SUMMARY
Ankyrin B (AnkB/LegAU13) is a translocated F box effector essential for the intracellular replication of the pathogen Legionella pneumophila. AnkB co-opts a host ubiquitin ligase to decorate the pathogen-containing vacuole with K48-linked polyubiquitinated proteins and degrade host proteins as a source of energy. Here, we report that AnkB commandeers the host ubiquitin-proteasome system through mimicry of two eukaryotic protein domains. Using X-ray crystallography, we determined the 3D structure of AnkB in complex with Skp1, a component of the human SCF ubiquitination ligase. The structure confirms that AnkB contains an N-terminal F box similar to Skp2 and a C-terminal substrate-binding domain similar to eukaryotic ankyrin repeats. We identified crucial amino acids in the substrate-binding domain of AnkB and showed them to be essential for the function of AnkB in L. pneumophila intracellular proliferation. The study reveals how Legionella uses molecular mimicry to manipulate the host ubiquitination pathway and proliferate intracellularly.
In Brief
Wong et al. describe the structure of the essential Legionella effector, AnkB, in complex with human Skp1, identifying important residues for the intracellular replication of Legionella pneumophila. This reveals how the pathogen uses molecular mimicry to manipulate host proteostasis and evade host defenses.
INTRODUCTION
Legionnaire’s disease is an atypical form of pneumonia with a fatality rate of up to 34% (Phin et al., 2014). The causative agent, Legionella pneumophila, is a Gram-negative bacterium found naturally in aquatic environments (McDade et al., 1977). It infects alveolar macrophages when contaminated aerosol is inhaled.L. pneumophila then uses the Dot/Icm type IV secretion system to translocate bacterial proteins, termed effectors, into the host cell where they manipulate eukaryotic processes to create a replicative niche termed the Legionella-containing vacuole (LCV) and avoid lysosomal fusion (Vogel et al., 1998). Approximately 300 effectors are injected, many of which are redundant (Burstein et al., 2009; Luo and Isberg, 2004). The L. pneumophila genome codes for a high number of eukaryotic-like proteins that interfere with the host through molecular mimicry. Ankyrin B (AnkB) is one of very few effectors essential for bacterial replication within human macrophages and ameba (Al-Khodor et al., 2008) and is conserved across the sequenced L. pneumophila genomes (Chien et al., 2004; Cazalet et al., 2004; Glockner et al., 2008; Burstein et al., 2016). Bioinformatic analysis predicts that AnkB of strain AA100/130b (lpg2144) contains an N-terminal F box domain, a two-repeat ankyrin domain (Cazalet et al., 2004; de Felipe et al., 2005), and a C-terminal CaaX farnesylation motif (Ivanov et al., 2010; Price et al., 2010) (Figures 1A and 1B).
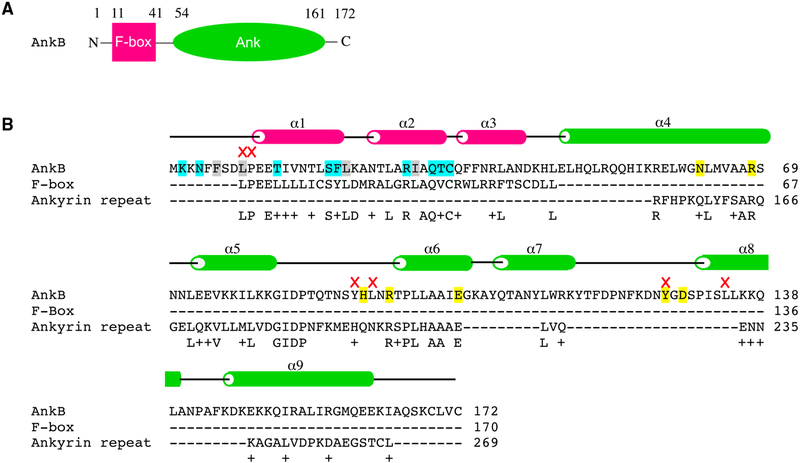
Figure 1. Domain Organization.
(A) AnkB is composed of two domains: an F box (pink) and ankyrin repeats (green).
(B) Sequence alignment of AnkB with human proteins containing an F box [EAW49753.1] and ankyrin repeats [AAH11608.2]. The secondary structure elements and important residues in AnkB are highlighted: cyan, hydrogen bonds with Skp1; gray, hydrophobic interactions with Skp1; yellow, putative substrate-binding residues in ankyrin repeats; red crosses, mutations that prevent intracellular growth; α, α helix.
F box-containing proteins are part of Skp1-Cullin-F box (SCF) E3 ubiquitin ligase complexes, which transfer ubiquitin from an E2 ubiquitin-conjugating enzyme to a target protein (Lyapina et al., 1998; Skowyra et al., 1997). The F box mediates interactions with Skp1, which in turn attaches to Cullin and the E2 enzyme. In eukaryotes, the F box is typically paired with a protein-protein interaction domain that confers substrate specificity. These domains are typically either tryptophan-aspartate (WD) or leucine-rich (LRR) repeats (Price and Kwaik, 2010; Zheng et al., 2002; Uro-Coste et al., 1998). F box proteins without an extra interaction domain exist as the FBXO (F box only) type (Kipreos and Pagano, 2000). In AnkB, the F box is paired with ankyrin repeats, a ~33 residue helix-turn-helix repeat motif that mediates protein interactions (Li et al., 2006). The association of an F box domain and ankyrin repeats is unusual and not found in proteins in metazoans.
