Abstract
Background:
We have previously reported a single nucleotide polymorphism (P-5, G-384A) in the proximal promoter of the gene for G protein receptor kinase 3 (GRK3) that was associated with bipolar disorder in two independent samples. In this study, we examined whether the G-384A variant has a functional effect on GRK3 transcription.
Methods:
Electrophoretic mobility shift assays were conducted using nuclear extracts from both Hela cells and adult mouse cortex. Transcriptional function was also examined using a dual luciferase reporter system transfected into in vitro human neuroblastoma cells and cultured mouse cortical neurons.
Results:
The G-384A variant abolished or reduced the formation of DNA-protein complexes using nuclear extract from both HeLa cells and adult mouse cortical neuron cells. However, gene expression was significantly enhanced by G-384A in both in vitro human neuroblastoma cells and cultured mouse cortical neurons.
Conclusions:
These data suggest that the G-384A SNP in the promoter of human GRK3 gene represents an important functional variant. The G-384A variant may alter binding of Sp1/Sp4 transcription factors resulting in an increase in gene transcription and an increase in vulnerability to bipolar disorder.
Keywords: Bipolar disorder, cortical neuron, EMSA, G-protein receptor kinase 3, gene expression, Sp1 family transcription factors, SNP
G-protein-coupled receptors (GPCRs) are the most numerous superfamily of cell surface receptors and play essential roles for a variety of physiologic functions. Most of the GPCRs are expressed in brain, which mediate the signal transduction of various neuromodulators such as dopamine, noradrenaline, serotonin, glutamate, γ-aminobutyric acid, and others (1). Neuropharmacologic studies have demonstrated that administration of agonists and antagonists for GPCRs in brain result in a range of animal behavioral abnormalities that mimic some human psychiatric disorders such as mania and depression (2–4). Signaling of GPCRs is sensitive and dynamic, in part because of the modulation of receptor function by homologous desensitization mediated by a family of G protein receptor kinases (GRKs) (5). The GRKs can discriminate between the inactive and agonist-activated states of the receptors, and phosphorylate the activated receptors for subsequent receptor internalization. The GRKs thus serve as gatekeepers for receptors, modulating their signaling pathways. GRK3, one of the six known GRKs, is abundantly expressed in several brain regions including cortex, hippocampus, and ventral striatum, suggesting an important role for the GRK3 gene in the modulation of neurotransmission signaling in these regions (6,7).
Two independent genomewide linkage analyses, one including 20 extended families from our lab and one including Wave 1 families from the National Institutes of Mental Health Bipolar Consortium, have suggested a susceptibility locus for bipolar disorder located on chromosome 22q12 near the GRK3 gene (8,9) around the microsatellite markers D22S419 and D22S533. These markers are in the immediate vicinity of G-protein receptor kinase 3. A study of two inhibitory electrophysiologic endophenotypes in families with schizophrenia also found genome-wide maximum evidence of linkage at a marker (D22S315) in intron 1 of the GRK3 gene (10). After conducting a detailed examination of GRK3 in bipolar subjects in search of potential pathogenic mutations and examining several resulting single nucleotide polymorphisms (SNPs) for association to bipolar disorder, we found that two (P-5 and P-6) of six SNPs identified in the proximal promoter of the GRK3 gene displayed association with the disease in transmission disequilibrium tests in two independent sets of family triads (11). Interestingly, the P-5 variant GaGAGGG, which is [H11002]384 bp upstream from the ATG, appears to disrupt the Sp1/Sp4 binding consensus sequence GGGAGGG found in the normal GRK3 allele. For clarity, the variant designated P-5 in our earlier article is referred to as G-384A here. To examine whether the G-384A variant indeed interrupted the binding of Sp1 family transcription factors, we conducted electrophoretic mobility shift assays (EMSA) by using nuclear extracts from both HeLa cells and adult mouse cortex. Dramatic reduction of a binding complex was observed for G-384A variant. The function of the G-384A variant in the promoter of human GRK3 gene was finally analyzed in human neuroblastoma cells and cultured mouse cortical neurons.
