), erm(A) (3;30), erm(C) (1;10), msr(A) (9;90), msr(B) (5;50), ant(4΄)-Ia (10;100), tet(M) (10;100), aac(6΄)-Ie/aph(2˝) (10;100), mupA (6;60) | PG, MUP, GM, AK, CIP, T, K, CRO, TS (5;50) | 10 (8.3) |
| GM, TN, E, CD, CRO, SYN (1;10) | |||||
| PG, K, AK, CRO (1;10) | |||||
| PG, CIP, T, E, CRO, MUP (3;30) | |||||
| ST15- SCCmecIV/t084 | II | pvl (5;55.6), tst (8;88.9), eta (4;44.4) | mecA(9;100), aac(6΄)-Ie/aph(2˝)(9;100), ant(4΄)-Ia(9;100), msr(A) (3;33.3), tet(M) (5;55.6), mupA (4;44.4) | PG, TN, K, CD, GM, CIP, T, AK, E (2;22.2) | 9 (7.5) |
| PG, CIP, T, E, CRO, MUP (1;11.1) | |||||
| PG, CRO, K, AK, GM, CIP, T, MUP, TS (6;66.7) |
Discussion
In consistent with the results of a multicenter study made by Ko et al. (20), a relatively high resistance to gentamicin (75%), amikacin (68.3%), kanamycin (77.5%), and tobramycin (59.2%) was reported in this study which was higher than the resistance rate reported by Goudarzi et al (12). In contrast to the findings of Marghaki et al `s study (6) who reported high frequency of aac(6′)/aph(2′′) gene (40.3% ) in comparison with other AME genes, in the present study, ant(4΄)-Ia (90%) was the most frequent AME gene in S. aureus isolates. However, in this study, aph(3΄)-IIIa gene frequency rate was higher than the study reported by Ida et al. (8.9%)(21) and Marghaki et al (15.7%) (6).
Mupirocin as an important agent in the control of MRSA outbreaks and eradicating MRSA colonization was used for the treatment of different types of staphylococcal skin infections. Long period widespread use of mupirocin may lead to the emergence of mupirocin resistance S. aureus strains (8). In this survey, 30 isolates (25%) presented mupirocin resistance phenotype and 14 (11.7%) isolates were confirmed as HLMUPR-MRSA which is relatively lower than study in Iran (40%) (22) and higher than study in India (5%) (23) and Jordan (2.6%) (24). However, as a result of proper mupirocin prescription in clinic, in our study mupirocin resistant S. aureus isolates were found to be lower. We detected the resistance gene mupA in 10 isolates (8.3%) which is lower than 12.6% (25) and 25% in Iran (22).
In accordance with the results of our previous study (25) a high tetracycline resistance was seen in the present work (70%). In consistent with others (25), in our study the tet(M) gene that may cause tetracycline resistance was detected in 55 (45.8%) isolates.
In this study, 58 isolates (48.3%) presented cMLSB phenotype while the frequency of iMLSB phenotype was 10% (12/120). In consistent with our findings, Schreckenberger et al. (7%) reported low frequency of iMLSB phenotype among S. aureus isolates as well (26). According to the literature, there are discrepant rates of inducible clindamycin resistance in different geographic area. Rashid Nezhad et al. performed a study in seven Iranian teaching hospitals (25). They found that the frequency of cMLSB, iMLSB and MLSB phenotypes was 52.6%, 12.6%, and 5.3% respectively. Similarly, Fiebelkorn et al. in USA (27) reported that of 114 S. aureus isolates resistance to erythromycin, 34% and 29% indicated constitutive and inducible resistance pattern respectively. In a Canadian survey (28), the frequency of iMLSB and cMLSB phenotypes was found to be 64.7% and 35.3% respectively. In current work, the frequency of constitutive resistance was found to be higher than inducible resistance which is in line with the findings reported by Rashidi Nezhad et al (25).