The function of AnkB in cells and the reason it is required for Legionella intracellular growth are not clear. A null mutant of ankB exhibits severe intracellular defect in the protozoan host Acanthamoeba polyphaga, and human macrophages (Al-Khodor et al., 2008). Mutagenesis studies have shown that both the F box and farnesylation motif are required for AnkB function of strain A100/130b (Price et al., 2009, 2010; Ivanov et al., 2010; Al-Quadan et al., 2011). Here, we present the crystal structure of AnkB in complex with human Skp1, revealing the specific host-pathogen interactions by which AnkB takes control of the host ubiquitin-proteasome system. We identify a protein-protein interaction site in the ankyrin domain for putative substrates and use mutagenesis and in vivo functional assays to show the ankyrin repeats are critical for poly-ubiquitination of the LCV and pathogen survival.
RESULTS
Structure of AnkB/Skp1
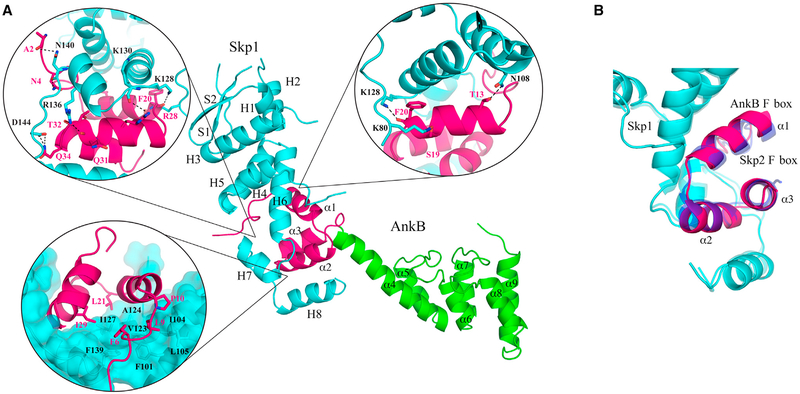
The structure of the AnkB effector was determined in complex with its host partner, Skp1. It contains one molecule in the asymmetric unit with interpretable electron density for Pro2-Cys160 of Skp1 and Lys2-Ala165 of AnkB. The structure of AnkB resembles a step stool, with the F box clasped into a groove formed by helices 5–8 of Skp1, and the ankyrin domain forming the next step (Figure 2A). The F box adopts a typical fold, with three α helices in a right-handed superhelical organization. An overlay of the F box of AnkB with other F boxes reveals high similarity, a root-mean-square deviation (RMSD) of 0.7Å over 33 Cα atoms with a Skp1-Skp2 complex (PDB: 1FQV) (Figures 2B and S1) (Schulman et al., 2000).
Figure 2. Structure of AnkB as a Component of the E3 Ubiquitin Ligase Complex.
(A) Co-crystal of AnkB (pink, F box; green, ankyrin repeats) and Skp1 (cyan). Hydrogen bonds between AnkB and human Skp1 are highlighted. Residues involved in the hydrophobic interaction surface between Skp1 and F box are labeled. In addition to the four AnkB residues involved, Pro10 is also shown. Leu9 and Pro10 are highly conserved between F boxes and abolish binding when mutated to alanine (Price et al., 2009). Skp1 secondary structure elements are labeled: H, α helix; S, β sheet. AnkB secondary structure elements are labeled: α, α helix.
(B) Comparison of AnkB (pink) and the human F box protein Skp2 (blue) binding to Skp1 (cyan) (PDB: 1FQV) yields an RMSD of 0.7Å over the F box Cα atoms. See also Figure S1.
We also solved the structure of the isolated ankyrin domain (residues 54–168) to close to 1Å resolution. The domain is composed of three ankyrin repeats, one more than originally predicted, a short middle repeat (Pro97 to Lys116) flanked by two longer repeats (Ile54 to Lys81 and Pro131 to Glu161). Each repeat adopts a helix-turn-helix fold with connecting loops forming an L-shaped interaction surface typical of other ankyrin repeats (Figures 1B and 2A) (Parra et al., 2015). The majority of ankyrin domains contain four to seven repeats, while up to 34 repeats have been reported (Li et al., 2006). The largest sequence differences generally occur in the loop regions and confer binding specificity. Refinement statistics for both structures are shown in Table 1.
Table 1.