Methods and Materials
Electrophoretic Mobility Shift Assays
Two sets of complementary 29mer oligonucleotides, containing either control or G-384A variant, were synthesized. The complementary oligonucleotide pairs were annealed to form the double-stranded DNA and labeled with Digoxigenin by using DIG Gel Shift Kit (DIG Gel Shift Kit, 2nd Generation, cat. No. 3353591; Roche Applied Science, Indianapolis, Indiana); 100 ng of each double-stranded DNA was used for a labeling reaction, and the labeling efficiency was then determined according to the manufacturer’s protocol. We used .4 ng of labeled DNA for incubation with the nuclear extract from either HeLa cell (Promega, Madison, Wisconsin) or mouse cortex. The binding reactions were separated by electrophoresis in nondenaturing 5% polyacrylamide gel (Criterion Precast Gel, cat. 345–0048; Bio-Rad, Hercules, California). After electrophoresis, the samples were transferred to NYTRAN nylon membranes (Schleicher & Schuell, Dassel, Germany) by contact blotting for at least 3 hours. After ultraviolet cross-linking, the DNA-protein complexes were visualized by chemiluminescent detection. For competition experiments, several transcription factor consensus oligonucleotides (AP1, AP2, CREB, SP1, OCT1) were obtained from Promega (Promega). In addition, double-stranded DNA containing GAGA factor binding site (5´ = GAGAGAGAGAGAGAGAGAGAGAG 3´), poly-(AT) (5´ ATATATATATATATATATATAT 3’) were also synthesized. Rabbit polyclonal antibodies for Sp1 (Upstate, Lake Placid, New York) and Sp4 (Santa Cruz Biotechnology, Santa Cruz, California) were used for supershift in the EMSA.
Nuclear Extract Preparation from Adult Mouse Cortex
The cortex was dissected from adult mouse with mixed S129/Black Swiss genetic background and immediately disrupted in homogenization medium (25 mmol/mL Sucrose, 25 mmol/mL KCl, 5 mmol/mL MgCl2, 20 mmol/mL Tris-Cl, pH 7.5) by using a Dounce Homogenizer (cat # KT885300–0002; VWR, West Chester, Pennsylvania) Pestle A (clearance for disruption of tissue) on ice. After 20 strokes, the homogenized mixture was placed on ice for 1 hour. Pestle B (clearance for the disruption of the cell, with the nuclei left intact) was then used to homogenize the mixture for about 20 strokes to release nuclei. Six volumes of 50% NycoPrep (50% NycoPrep, 25 mmol/mL KCl, 5 mmol/mL MgCl2, 20 mmol/mL Tris-Cl, pH 7.5) were added to four volumes of cell lysis and were well mixed and centrifuged at 13,000 rpm for 2 min to pellet nuclei. One volume of Buffer A (10 mmol/mL 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.9, 10 mmol/mL KCl, 1.5 mmol/mL MgCl2, 5 mmol/mL DTT, .5 mmol/mL NaF, .5 mmol/mL Na3VO4, 1X proteinase inhibitor cocktail (Sigma, St. Louis, Missouri) was used to suspend nuclei pellet, and 3 volumes of Buffer B (20 mmol/mL HEPES, pH 7.9, 420 mmol/mL NaCl, 1.5 mmol/mL MgCl2, .2 mmol/mL ethylenediamine tetraacetate, 5 mmol/mL dithiothreitol, .5 mmol/mL NaF, .5 mmol/mL Na3VO4, 1X proteinase inhibitor cocktail (Sigma) 20% Glycerol) was then added to release nuclear proteins, and the samples were incubated on ice for 30 min. After brief centrifugation, the nuclear extract was used immediately or frozen in liquid nitrogen and stored in −70°C.
Construction of Luciferase Reporter Genes
The promoter of human GRK3 gene is a GC-rich region, which is difficult to amplify. Two sets of primers were instead designed to amplify two overlapping fragments from both control and bipolar disorder patient DNA with G-384A variant, respectively. The two sets of primer pairs are as follows: set 1, forward: 5’ CCCATAACCCCTTGGGTTGTGGA 3’, reverse: 5’ CTGCGGGGAGCGCGCGTCCAACGGTCA 3’; set 2, forward:5’ GGTCGGGAGGCGCCGGCCCAGCGA 3’, reverse: 5’ GGCGGCGGTTACTCCGGACCTGGAC 3’. The two overlapping ments were then recombined frag-using polymerase chain reaction (PCR) into full-length human GRK3 promoters. The final putative promoters, the 550 bp DNA fragments from −580 to −30 bp upstream of the start codon of human GRK3 gene were cloned upstream of firefly luciferase reporter gene in pGL3 basic vector (Promega).
Cell Culture and Transfection Experiments
HeLa cells (ATCC, CCL-2) were cultured in Dulbecco’s Modified Eagle Medium with 10% fetal bovine serum and ampicilin/streptomycin. The phRL-TK (Promega) expressing Renilla lucif-erase was used as an internal control for normalization of transfection experiments. One or two micrograms of a firefly luciferase DNA reporter construct mixed with .1 μg phRL-TK constructs (ratio 10:1 or 20:1) were used to transfect cells at 70% confluence with Superfect (Invitrogen, Carlsbad, California) according to manufacturer’s protocol. Six replicate 35-mm dishes were used for each firefly luciferase construct. The transfected cells were harvested 48 hours after transfection and lysed to measure both Renilla and firefly luciferase activities using a Dual Luciferase Kit (Promega).