As previously mentioned, resistance to macrolides is encoded by genes often carried on plasmids (erm(C)) or transposons (erm(A) and erm(B)) and msr genes expressing active efflux pumps mainly msr(A)) (9). Our results revealed the frequency of msr(A), erm(A), msr(B), erm(C), and erm(B) genes to be 40.8%, 26.7%, 14.2%, 11.7%, and 10.8% respectively. In contrast to Rashidi Nezhad’s study (25) who reported erm(A) gene as the predominant gene among the isolates with inducible phenotype and erm(C) among the isolates with the constitutive phenotype, our finding revealed that the msr(A) gene was the most common gene among strains with the constitutive phenotype (35; 29.2%), followed by erm(A) (21; 17.5%), erm(C) (11; 9.2%), erm(B) (10; 8.3%), msr(B) (10; 8.3%) while erm(A) (4; 3.3%), erm(B) (2; 1.7%), erm(C) (1; 0.83%), msr(A) (5; 4.2%) and msr(B) (1; 0.83%) were much more common among the isolates with inducible phenotype. It is worth mentioning that the frequency of erm and msr genes depends on the bacterial species as well as the geographic region in which the study is carried on.
Type III was the most frequently found SCCmec (77.6%), according to the results of SCCmec typing, associated with an MDR pattern among MRSA isolates. This is in consistence with the results reported by Japoni and colleagues (10). The nosocomial origin of the samples was confirmed by the high frequency of SCCmec type III.
A significant relationship was found between the expression of virulence factors and specific agr locus. In consistence with our previous study, the most common agr types were agr type I (75.8%), type III (16.7%), and type II (7.5%), respectively. Goudarzi et al. (12) reported agr type III to be the most frequent type of SCCmec in Iran. According to several studies, the frequency of toxin and adhesion molecules was higher in isolates harboring agr type I gene in comparison with those harboring agr type III; this is in agreement with our study. Therefore, it can presume that regulation of staphylococcal adhesion molecules and toxins is associated with agr type I.
According to the spa typing results, spa t037 was recognized as the most common spa type (23.3%). This spa type was reported from Saudi Arabia, China, Iran as well as among HA-MRSA isolates found in Europe, America and other regions of Asia (12, 29, 30).
In present study spa type t388 was estimated at 22.5%. This spa type has also been reported in a study in Iran (31). Similarly, in a study performed in Taiwan, Ho et al described spa type t388 in MRSA strains recovered from blood cultures in different medical centers (32). It seems that the prevalence of t388 is progressively increased and has been successfully established in our healthcare settings. In our study, the frequency of t713 and t924, earlier reported in UAE and Iran (12), were found to be 16.8% and 8.3% respectively.
In contrast with previous study conducted in Iran (12), in this study, low frequency of t790 and t084 spa types among our isolates, was also demonstrated. For the first time we are reporting t196, and t171 spa types in MSSA strains from Iran.
Using various MRSA typing methods, the isolates were attributed to six different clones, namely ST239-SCCmec III/t388, ST22-SCCmec IV/t790, ST239-SCCmec III/t037, ST15-SCCmec IV/t084, ST585-SCCmec III/t713, and ST239-SCCmec III/t924. In line with previous study from Iran (12), ST239-SCCmec III/t037 clone is currently more frequent in our hospitals (23.3%). The review of the literature reveals that the multiresistant ST239 clone is responsible for at least 90% of HA-MRSA infections in Europe, United States, and some Asian countries, including Kuwait and Malaysia (29). Therefore, the presence of ST239-SCCmec III/t037 in our healthcare setting might be attributed to neighboring regions.
ST239-SCCmec III/t388 (22.5%) was the second most commonly detected MRSA clone. A similar result was reported by Ohadian Moghadam et al from Iran in which major universal MRSA clones were described as ST239, ST291, and ST30 (31).
On the other hand, the third most commonly detected clone was ST22-SCCmec IV/t790 (8.3%). This clone was associated with high resistance to mupirocin, carrying resistance genes, including mecA, msr(A), ant(4΄)-Ia, msr(B), tet(M), and mupA. In our study, all ST22-SCCmec IV/t790 strains contained pvl genes. There are reports of S. aureus ST22 harboring pvl gene from Iran (12), England (33), Saudi Arabia (30), and Kuwait (34). In this regard, a study on PVL-positive MDR-MRSA isolates by Ellington et al (33) from England reported ST5, ST22, ST772, ST80, ST8, and ST59 strains, as later confirmed by Nadig and colleagues (35).