Data Collection and Refinement Statistics for AnkB
| Data Collection | 54–168 | 1–168/Skp1(1–163) |
|---|---|---|
| Space group | C2221 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 54.32, 80.49, 54.08 | 53.58, 57.04, 150.90 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–1.15 (1.17–1.15)a | 50–2.85 (2.90–2.85) |
| Rsym | 0.104 (0.435) | 0.113 (0.622) |
| I/σI | 22.3 (3.8) | 48.5 (6.42) |
| Completeness (%) | 98.6 (97.9) | 100 (100) |
| Redundancy | 3.6 (3.6) | 14.2 (14.5) |
| CC1/2 in highest shell | 0.842 | 0.920 |
| Refinement | ||
| Resolution (Å) | 45.0–1.15 | 75.45–2.85 |
| No. reflections | 39,731 | 10,735 |
| Rwork/Rfree | 0.173/0.191 | 0.219/0.275 |
| No. atoms | ||
| Protein | 941 | 2,498 |
| Water | 166 | 5 |
| B-factors | ||
| Protein | 12.8 | 40.4 |
| Water | 24.5 | 48.7 |
| RMSDs | ||
| Bond lengths (Å) | 0.016 | 0.008 |
| Bond angles (°) | 1.697 | 1.253 |
| Ramachandran statistics (%) | ||
| Most favored regions | 100.0 | 95.7 |
| Additional allowed regions | 0.0 | 4.3 |
Highest resolution shell is shown in parentheses.
Structural Basis of AnkB-Skp1 Binding
Full-length AnkB was insoluble when expressed without Skp1. The structure of the complex explains this phenomenon as the F box is unlikely to fold without Skp1. A large hydrophobic surface formed by the N-terminal tail and helices 1 and 2 of AnkB interacts with helices 5, 6, and 7 of Skp1 (Figure 2A). Multiple hydrogen bonds with helix 7 and its surrounding loops of Skp1 also stabilize the interaction. AnkB is insoluble without Skp1 to shield the hydrophobic surfaces and provide polar contacts.
A previous mutagenesis study showed that a mutation in the AnkB F box domain leads to a defect in intracellular bacterial proliferation (Price et al., 2009). The L9A P10A mutant is unable to interact with host Skp1 and fails to decorate the LCV with polyubiquitinated proteins, a crucial source of carbon and energy for intracellular proliferation (Price et al., 2009). The leucine forms part of the hydrophobic interaction surface with Skp1, while the proline is responsible for initiating the first F box α helix (Figure 2A). Both residues are highly conserved among F box domains (Kipreos and Pagano, 2000) and their mutation to alanine likely prevents proper folding of the AnkB F box domain.
Identification of the Substrate-Binding Site on AnkB
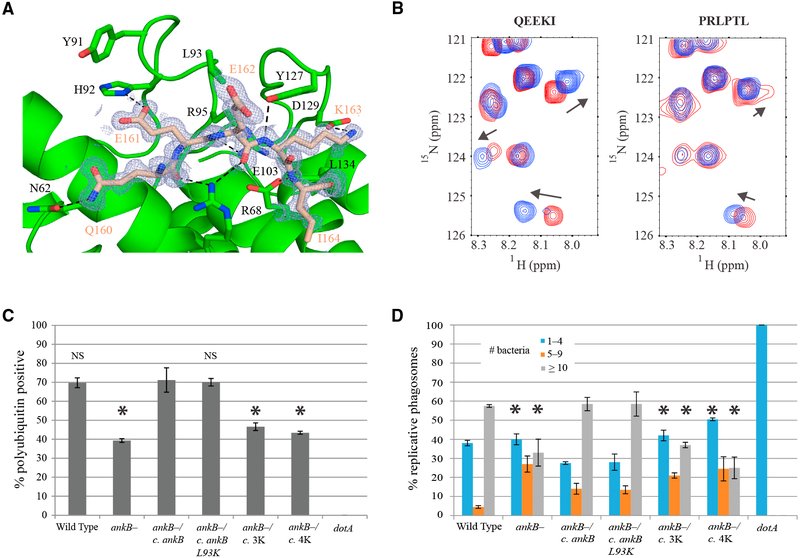
We observed unusually well-ordered crystal contacts between the C-terminal tail of AnkB and the ankyrin domain of another molecule (Figure 3A). The contacts also occurred in the crystals of the AnkB-Skp1 complex that adopt a different space group (Figure S2). In the ankyrin domain crystal, a total of nine hydrogen bonds are formed between the backbone of the C-terminal tail, Q160EEKI, and the putative AnkB substrate-binding site. Additional side chain polar contacts contribute to the structuring of the peptide in the groove formed by the first two ankyrin repeats.
Figure 3. Substrate Binding by the Ankyrin Repeats.
(A) Crystal contacts in the AnkB ankyrin repeats mimic substrate binding. The C-terminal tail (residues 160–164, QEEKI) of one molecule (wheat) binds to the ankyrin repeats of another (green). Hydrogen bonds are indicated by dashed black lines. Residues involved in contacting the peptide and residues that were mutated for further functional studies are labeled in black. An omit map of the substrate is colored and labeled in wheat. See also Figure S2.
(B) Downfield region of HSQC spectra of the 15N-labeled ankyrin domain show chemical shifts upon titration with the QEEKI peptide (C-terminal tail) and weaker binding upon titration with the PRLPTL peptide (negative control) at 0 mM (red) and 8 mM (blue).
(C) Percentage of LCVs colocalizing with polyubiquitinated proteins by confocal microscopy at 2 hr post-infection. Human monocytes-derived macrophages (hMDMs) were infected with wild-type L. pneumophila, ankB mutant, or the ankB mutant complemented with either a wild-type copy of ankB or ankB containing the indicated single or multiple mutations. The data are representative of three independent experiments and are based on analysis of 100 infected cells per strain with each strain analyzed in duplicate. Error bars indicate ± 1 SD. Abbreviations: 3K, Y91K/L93K/Y127K; 4K, Y91K/L93K/Y127K/L134K. *p < 0.02 compared with ankB mutant complemented with wild-type ankB. NS, not significant. See also Figure S3.