Human neuroblastoma BE(2)-C cells were cultured in a 1:1 mixture of Eagle’s minimal essential medium with nonessential amino acids and Ham’s F12 medium containing 10% fetal bovine serum with ampicilin/streptomycin. Two micrograms of a firefly luciferase DNA reporter construct mixed with .2 μg phRL-TK constructs (ratio 10:1) were used to transfect cells at 70% confluence with Superfect (Invitrogen) as described earlier. Two double stranded decoy oligodeoxynucleotides (ODN), designated as G3 and P-5 decoy ODNs, were synthesized with the capping of both 3’ and 5’ ends of the two probes (Figure 1A) by phosphorothioate linkages to study the function of the G-384A variant in BE(2)-C cells. For decoy competition experiments, 2 μg of either G3 or P-5 ODNs were mixed with 2 μg of a firefly luciferase DNA reporter construct for the transfection of each well of 12-well plates. Five replicate wells were used for each treatment, and each experiment was repeated. The transfected cells were harvested 48 hours after transfection and lysed to measure both Renilla and firefly luciferase activities using a Dual Luciferase Kit (Promega).
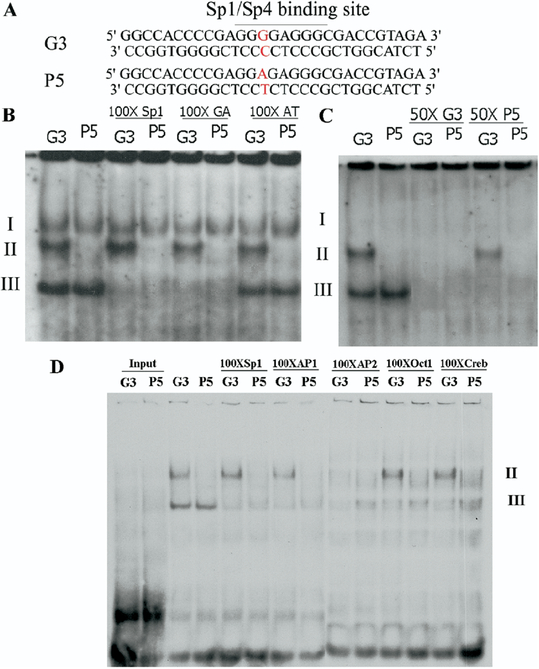
Figure 1.
Alteration of in vitro transcription factor binding by the G-384A SNP in HeLa nuclear extract. (A) Used as the probe for EMSA were 29bp DNA fragments from the promoter of human GRK3 gene. The putative Sp1/Sp4 binding site was identified in the control (G3), and the G-384A (P-5) single nucleotide polymorphisms disrupted the Sp1/Sp4 consensus binding site (P-5). (B) The HeLa nuclear extract was incubated with labeled G3 and P-5 probes, respectively. One-hundred-fold excessive unlabeled Sp1, GA, and AT binding sequences were used for competition experiments. (C) Fifty-fold excessive unlabeled G3, but not P-5, effectively competed out all the binding in HeLa nuclear extract. (D) The consensus binding oligonucleo-tides for five transcription factors were used in the competition binding reaction with HeLa nuclear extract. Only AP2 consensus binding oligonucleotide effectively disrupted complex II.
Primary mouse cortical neurons were established form C57 BL/6 mouse embryos at E16 to E18 (Harlan, Indianapolis, Indiana). The entire cerebral cortex were dissected and cut into small pieces in Neurobasal A medium on ice. The tissue was titrated 10 times with a fire polished 9-inch Pasteur pipette and allowed to settle on ice for 1 min. The supernatant were then transferred to a new tube and centrifuged gently at 600–700 rpm for 5 min to pellet the cells. The cells were resuspended in B27/Neurobasal medium (B27/Neurobasal with .5 mmol/mL glutamine, no glutamate, 5 ng/mL FGF2) and counted. About 20 million cells were seeded in each well (12-well plates) coated with poly-D-lysine. The half of culture medium was changed every 3 days. The transfection experiments were performed at the sixth day of cortical neuron culture. Three micrograms of each firefly reporter construct per well, mixed with 1 [H9262]g of phRL-TK construct, was transfected using the NeuroPorter transfection system (Sigma) for the transfection of the primary cultured cortical neurons in each well. For each firefly DNA construct, five replicate wells were used for transfection. After overnight incubation, half of the medium was replaced. The cells were harvested 48 hours later for measurement of dual luciferase activities, and the firefly luciferase activity was normalized with the activity of Renilla luciferase. The Student’s t test was used for statistical analysis.