Based on the findings, in clinical MRSA strains, ST15-SCCmec IV/t084 (7.5%) was the fourth most commonly detected clone. The low frequency of this clone has been previously reported in 16 European countries (36). PVL-carrying ST15 isolates were identified in a study by Rasigade and colleagues on 211 S. aureus strains from 19 different countries (37). Five (4.2%) ST15-SCCmec IV/t084 isolates carried pvl genes in our study. Previously, PVL-positive ST15 was reported by Japoni-Nejad and colleagues in Iran (10). In contrast to several studies in which ST15 was reported to be prevalent among CA- and HA-MSSA isolates, all ST15 isolates belonged to MRSA strains in our study. We reported the presence of ST585-SCCmec III/t713 in 5% of isolates. In another study by Goudarzi et al the molecular features of MRSA isolates were identified, and ST585-SCCmec III/t713 was reported in 12% of blood samples from bacteremia patients (12).
To sum up, the present findings showed that MRSA isolates have various genetic backgrounds in our hospitals and involve six major clones. Certain molecular types were associated with some resistance and virulence genes (e.g., eta with ST22-SCCmec IV/t790, ST15-SCCmec IV/t084, and ST239-SCCmec III/t037; mupA with ST15-SCCmec IV/t084 and ST22-SCCmec IV/t790; pvl with ST15-SCCmec IV/t084 and ST22-SCCmec IV/t790; etb with ST585-SCCmec III/t713 and ST22-SCCmec IV/t790). The presence of eight different spa types, i.e., t037, t388, t713, t924, t790, t196, t084, and t171, was also confirmed. For the first time in Iran, STs 182 and 123, as well as spa types t196 and t171, were detected, which might be indicative of the emergence of new clones. Further studies on other neighboring regions, focusing on the emergence of new circulating clones, are necessary to reach an overall understanding of dynamic MRSA clones in Iran and the Middle East.
Acknowledgment
This study was supported financially by the grant (No 11417) from Research Deputy of Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Conflicts of Interest
The authors declare that they have no conflicts of interest with the content of this article.
References
- 1.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dulon M, Haamann F, Peters C, Schablon A, Nienhaus A. MRSA prevalence in European healthcare settings: a review. BMC Infect Dis. 2011;11:138–151. doi: 10.1186/1471-2334-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boye K, Bartels MD, Andersen IS, Moeller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect. 2007;13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 5.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, et al. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Seyedi Marghaki F, Kalantar-Neyestanaki D, Safaari F, Fasihi Y, Moradi M. Frequency of aminoglycoside-resistance genes in methicillin resistant Staphylococcus aureus isolated from clinical specimens. J Mazandaran Univ Med Sci. 2017;27:112–117. [Google Scholar]
- 7.Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. The future of aminoglycosides: the end or renaissance? ChemBioChem. 2010;11:880–902. doi: 10.1002/cbic.200900779. [DOI] [PubMed] [Google Scholar]
- 8.Upton A, Lang S, Heffernan H. Mupirocin and Staphylococcus aureus: a recent paradigm of emerging antibiotic resistance. J Antimicrob Chemother. 2003;51:613–617. doi: 10.1093/jac/dkg127. [DOI] [PubMed] [Google Scholar]
- 9.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43:1716–1721. doi: 10.1128/JCM.43.4.1716-1721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Japoni-Nejad A, Rezazadeh M, Kazemian H, Fardmousavi N, van Belkum A, Ghaznavi-Rad E. Molecular characterization of the first community-acquired methicillin-resistant Staphylococcus aureus strains from Central Iran. I International J Infect Dis. 2013;17:949–954. doi: 10.1016/j.ijid.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; 27th Informational Supplement. 2017 CLSI document M100-S27 (ISBN 1-56238-805-3) [Google Scholar]
- 12.Goudarzi M, Seyedjavadi SS, Nasiri MJ, Goudarzi H, Nia RS, Dabiri H. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb Pathog. 2017;104:328–335. doi: 10.1016/j.micpath.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 13.Alfatemi SMH, Motamedifar M, Hadi N, Saraie HSE. Analysis of virulence genes among methicillin resistant Staphylococcus aureus (MRSA) strains. Jundishapur J Microbiol. 2014;7:e10741. doi: 10.5812/jjm.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol Res. 2006;161:49–54. doi: 10.1016/j.micres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Martineau F, Picard FJ, Lansac N, Ménard C, Roy PH, Ouellette M, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–238. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lina G, Quaglia A, Reverdy M-E, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udo E, Jacob L, Mathew B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high-and low-level mupirocin resistance. J Med Microbiol. 2001;50:909–915. doi: 10.1099/0022-1317-50-10-909. [DOI] [PubMed] [Google Scholar]