(D) hMDMs were infected with wild-type, dotA mutant, ankB mutant, or the ankB mutant complemented with either wild-type ankB or ankB containing the indicated mutations at an MOI of 1 followed by 1 hr treatment with gentamicin to kill extracellular bacteria. After 10 hr, 100 infected cells were analyzed by confocal microscopy and the number of bacteria per cell was determined. The data are representative of two independent experiments with each strain analyzed in duplicate. Error bars indicate ± 1 SD. Abbreviations: 3K, Y91K/L93K/Y127K; 4K, Y91K/L93K/Y127K/L134K. *p < 0.05 compared with ankB mutant complemented with wild-type ankB. See also Figure S4.
To validate the identification of the substrate-binding site, we 15N-labeled the ankyrin domain and acquired nuclear magnetic resonance (NMR) 15N-1H correlation spectra following a stepwise addition of peptides. Titrations of the ankyrin domain with a pentapeptide QEEKI derived from the AnkB C-terminus, resulted in several chemical shift changes, indicating weak but significant binding. We also tested the effects of N-terminal acetylation and C-terminal amidation and single amino acid substitutions to alanine but these had no significant impact on binding. Titration with a second peptide, PRLPTL, which binds to the ankyrin domain of ANKRA2 (Xu et al., 2012), showed smaller shifts suggestive of weaker binding (Figure 3B).
Residues within the Ankyrin Domain of AnkB Are Essential for Recruitment of Polyubiquitinated Proteins to the LCV
We selected four residues for mutagenesis that are predicted to be involved in substrate binding based on the AnkB crystal structure. Tyr91, Leu93, and Tyr127 form a hydrogen-bonding network connecting and stabilizing the loop residues. We also observed Leu134, located on the first helix of the last ankyrin repeat, is solvent-exposed and potentially provides hydrophobic interactions with substrates.
To validate our structural prediction that Tyr91, Leu93, Tyr127, and Leu134 are important in the biological function of AnkB during infection, the residues were substituted with lysine. To verify that the lysine mutants were still correctly folded, we acquired 1D NMR spectra of the mutants and wild-type ankyrin domain. The mutant spectra are similar to that of the native domain, indicating proper folding (Figure S3). Human monocyte-derived macrophages (hMDM) were then infected with the wild-type strain (AA100/130b), the ankB null mutant, or the ankB mutant complemented with either a wild-type copy of ankB or one of the ankB mutant constructs. At 2 hr post-infection, the function of the AnkB variants was evaluated by assessment of recruitment of polyubiquitinated proteins using confocal microscopy. The data showed that approximately 70% of the LCVs of the wild-type strain-infected cells stained positively for polyubiquitin, whereas only 39% of the LCVs harboring the ankB null mutant were positive. Complementing the ankB mutant with a wild-type copy of ankB fully restored recruitment of polyubiquitinated proteins to the LCV with approximately 70% of the LCVs staining positively (Figure 3C). In contrast, the Y91K L93K Y127K triple mutant and the Y91K L93K Y127K L134K quadruple mutant were defective in recruitment of polyubiquitinated proteins at levels similar to the ankB null mutant. In contrast, the single lysine mutants and the Y91K L93K double mutant were functionally similar to wild-type ankB in the intracellular growth kinetics in U937 macrophages (Figure S4). This confirms the importance of the substrate-binding site on the ankyrin domain for the recruitment of polyubiquitinated proteins to the LCV (Bruckert and Abu Kwaik, 2015a).
The Ankyrin Domain Is Required for Intracellular Replication
We determined if mutations of these four residues resulted in a replication defect of the bacteria within the LCV. hMDMs were infected with wild-type L. pneumophila, its isogenic dotA or ankB mutants, or the ankB mutant complemented with a wild-type or mutated copy of ankB. At 10 hr post-infection, the dotA null mutant showed no replication and the ankB null mutant was markedly compromised compared with the wild-type strain (Figure 3D). Complementation of the ankB mutant with a wild-type copy of ankB restored replication to wild-type levels. The single mutants Y91K, L93K, Y127K, L134K, and double mutant Y91K L93K were also effective in restoring growth. In contrast, complementation with the triple and quadruple mutations in the ankyrin domain showed a significant defect in replication. Similar results were obtained with the U937 macrophage cell line (Figure S4). These data are in agreement with the decreased ubiquitination of the LCV observed in bacteria expressing ankB with the same triple and quadruple mutations. Residues Tyr91, Leu93, Tyr127, and Leu134 within the ankyrin repeats of AnkB are critical both for recruitment of ubiquitinated proteins to the LCV and for replication within hMDMs and U937 macrophages.