Results
Attenuated Formation of DNA-Protein Complex by G-384A Variant
To examine the potential function of the G-384A variant in the promoter of human GRK3 gene, we conducted EMSA to investigate whether the G-384A variant altered the binding of any transcription factors. Two double-stranded DNA probes, differing only by the presence of the G-384A variant, were used for the formation of DNA-protein complexes with nuclear extracts from HeLa cells (Figure 1A). Three distinct complexes (I, II, III) were formed with the control probe (G3). However, there were only two complexes (I and III) formed when the G-384A variant (P-5) probe was used, suggesting that the G-384A variant prevented the formation of complex II (Figure 1B). Complex I did not show up in the next several EMSAs, suggesting that it is unstable. To confirm these DNA-protein complexes, we conducted competition experiments. Addition of 50-fold excess unlabeled G3 oligonucleotide completely abolished the formation of both complexes II and III. In contrast, addition of 50-fold excess unlabeled P-5 oligonucleotides only abolished the complex III, whereas the complex II remained unchanged (Figure 1C). Because the G-384A variant disrupts the consensus Sp1 binding sequence and HeLa nuclear extract contains abundant Sp1 proteins, we performed competition experiments by including 100-fold excessive Sp1 consensus binding sequence in the binding reaction. Complex II remained intact, suggesting that complex II did not result from the binding of Sp1 family transcription factors. In contrast, complex III was abolished by the addition of 100-fold excess Sp1 consensus sequence; however, it was also abolished by the addition of 100-fold excessive GAGA binding sequence (Figure 1B). To determine further which transcription factor was involved in the formation of complex II, we performed competition experiments with several candidate transcription factor consensus oligonucleo-tides (Figure 1D). Complex III was effectively abolished by all the oligonucleotides. However, complex II could only be abolished by the AP2 consensus oligonucleotide, suggesting that the AP2 transcription factor was responsible for the formation of complex II in HeLa nuclear extract.
Because the GRK3 gene is highly expressed in brain, we studied whether the G-384A variant might also have a different binding affinity for transcription factors from neuronal cells. Nuclear extract from adult mouse cortex was then prepared for binding with both G3 and P-5 probes. There was only a single complex formed for both the control (G3) and the G-384A variant (P-5) probes. However, this complex had much less intensity for the P-5 probe than for the G3 probe. To determine whether Sp1 family transcription factors were involved in the formation of this complex, we conducted competition experiments by including 100-fold excessive Sp1 consensus binding sequence in the binding reaction. The complex was totally abolished (Figure 2A). To examine the sequence binding specificity, two other non-Sp1 consensus binding sequences (GAGA and AT) were included in the binding reactions, respectively. Neither of the two sequences was capable of disrupting the formation of the complex. Taken together, the competition experiments suggested that the complex is likely formed by the Sp1 family transcription factors. To differentiate which member of the Sp1 family of transcription factors contribute to the formation of the complex, we performed supershift experiments by adding either Sp1 or Sp4 antibodies to the binding reactions. Both antibodies abolished the formation of the complex instead of supershifting the complex (Figure 2B). As expected, the addition of 50-fold excessive unlabeled G3 probe completely abolished the formation of complex, whereas the addition of 50-fold excessive unlabeled P-5 probe was unable to abolish the formation of the complex.
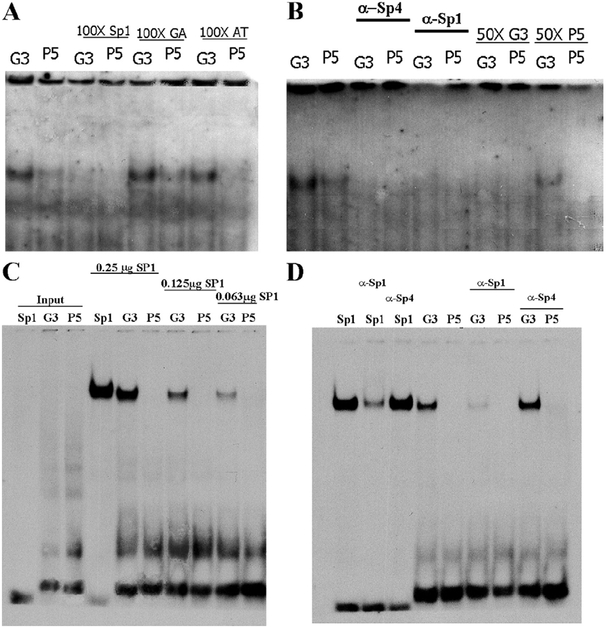
Figure 2.