- 18.Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40:4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko KS, Lee J-Y, Suh JY, Oh WS, Peck KR, Lee NY, et al. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J Clin Microbiol. 2005;43:421–426. doi: 10.1128/JCM.43.1.421-426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbiol. 2001;39:3115–3121. doi: 10.1128/JCM.39.9.3115-3121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahsavan S, Emaneini M, Khoshgnab BN, Khoramian B, Asadollahi P, Aligholi M, et al. A high prevalence of mupirocin and macrolide resistance determinant among Staphylococcus aureus strains isolated from burnt patients. Burns. 2012;38:378–382. doi: 10.1016/j.burns.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Gadepalli R, Dhawan B, Mohanty S, Kapil A, Das BK, Chaudhry R, et al. Mupirocin resistance in Staphylococcus aureus in an Indian hospital. Diagn Microbiol Infect Dis. 2007;58:125–127. doi: 10.1016/j.diagmicrobio.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Aqel A, Ibrahim A, Shehabi A. Rare occurrence of mupirocin resistance among clinical Staphylococcus isolates in Jordan. Acta Microbiol Immunol Hung. 2012;59:239–247. doi: 10.1556/AMicr.59.2012.2.8. [DOI] [PubMed] [Google Scholar]
- 25.Nezhad RR, Meybodi SM, Rezaee R, Goudarzi M, Fazeli M. Molecular characterization and resistance profile of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients in intensive care unit, Tehran-Iran. Jundishapur J Microbiol. 2017;10:e41666. [Google Scholar]
- 26.Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42:2777–2779. doi: 10.1128/JCM.42.6.2777-2779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiebelkorn K, Crawford S, McElmeel M, Jorgensen J. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740–4744. doi: 10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavallée C, Rouleau D, Gaudreau C, Roger M, Tsimiklis C, Locas M-C, et al. Performance of an agar dilution method and a Vitek 2 card for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2010;48:1354–1357. doi: 10.1128/JCM.01751-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcusaureus. PloS one. 2011;6 doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, Hakawi A, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 2012;12:146–155. doi: 10.1186/1471-2180-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohadian Moghadam S, Pourmand MR, Mahmoudi M, Sadighian H. Molecular characterization of methicillin-resistant Staphylococcus aureus: characterization of major clones and emergence of epidemic clones of sequence type (ST) 36 and ST 121 in Tehran, Iran. FEMS Microbiol Lett. 2015;362:fnv043. doi: 10.1093/femsle/fnv043. [DOI] [PubMed] [Google Scholar]
- 32.Ho C-M, Ho M-W, Li C-Y, Lu J-J. Fine typing of methicillin-resistant Staphylococcus aureus isolates using direct repeat unit and staphylococcal interspersed repeat unit typing methods. J Microbiol Immunol Infect. 2015;48:370–375. doi: 10.1016/j.jmii.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 33.Ellington MJ, Ganner M, Warner M, Cookson BD, Kearns AM. Polyclonal multiply antibiotic-resistant methicillin-resistant Staphylococcus aureus with Panton–Valentine leucocidin in England. J Antimicrob Chemother. 2009;65:46–50. doi: 10.1093/jac/dkp386. [DOI] [PubMed] [Google Scholar]
- 34.Udo E, O’brien F, Al-Sweih N, Noronha B, Matthew B, Grubb W. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J Clin Microbiol. 2008;46:3514–3516. doi: 10.1128/JCM.00966-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadig S, Raju SR, Arakere G. Epidemic meticillin-resistant Staphylococcus aureus (EMRSA-15) variants detected in healthy and diseased individuals in India. J Med Microbiol. 2010;59:815–821. doi: 10.1099/jmm.0.017632-0. [DOI] [PubMed] [Google Scholar]
- 36.Rolo J, Miragaia M, Turlej-Rogacka A, Empel J, Bouchami O, Faria NA, et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PloS one. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasigade J-P, Laurent F, Lina G, Meugnier H, Bes M, Vandenesch F, et al. Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981–2007. J Infect Dis. 2010;201:1589–1597. doi: 10.1086/652008. [DOI] [PubMed] [Google Scholar]