DISCUSSION
Here, we present the first structure of a bacterial F box protein, the targeting subunits of SCF ligases. There are close to 70 F box proteins in humans that are involved in a wide range of diseases. These proteins are composed of an F box domain and a variable targeting domain which belongs to three main classes: WD40 domains, leucine-rich repeats, and other domains. AnkB represents a unique association of an F box and ankyrin repeats that appears to be unique to a small number of lower eukaryotes, bacteria, and viruses (Herbert et al., 2015). Two other F box effectors exist in the Legionella genome that could interact with the host SCF complex, but do not contain an ankyrin domain. One has a coiled coil domain (lpp2486) and the other consists of only an F box (lpp0233). Previous mutagenesis studies have shown the importance of the AnkB F box for acquisition of polyubiquitinated proteins to the LCV and bacterial proliferation (Price et al., 2009).
The ankyrin domain of AnkB is likely involved in recruiting substrates for ubiquitination. The structures of a large number of ankyrin protein complexes have been determined and reveal a wide range of types of interactions. Generally, ankyrin domains use the inter-repeat loops and inner row of α helices to bind other proteins; however, there is no consensus for the structure of the bound partner (Parra et al., 2015). Ankyrin repeats can bind discontinuous protein surfaces, α helices, and extended strands as observed for AnkB. The interactions of AnkB with its C-terminal tail most closely resemble the complex of ANKRA2 with a PxLPxI/L motif found in some histone deacetylases and other proteins (Xu et al., 2012); we observed low affinity binding of AnkB to the peptide PRLPTL.
The ankyrin domain of AnkB shows broad specificity. This is typical of ankyrin domains that bind unfolded or extended peptide sequences (Parra et al., 2015) and likely reflects the preponderance of AnkB interactions with the backbone atoms in the bound peptide. Fitting of the NMR titration curve suggested a dissociation constant (Kd) greater than 8 mM for the QEEKI peptide (Figure 3B). Single amino acid substitutions in the peptide did not result in significant changes in the titration behavior, which is consistent with low specificity and a distributed binding interface. Similarly, single point mutations in the AnkB ankyrin domain did not perturb its function in poly-ubiquitination of LCVs and promoting Legionella proliferation (Figures 3C and 3D).
In cells, AnkB is unlikely to bind its own tail or that of another AnkB molecule. The QEEKI motif extends from the final helix in the third ankyrin repeat and is unable to bind to the first two repeats. The interactions between two AnkB molecules is also unlikely as the QEEKI motif is separated by only four residues from the AnkB CaaX farnesylation site. The tethering of AnkB to the membrane would block access of the QEEKI of one molecule to the ankyrin domain of another. Sequence alignment of the AnkB gene between different strains reveals high similarity, and a conservation of the substrate-binding site (Figure S5A). The Paris strain homolog is a truncated version of the AnkB structure presented in this paper. While the last α helix of the last ankyrin repeat is absent, the Paris homolog retains the last loop and half of the last repeat. From analysis of the crystal structures, this would suggest that the Paris homolog would still have a functional substrate-binding interface.
To date, two interacting partners of AnkB have been identified. Parvin B (ParvB), a target of AnkB ubiquitination, was identified by a yeast two-hybrid screen and co-immunoprecipitation (Lomma et al., 2010). ParvB functions in regulating the actin cytoskeleton for cell adhesion and migration (Legate et al., 2006). Overexpression of AnkB competed with endogenous ubiquitin ligase for ParvB interaction and decreased ParvB ubiquitination (Lomma et al., 2010). Solubility issues prevented us from detecting direct binding of ParvB to the purified AnkB ankyrin domain. More recently, TRIM21 was identified by coimmunoprecipitation as a partner of AnkB. TRIM21 attaches Lys11-linked polyubiquitin chains on Lys76 of AnkB without affecting AnkB stability (Bruckert and Abu Kwaik, 2015b).
Studies have elucidated two roles of AnkB in Legionella virulence. Following phagocytosis, Legionella injects effector proteins into the host cell cytosol via the Dot/Icm secretion system (Figure 4). Considerable redundancy exists between effectors and loss of the dotA gene (equivalent to a knockout of all 300 effectors) gives rise to a much stronger ubiquitination and replication deficiency than the loss of only ankB (Figures 3C and 3D). Nonetheless, AnkB is effectively essential for virulence and acts as a linker to recruit the SCF complex to the LCV. Farnesylation of AnkB appears to be essential for its function (Price et al., 2010; Al-Quadan et al., 2011); however, there is strain specificity as AnkB from the Legionella strain Paris lacks the CaaX motif but retains function (Lomma et al., 2010). By co-opting the host SCF complex, AnkB redirects host ubiquitination to the LCV and substrates selected by AnkB. We have built a model of AnkB in context of the SCF ubiquitination complex and the connected UbcH7 (E2 conjugating enzyme), by aligning AnkB onto the F box of a Cul1-Rbx1-Skp1-Skp2 (PDB: 1LDK) and docking UbcH7 onto the Rbx1 RING domain based on a c-Cbl-UbcH7 structure (PDB: 1FBV) (Figure S5B) (Zheng et al., 2000, 2002). In the model, the active cysteine (Cys86) of the E2 points toward the putative substrate-binding site of the AnkB ankyrin repeats, positioning the substrate to receive ubiquitin. Lys48-linked polyubiquitination of the LCV is a critical step in the maturation of the LCV and required to prevent fusion with the lysosome. We observed a strong correlation between loss of ubiquitination activity and loss of Legionella proliferation for the AnkB mutations tested. Legionella hijacks members of the secretory pathway to fuse endoplasmic reticulum-derived vesicles to the LCV. As Lys48-linked polyubiquitin chains are also associated with recruitment of the autophagy machinery, the reported association of AnkB with E3 ligases containing different chain specificities, such as TRIM21, is particularly interesting (Bruckert and Abu Kwaik, 2015b).