Attenuation of Sp1 family transcription factor binding by the G-384A single nucleotide polymorphism in mouse cortical nuclear extract. (A)The mouse cortical nuclear extract was incubated with labeled G3 and P-5 probes respectively. One-hundred-fold excessive unlabeled Sp1, GA, and AT binding sequences were used for competition experiments. Only Sp1 consensus binding oligonucleotide effectively competed away the binding complex. (B) Polyclonal antibodies against Sp1 and Sp4 were included in the binding reactions to identify the binding transcription factors responsible for the formation of the complex. Fifty-fold excessive unlabeled G3 and P-5 oligonucleotides were also tested in the competition experiments. Only the G3 oligonucleotides effectively competed out the formation of the complex. (C) The disruption of recombinant human SP1 transcription factor by the G-384A SNP. The Sp1 consensus binding G3 and P-5 oligonucleotides were labeled and incubated with purified recombinant human SP1 transcription factors. Strong binding activities were observed with both Sp1 consensus and G3 oligonucleotides, but not P-5. (D) The Sp1 polyclonal antibodies dramatically decreased the formation of the complex, instead of supershifting the complexes in either Sp1 or G3 probes. The Sp4 polyclonal antibodies have no effect on the formation of SP1-DNA complex.
To determine whether the G-384A SNP indeed disrupted the binding of SP1 family transcription factors, we conducted EMSA using purified recombinant human SP1 transcription factor. The recombinant SP1 transcription factors readily bound both Sp1 consensus and G3 oligonucleotides (Figure 2C). However, no detectable SP1 binding was observed for the P-5 oligonucleo-tides, suggesting that the G-384A SNP disrupts the binding of SP1 transcription factor. In our EMSA using nuclear extract from mouse cortex, we observed that the addition of either Sp1 or Sp4 antibodies failed to supershift the complex but disrupted the formation of the complex. To clarify the specificity of antibodies in the EMSA experiments, we performed supershift experiments using purified recombinant human SP1 transcription factor. A DNA-protein complex was readily formed between the recombinant SP1 transcription factor and the labeled Sp1 consensus oligonucleotides. However, the complex was dramatically disrupted by the coincubation of Sp1 antibodies, but not Sp4 antibodies (Figure 2D). When the G3 probe was used for the supershift experiments, the same disruption was observed by using anti-SP1 antibodies, but not anti-SP4 antibodies. Therefore, it is clear that the antibodies we used in EMSA specifically disrupted the formation of complex in our EMSA reactions instead of supershifting the complex. It has been reported that in some cases antibodies disrupted the formation of DNA-protein complex in EMSA (12). However, it is unclear why both Sp1 and Sp4 antibodies could abolish the complex in the EMSA using nuclear extract from mouse cortex. The potential cross reactions between anti-SP1 and anti-SP4 polyclonal antibodies were not supported by our experiments with purified SP1 binding experiments. Therefore, it is possible that the complex may contain both Sp1 and Sp4 proteins.
Functional Analysis of the G-384A Variant in Cell Culture Experiments
Our in vitro binding experiments indicated that the G-384A variant could play an important role in the regulation of human GRK3 expression. To study the function of the G-384A variant, we first cloned the putative promoters, 550bp DNA fragments from −580 to −30 upstream of the start codon of human GRK3 gene, from both control and patient DNA. The promoters were then sequenced to confirm that no additional mutations were introduced except the G-384A variant (Figure 3A). The putative promoters were further cloned upstream of firefly luciferase reporter gene in pGL3-Basic vector (Promega). The two final expression constructs (G3 and P-5) were identical except the G-384A variant in the promoter of the P-5 construct (Figure 3B).
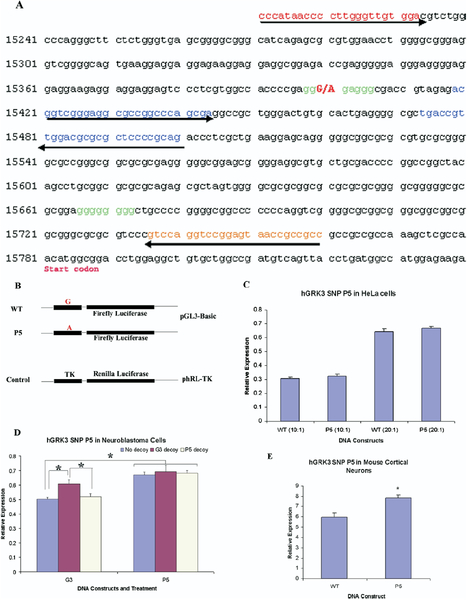
Figure 3.
Functional characterization of the G-384A single nucleotide polymorphisms. (A) The putative promoter of human GRK3 gene was amplified from control and patient DNA, respectively. The subsequent DNA fragments were confirmed by double-strand DNA sequencing. (B) The two promoters were cloned upstream of firefly Luciferase Gene in the pGL3-Basic vectors. The TK-Renilla luciferase construct was used as an internal control for the transfection experiments. Relative activities of both wildtype and G-384A promoters in HeLa cells (C), BE(2)-C cells (D), and primary cultured mouse cortical neurons (E) after the normalization of the cotransfected Renilla luciferase construct. * p<.05.