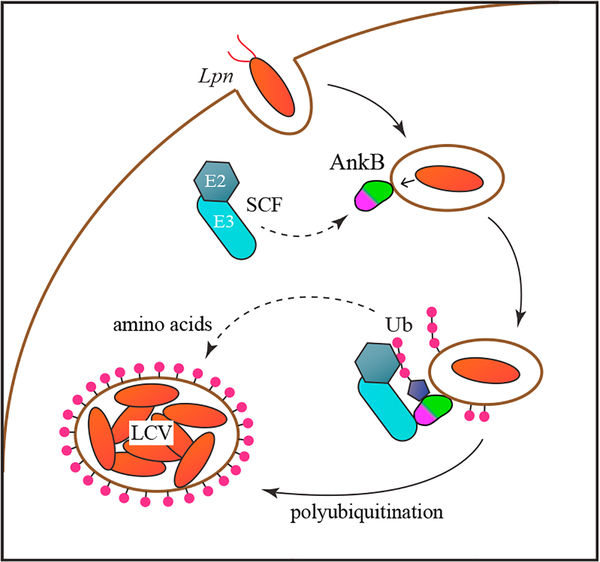
Figure 4. Role of AnkB in LCV Maturation.
After phagocytosis, L. pneumophila secretes AnkB which remains anchored to the vacuole via its farnesylation motif. AnkB then recruits the SCF E3 ubiquitin ligase complex through its F box. Increased ubiquitination of host proteins leads to increased free amino acid production and the accumulation of ubiquitin on the mature LCV to prevent fusion with lysosomes. See also Figure S5.
AnkB also plays a role in enriching the cytosolic pool of free amino acids through triggering Lys48-linked poly-ubiquitination and increased protein turnover (Price et al., 2011). The levels of amino acids in the infected host cell are insufficient sources of carbon, nitrogen, and energy for L. pneumophila (Price et al., 2014). AnkB promotes intra-vacuolar proliferation by ubiquitinating host proteins for their degradation into free amino acids (Price et al., 2011; Bruckert and Abu Kwaik, 2015a). The growth defect of the ankB null mutant in both protozoan and eukaryotic cells can be rescued by supplementation with a mixture of free amino acids (Price et al., 2011; Bruckert et al., 2014). AnkB likely functions by directly recruiting substrate proteins through the ankyrin domain. Mutating either the F box or the ankyrin domain of AnkB results in the same phenotype, suggesting that the ability of Legionella to co-opt host E3 ubiquitin ligases through molecular mimicry plays a key role in pathogenesis.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The human gene Skp1 (residues 1–163) was first cloned into pRSFDuet-1 between NdeI and AvrII restriction sites. The gene AnkB (lpg2144, residues 1–168) from L. pneumophila strain Philadelphia was then cloned into the same vector between BamHI and NotI as an N-terminal His-tagged fusion protein. The C-terminal ankyrin domain (residues 54–168) was cloned into pET15b as an N-terminal His-tagged fusion and pET29a as a C-terminal His-tagged fusion. Mutagenesis was performed using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies). All constructs were verified by DNA sequencing and transformed into a BL21 Escherichia coli strain. The cells were grown at 37°C in Luria Broth (LB) to an optical density of 0.8, and expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside at 30°C for 4 hr or 16°C overnight. After centrifuging the cells, the pellets were resus-pended in buffer A (50 mM HEPES, 500 mM NaCl, 5% (w/v) glycerol, pH 7.6), containing 1 mM phenylmethylsulfonyl fluoride and 0.1 mg/mL lysozyme, and lysed by sonication. Cell debris was removed by centrifugation, and the fusion protein was bound to Ni-NTA Agarose (QIAGEN) beads, washed with buffer A containing 30 mM imidazole, and eluted with buffer A containing 250 mM imidazole. The protein was further purified by size-exclusion chromatography on a Superdex75 column (GE Healthcare) in buffer B (10 mM HEPES, 100 mM NaCl [pH 7.0]) before crystallization trials. The His-tag in the pET15b constructs was cleaved with thrombin before injecting the protein into a size-exclusion column.
For selenomethionine labeling, the plasmid was transformed into a methionine auxotroph, E. coli DL41 (DE3), and the cells were grown in LeMaster medium supplemented with selenomethionine. For 15N labeling, the cells were grown in M9 minimal medium supplemented with 15N-ammonium chlo-ride as the sole source of nitrogen. The expression and purification protocols were the same as for the native protein.