To test whether the 550bp DNA fragments upstream of human GRK3 start codon function as putative promoters, we transfected these constructs into HeLa cells. After 48 hours, we examined the expression of both firefly luciferase and Renilla luciferase genes. After normalization with cotransfected internal control Renilla luciferase gene, no significant difference was found between G3 (WT) and P-5 (G-384A) constructs (the experiments were repeated three times, and each experiment consisted of six replicates; Figure 3C). Because different cells express different sets of transcription factors, which may in turn form different DNA-protein complexes, it is critical to study the most relevant cell system to understand the effect of G-384A on transcriptional regulation.
The expression of the endogenous GRK3 gene has been demonstrated in human neuroblastoma BE(2)-C cells. The activation of adrenegeric receptors was suggested to regulate the expression of GRK3 gene (13). To examine the function of the G-384A variant, we transfected a firefly luciferase DNA reporter construct with an internal control phRL-TK in BE(2)-C cells and measured the activities of both firefly and Renilla luciferases. After normalization with the activity of Renilla luciferases, the G-384A variant significantly increased the expression of the downstream firefly luciferase reporter gene (p value [H11005] .0001; Figure 3D). To confirm the regulatory function for the G-384A variant, either G3 or P-5 decoy ODN was included in the transfection of BE(2)-C cells. The G3 decoy ODN significantly increased the expression of the firefly luciferase reporter gene, suggesting that the G3 decoy ODN sequestered transcription repressors from binding to the G3 site in the GRK3 promoter. In contrast, there was no effect for the P-5 decoy ODN in the regulation of the cotransfected luciferase DNA reporter gene. There were no significant effects for either G3 or P-5 decoy ODN on the regulation of the GRK3 promoter containing the G-384A variant. Taken together, these data suggest that the G-384A variant prevents the binding of certain transcription factors which might function as repressors in BE(2)-C cells.
Primary cultured mouse cortical neurons, in which GRK3 is abundantly expressed, were also studied as a system that would be more informative regarding the effect of the G-384A variant on GRK3 expression in brain. Mouse embryonic cortical neurons were isolated and cultured for a week before transfection, when the neurons appeared well differentiated. Two days after transfection, the cells were harvested for the measurement of activities of the dual luciferases. Surprisingly, the G-384A variant significantly increased the expression of the downstream reporter gene after normalization of cotransfected Renilla luciferase gene (each experiment consists of five replicate wells for each DNA construct, Student t test p value = .013; Figure 3E). The experiments were repeated three times and yielded the same result that the G-384A variant increased the activity of human GRK3 gene in the primary cultured mouse cortical neurons. Because the G-384A variant decreases the binding of Sp1/Sp4, further studies of Sp1/Sp4 as potential repressors in the regulation of human GRK3 expression are warranted.
Discussion
Our previous human linkage analyses have suggested that GRK3 gene is a strong candidate gene on chromosome 22q12 for bipolar disorder (11). Further sequence analysis revealed the significant association of one SNP (P-5, G-384A), disrupting the Sp1/Sp4 consensus binding site, in the proximal promoter of the GRK3 gene with the disease. This study provides biological evidence for the potential functional role of the G-384A variant. Our in vitro EMSA demonstrated that the G-384A variant attenuated the binding of Sp1/Sp4 transcription factors in the nuclear extract made from adult mouse cortex. Our subsequent functional analysis revealed an increased transcriptional activation by the G-384A variant in both human neuroblastoma BE(2)-C cells and primary cultured mouse cortical neurons. Potential repression mediated by the Sp1 family transcription factors has been reported in the regulation of several genes (14,15). Therefore, the G-384A variant may represent a functional SNP for susceptibility for bipolar disorder. However, it may need to interact with Sp1/Sp4 transcriptional factors to exert its regulatory role in the transcription of human GRK3 gene in cortical neurons.
The G-384A variant may affect the binding of more than just Sp1 family transcription factors. Other transcription factors, which share overlapping consensus binding sequences with Sp1 family transcription factors, may also be affected by the G-384A variant, as shown by the EMSA experiments with HeLa cell nuclear extract. DNA competition experiments indicated that the AP2 family transcription factors may be disrupted by the G-384A SNP in HeLa cells. To examine the potential functional consequence, we conducted transient expression experiments. No differential expression was found, suggesting that the binding of AP2 transcription factor was not essential for the baseline expression of human GRK3 expression in HeLa cells. However, it remains to be understood whether the SNP G-384A, by disrupting AP2 binding, may be involved in the regulation of GRK3 expression under certain conditions. Therefore, the effect of the G-384A variant could vary by cell type. The potential involvement of the G-384A variant in the regulation of GRK3 expression in nonneuronal cells could not be ruled out.
The aberrant expression of GRKs has been found in several human psychiatric disorders (16–18). Upregulation of GRK2 in the prefrontal cortex was found to associate with major depression and antidepressant treatment suppressed that overexpression (19). Therefore, it would be interesting to know whether the upregulation of the GRK3 promoter mediated by G-384A SNP in cortical neurons would play a similar role in the pathophysiology of human bipolar disorder, considering that both GRK2 and GRK3 are functionally partially redundant.