Crystallization and Structure Determination
Crystallization was performed by the hanging drop vapor diffusion method at 293K using the Classics II commercial screen (QIAGEN). Native AnkB (54–168) concentrated to 7.6 mg/mL crystallized in a 1:1 mixture with the reservoir buffer (0.2 M lithium sulfate, 0.1 M HEPES [pH 7.5], 25% [w/v] polyethylene glycol [PEG] 3350). Crystals of the SeMet-labeled C-terminal domain were obtained at 10 mg/mL with the mother liquor (0.2 M lithium sulfate, 0.1 M Bis-Tris [pH 6.5], 25% [w/v] PEG 3350). AnkB (1–168) in complex with Skp1 was concentrated to ~4.5 mg/mL and crystals were obtained from a condition containing 0.2 M trimethylamine N-oxide, 0.1 M Tris (pH 8.5), and 20% (w/v) PEG 2000 monomethylether.
The ankyrin domain and complex crystals were cryoprotected with 20% glycerol and 20% sucrose, respectively, and flash-cooled in a N2 cold stream. X-ray diffraction data were collected at beamlines A1 and F1 of Cornell High-Energy Synchrotron Source (CHESS) using an ADSC Quantum 210 CCD detector. Data processing and scaling were performed with HKL-2000 (Otwinowski and Minor, 1997).
The diffraction data of the ankyrin domain were phased using anomalous signal from selenium atoms by the single-wavelength anomalous dispersion method, with the program SHELX (Sheldrick, 2008). The initial model was built with ARP/wARP (Langer et al., 2008) and refined with Refmac5 (Murshudov et al., 2011). Full-length AnkB in complex with Skp1 was determined by molecular replacement using Skp1 and F box from a deposited SCF complex structure (PDB: 1LDK) and AnkB (54–168) as the search model (Zheng et al., 2002). The model was built by ARP/wARP (Langer et al., 2008), completed with Coot (Emsley and Cowtan, 2004), and improved by several cycles of refinement using Refmac5 (Murshudov et al., 2011). Water molecules were added in the last stage of refinement.
The refinement statistics are shown in Table 1. The final ankyrin domain and complex structures respectively have 0 and 1 outlier in the Ramachandran plot computed using MolProbity (Chen et al., 2010).
NMR Spectroscopy
15N-1H heteronuclear single quantum correlation spectroscopy and 1D experiments were performed at 25°C on a Bruker 600 MHz spectrometer. Samples of the AnkB (54–168) were prepared at 0.28 mM in 90% buffer B and 10% D2O. Titrations were performed by the stepwise addition of QEEKI, PRLPTL, AcQEEAINH2, AcQAEKINH2, AcAEEKINH2, AcQEERINH2, and AcQEEYINH2 peptides (Bio Basic). The highest peptide concentration was 8 mM. NMR spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with SPARKY (Goddard and Kneller, 2008).
Bacterial Strains, Cell Cultures, and Infections
L. pneumophila strain AA100/130b (ATCC BAA-74), its isogenic dotA and ankB mutants, and complemented mutants were grown on buffered charcoal yeast extract agar plates for 3–4 days at 37°C prior to infection as previously described (Al-Khodor et al., 2008). When required, antibiotics were used at a concentration of 50 μg/mL for kanamycin and 5 μg/mL for chloramphenicol. The E. coli strain DH5α was used for cloning. E. coli was grown in LB and antibiotics were used at a concentration of 100 μg/mL for ampicillin and 40 μg/mL for chloramphenicol.
Purification and preparation of human monocyte-derived macrophages (hMDMs) was performed as previously described (Habyarimana et al., 2008). Monocytes were isolated from whole blood of healthy donors and then allowed to adhere to 6-well low adherence cell culture plates for 3 days at 37°C and 5% CO2 in RPMI 1640 supplemented with 20% fetal bovine serum (FBS). Monocytes were then counted and resuspended in RPMI 1640 supplemented with 10% FBS and plated on coverslips at a density of 3 × 105 cells per well of a 24-well cell culture plate and incubated for a further 2 days. The cell culture media was then replaced with RPMI 1640 supplemented with 5% FBS for 1 day, and then with RPMI 1640 supplemented with 1% FBS for 1 day. The resulting hMDMs were then used for infection. Maintenance of U937 macrophages was performed as described previously (Habyarimana et al., 2008).
Infection of hMDMs or U937 cells was performed as previously described (Habyarimana et al., 2008). Bacteria were suspended in RPMI 1640 with 10% FBS and macrophages were infected in duplicate for 1 hr at an MOI of 50. Plates were centrifuged at 200 × g for 5 min to synchronize the infection. Infected cells were treated with 50 μg/mL gentamicin for 1 hr to kill extracellular bacteria. Following gentamicin treatment, cells were washed three times with Hank’s buffered saline solution and then RPMI containing 10% FBS was added. At 10 hr post-infection, cells were fixed in 100% cold methanol and processed for confocal microscopy.
Confocal Microscopy
Processing of infected cells for confocal microscopy was performed as we described previously (Price et al., 2009). Rabbit polyclonal anti-L. pneumophila was used at a dilution of 1/1,000 and detected by Alexa-Fluor 488-conjugated donkey anti-rabbit IgG (Invitrogen). Polyubiquitinated proteins were detected using mouse anti-polyubiquitin FK1 antibody at a dilution of 1/50 (Enzo Life Sciences), followed by Alexa-Fluor 647-conjugated goat anti-mouse immunoglobulin M (Invitrogen). An Olympus FV1000 laser scanning confocal microscope was used to examine cells as we described previously (Price et al., 2009). One hundred cells were examined in duplicate for each strain for both ubiquitin recruitment and replication.