The physiological function of the Grk3 gene was implicated by recent studies on mouse Grk3 null mutants. Contrary to the expected supersensitive response to dopaminergic agonists, assuming that lack of GRK3 would undermine the desensitization of dopamine receptors, the mutant mice instead displayed less sensitivity to the administration of either cocaine or apomorphine in open field tests (1). The expression of Grk3 gene in presynaptic dopaminergic neurons may contribute to the unexpected behavioral abnormalities in which presynaptic dopamine receptor D3 was suggested to be desensitized by the GRK3 to modulate the release of dopamine (20). The high expression of mouse Grk3 mRNA in substantia nigra supports the potential role for GRK3 in the modulation of presynaptic dopamine release. Our studies of dopamine concentration in the caudate, inner-vated by the projection of dopaminergic neurons form substantia nigra, indicated significantly higher concentration of dopamine and 3,4-dihydroxy-phenylacetic acid in the Grk3 null mutants than the sibling wildtype control mice (data not shown). GRK3 may play important roles not only in the postsynaptic neurons presumably in cortex but also in presynaptic neurons in substantia nigra. Further studies of the physiologic functions of GRK3 in the modulation of neurotransmission involved in animal behaviors and its relevance to bipolar disorder are therefore warranted.
Sp1 family transcription factors are critical for the expression of genes directed by GC-rich promoters, most of which reside in CpG islands and are highly expressed in brain. Several SNP variants in Sp1 binding sites have been reported to be associated with schizophrenia in the promoters of N-methyl-D-aspartate receptor 2B (21) and quinone oxidoreductase2 gene (22). The importance of Sp1 family transcription factors has also been underscored in our previous studies of mice harboring a hypomorphic Sp4 gene, an Sp1 family member recognizing the same DNA binding motif as Sp1, that displayed robust deficits in memory and sensorimotor gating (23). These behavioral abnormalities are considered putative endophenotypes for several human psychiatric disorders including bipolar disorder (24,25). Our recent human genetic studies on the association of human SP4 gene with bipolar disorder suggested that the human SP4 gene be a susceptibility gene for bipolar disorder (data to be reported elsewhere). Therefore, the G-384A variant, by dramatically decreasing the binding of Sp1/Sp4 binding in the promoter of human GRK3 gene, may epistatically interact with SP4, another susceptibility gene, in the pathogenesis of bipolar disorder. Their genetic interactions merit further investigation in the future studies.
Acknowledgments
This work was supported by a Young Investigator Award to XZ from the National Alliance on Research in Schizophrenia and Affective Disorders (NARSAD); by grants to JRK from the Department of Veterans Affairs and the National Institute of Mental Health (NIMH) (Grant Nos. MH47612, MH59567, and MH68503), the University of California—San Diego General Clinical Research Center (M01 RR00827), and the VA VISN22 MIRECC; and to TBB from the NIMH (Grant Nos. MH067959 and MH071911). TBB is currently at the Department of Psychiatry, Portland VA Medical Center, Portland, Oregon.
Footnotes
JRK and TBB are founders and hold equity in Psynomics, Inc. The terms of this arrangement have been reviewed and approved by the University of California—San Diego in accordance with its conflict of interest policies. XZ reports no biomedical financial interests or conflicts of interest.
References
- 1.Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG (2004): Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci 27:107–144. [DOI] [PubMed] [Google Scholar]
- 2.Mamelak M (1978): An amphetamine model of manic depressive illness. Int Pharmacopsychiatry 13:193–208. [DOI] [PubMed] [Google Scholar]
- 3.Ong JC, Brody SA, Large CH, Geyer MA (2005): An investigation of the efficacy of mood stabilizers in rodent models of prepulse inhibition. J Pharmacol Exp Ther 315:1163–1171. [DOI] [PubMed] [Google Scholar]
- 4.Cryan JF, Kaupmann K (2005): Don’t worry “B” happy!: A role for GABA(B) receptors in anxiety and depression. Trends Pharmacol Sci 26:36–43. [DOI] [PubMed] [Google Scholar]
- 5.Premont RT (2005): Once and future signaling: G protein-coupled receptor kinase control of neuronal sensitivity. Neuromolecular Med 7:129–147. [DOI] [PubMed] [Google Scholar]
- 6.Erdtmann-Vourliotis M, Mayer P, Ammon S, Riechert U, Hollt V (2001): Distribution of G-protein-coupled receptor kinase (GRK) isoforms 2, 3, 5 and 6 mRNA in the rat brain. Brain Res Mol Brain Res 95:129–137. [DOI] [PubMed] [Google Scholar]
- 7.Arriza JL, Dawson TM, Simerly RB, Martin LJ, Caron MG, Snyder SH, Lefkowitz RJ (1992): The G-protein-coupled receptor kinases beta ARK1 and beta ARK2 are widely distributed at synapses in rat brain. J Neurosci 12:4045–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, et al. (2001): A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci U S A 98:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edenberg HJ, Foroud T, Conneally PM, Sorbel JJ, Carr K, Crose C, et al. (1997): Initial genomic scan of the NIMH genetics initiative bipolar ped igrees: Chromosomes 3, 5, 15, 16, 17, and 22. Am J Med Genet 74:238–246. [PubMed] [Google Scholar]
- 10.Myles-Worsley M, Coon H, McDowell J, Brenner C, Hoff M, Lind B, et al. (1999): Linkage of a composite inhibitory phenotype to a chromosome 22q locus in eight Utah families. Am J Med Genet 88:544–550. [PubMed] [Google Scholar]
- 11.Barrett TB, Hauger RL, Kennedy JL, Sadovnick AD, Remick RA, Keck PE, et al. (2003): Evidence that a single nucleotide polymorphism in the promoter of the G protein receptor kinase 3 gene is associated with bipolar disorder. Mol Psychiatry 8:546–557. [DOI] [PubMed] [Google Scholar]
- 12.Ross S, Tienhaara A, Lee MS, Tsai LH, Gill G (2002): GC box-binding transcription factors control the neuronal specific transcription of the cyclin-dependent kinase 5 regulator p35. J Biol Chem 277:4455–4464. [DOI] [PubMed] [Google Scholar]
- 13.Salim S, Standifer KM, Eikenburg DC (2007): Extracellular signal-regulated kinase 1/2-mediated transcriptional regulation of G-protein-coupled receptor kinase 3 expression in neuronal cells. J Pharmacol Exp Ther 321:51–59. [DOI] [PubMed] [Google Scholar]
- 14.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, et al. (1999): Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 19:5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng D, Kan YW (2005): The binding of the ubiquitous transcription factor Sp1 at the locus control region represses the expression of beta-like globin genes. Proc Natl Acad Sci U S A 102:9896–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sevilla JA, Escriba PV, Ozaita A, La Harpe R, Walzer C, Eytan A, Guimon J (1999): Up-regulation of immunolabeled alpha2A-adrenoceptors, Gi coupling proteins, and regulatory receptor kinases in the prefrontal cortex of depressed suicides. J Neurochem 72:282–291. [DOI] [PubMed] [Google Scholar]
- 17.Shaltiel G, Shamir A, Levi I, Bersudsky Y, Agam G (2006): Lymphocyte G-protein receptor kinase (GRK)3 mRNA levels in bipolar disorder. Int J Neuropsychopharmacol 9:761–666. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Sevilla JA, Ventayol P, Perez V, Rubovszky G, Puigdemont D, Ferrer-Alcon M, et al. (2004): Regulation of platelet α 2A-adrenoceptors, Gi proteins and receptor kinases in major depression: Effects of mirtaza-pine treatment. Neuropsychopharmacology 29:580–588. [DOI] [PubMed] [Google Scholar]
- 19.Grange-Midroit M, Garcia-Sevilla JA, Ferrer-Alcon M, La Harpe R, Huguelet P, Guimon J (2003): Regulation of GRK 2 and 6, beta-arrestin-2 and associated proteins in the prefrontal cortex of drug-free and antidepressant drug-treated subjects with major depression. Brain Res Mol Brain Res 111:31–41. [DOI] [PubMed] [Google Scholar]
- 20.Kim KM, Gainetdinov RR, Laporte SA, Caron MG, Barak LS (2005): G protein-coupled receptor kinase regulates dopamine D3 receptor signaling by modulating the stability of a receptor-filamin-beta-arrestin complex. A case of autoreceptor regulation. J Biol Chem 280:12774–12780. [DOI] [PubMed] [Google Scholar]
- 21.Miyatake R, Furukawa A, Suwaki H (2002): Identification of a novel variant of the human NR2B gene promoter region and its possible association with schizophrenia. Mol Psychiatry 7:1101–1106. [DOI] [PubMed] [Google Scholar]
- 22.Harada S, Tachikawa H, Kawanishi Y (2003): A possible association between an insertion/deletion polymorphism of the NQO2 gene and schizophrenia. Psychiatr Genet 13:205–209. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR (2005): Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol Psychiatry 10:393–406. [DOI] [PubMed] [Google Scholar]
- 24.Braff DL, Geyer MA, Swerdlow NR (2001): Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 156:234–258. [DOI] [PubMed] [Google Scholar]
- 25.Perry W, Minassian A, Feifel D, Braff DL (2001): Sensorimotor gating deficits in bipolar disorder patients with acute psychotic mania. Biol Psychiatry 50:418–424. [DOI] [PubMed] [Google Scholar]