Supplementary Material
Highlights.
The structure of AnkB contains an F box domain and an ankyrin domain
The F box domain interacts with the host SCF ubiquitin ligase complex
The ankyrin domain is essential for recruiting polyubiquitinated proteins
The ankyrin domain is essential for replication of Legionella pneumophila
ACKNOWLEDGMENTS
K.W. acknowledges a studentship from Canadian Institutes of Health Research (CIHR). This work was funded by a CIHR Genomics grant GSP-48370. Y.A.K is supported by Public Health Service Award 1R01AI120244 and 1R21AI116517 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund. Crystallographic data were acquired at the Macromolecular Diffraction (MacCHESS) facility at the Cornell High-Energy Synchrotron Source (CHESS) supported by NSF award DMR-0225180 and NIH/NCRR award RR-01646.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and diffraction data of the structures have been deposited with the PDB, www.pdb.org (PDB: 5K34 and 5K35 for AnkB residues 54–168, and AnkB-Skp1, respectively.)
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.12.015.
REFERENCES
- Al-Khodor S, Price CT, Habyarimana F, Kalia A, and Abu Kwaik Y (2008). A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol 70, 908–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Quadan T, Price CT, London N, Schueler-Furman O, and AbuKwaik Y (2011). Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol. 19, 573–579. [DOI] [PubMed] [Google Scholar]
- Bruckert WM, and Abu Kwaik Y (2015a). Complete and ubiquitinated proteome of the Legionella-containing vacuole within human macrophages. J. Proteome Res. 14, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckert WM, and Abu Kwaik Y (2015b). Lysine11-linked polyubiquitination of the AnkB F-Box effector of Legionella pneumophila. Infect Immun. 84, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckert WM, Price CT, and Abu Kwaik Y (2014). Rapid nutritional remodeling of the host cell upon attachment of Legionella pneumophila. Infect Immun. 82, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, and Pupko T (2009). Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5, e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA, Pupko T, Shuman HA, and Segal G (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet 48, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet 36, 1165–1173. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, et al. (2004). The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968. [DOI] [PubMed] [Google Scholar]
- De Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, and Shuman HA (2005). Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol 187, 7716–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Glockner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, Steinert M, Hacker J, and Heuner K (2008). Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol 298, 411–428. [DOI] [PubMed] [Google Scholar]
- Goddard TD, and Kneller DG (2008). SPARKY 3 (University of California; ). [Google Scholar]
- Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, Garcia MT, and Kwaik YA (2008). Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol 10, 1460–1474. [DOI] [PubMed] [Google Scholar]
- Herbert MH, Squire CJ, and Mercer AA (2015). Poxviral ankyrin proteins. Viruses 7, 709–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov SS, Charron G, Hang HC, and Roy CR (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem 285, 34686–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, and Pagano M (2000). The F-box protein family. Genome Biol. 1, REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, and Perrakis A (2008). Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc 3, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate KR, Montanez E, Kudlacek O, and Fassler R (2006). ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7, 20–31. [DOI] [PubMed] [Google Scholar]
- Li J, Mahajan A, and Tsai MD (2006). Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178. [DOI] [PubMed] [Google Scholar]
- Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, et al. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 12, 1272–1291. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, and Isberg RR (2004). Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, and Deshaies RJ (1998). Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95, 7451–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, and Dowdle WR (1977). Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med 297, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, and Vagin AA (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parra RG, Espada R, Verstraete N, and Ferreiro DU (2015). Structural and energetic characterization of the ankyrin repeat protein family. PLoS Comput. Biol 11, e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, Lortholary O, Zumla A, and Abubakar I (2014). Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 14, 1011–1021. [DOI] [PubMed] [Google Scholar]
- Price CT, and Kwaik YA (2010). Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-Box effectors. Front. Microbiol 1, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, and Kwaik YA (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5, e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Quadan T, Santic M, Jones SC, and Abu Kwaik Y (2010). Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med 207, 1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Quadan T, Santic M, Rosenshine I, and Abu Kwaik Y (2011). Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334, 1553–1557. [DOI] [PubMed] [Google Scholar]
- Price CT, Richards AM, and Abu Kwaik Y (2014). Nutrient generation and retrieval from the host cell cytosol by intra-vacuolar Legionella pneumophila. Front. Cell. Infect. Microbiol 4, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, and Pavletich NP (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2008). A short history of SHELX. Acta Crystallogr. A 64, 112–122. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, and Harper JW (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Uro-Coste E, Perret E, Fonta C, Mathieu M, Delisle MB, Caput D, and Imbert M (1998). The cell cycle gene SKP1 is regulated by light in postnatal rat brain. Brain Res. Mol. Brain Res. 56, 192–199. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, and Isberg RR (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876. [DOI] [PubMed] [Google Scholar]
- Xu C, Jin J, Bian C, Lam R, Tian R, Weist R, You L, Nie J, Bochkarev A, Tempel W, et al. (2012). Sequence-specific recognition of a PxLPxI/L motif by an ankyrin repeat tumbler lock. Sci. Signal 5, ra39. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, and Pavletich NP (2000). Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.