Abstract
The endoplasmic reticulum (ER) is an expansive, membrane-enclosed organelle that plays crucial roles in numerous cellular functions. We used emerging superresolution imaging technologies to clarify the morphology and dynamics of the peripheral ER, which contacts and modulates most other intracellular organelles. Peripheral components of the ER have classically been described as comprising both tubules and flat sheets. We show that this system consists almost exclusively of tubules at varying densities, including structures that we term ER matrices. Conventional optical imaging technologies had led to misidentification of these structures as sheets because of the dense clustering of tubular junctions and a previously uncharacterized rapid form of ER motion. The existence of ER matrices explains previous confounding evidence that had indicated the occurrence of ER “sheet” proliferation after overexpression of tubular junction–forming proteins.
The ER is a continuous, membranous network extending from the nuclear envelope to the outer periphery of cells; it plays vital roles in processes such as protein synthesis and folding, mitochondrial division, calcium storage and signaling, and lipid synthesis and transfer. In the cell periphery, the ER is thought to exist as an elaborate membrane system that makes contact with nearly every other cellular organelle. Prevailing models of its structure propose a complex arrangement of interconnected tubules and sheets, each of which is maintained by distinct mechanisms (1, 2). Numerous proteins are involved in maintaining this complex structural organization. Membrane curvature-stabilizing proteins, including members of the reticulon (RTN) and REEP families, contain hydrophobic hairpin domains that are thought to be responsible for promoting curvature in ER tubules via scaffolding and hydrophobic wedging. Members of the atlastin (ATL) family of dynamin-related guano-sine triphosphatases (GTPases) are thought to mediate the formation of tubular three-way junctions, giving rise to the characteristic polygonal tubular network (3). Meanwhile, an alternative complement of proteins is proposed to regulate the structure of ER sheets, with p180, kinectin, and CLIMP63 all thought to play a role in shaping, helicoidal stacking, and luminal spacing (3). Mutations in many of these ER-shaping proteins are connected to a variety of human disease conditions, most notably the hereditary spastic paraplegias (4). Thus, characterizing ER morphology is critical to understanding the basic biology of cells in both health and disease.
Determining the structure of the ER is challenging because of limitations in our ability to visualize the intricate nature of its morphology. The peripheral ER is particularly susceptible to this constraint, given its well-documented dynamic rearrangements and fine ultrastructure (5, 6). These characteristics impede attempts to derive functional information based on changes to ER structure. The recent development of various superresolution (SR) imaging approaches, however, offers an opportunity to examine ER structure and dynamism with substantially improved spatiotemporal resolution. Here, we used five different SR modalities, each having complementary strengths and weaknesses in the spatial and temporal domains, to examine ER structure and dynamics. A high-speed variation of structured illumination microscopy (SIM) allowed ER dynamics to be visualized at unprecedented speeds and resolution. Three-dimensional SIM (3D-SIM) and Airyscan imaging allowed comparison of the fine distributions of different ER-shaping proteins. Finally, lattice light sheet-point accumulation for imaging in nanoscale topography (LLS-PAINT) and focused ion beam scanning electron microscopy (FIB-SEM) permitted 3D characterization of different ER structures. Thoroughly probing the ER in this manner provides unprecedented information about the morphology and dynamics of this organelle, including the characterization of a previously underappreciated structure within the peripheral ER.
ER tubules and junctions undergo rapid motion in living cells
ER tubules are known to undergo rapid structural rearrangements, occurring over seconds or minutes, yet examination of these processes has typically been confined to the extension and retraction of tubules and the formation of tubular three-way junctions (5, 6). To obtain a more comprehensive picture of tubular motion, we used high-speed SIM with grazing incidence illumination (GI-SIM; see materials and methods) (7). This live SR imaging modality (resolution ~100 nm) uses light beams counterpropagating just above the sample substrate to image cellular features near the basal plasma membrane at frequencies up to 40 Hz. This translates to a factor of 4 to 10 increase in acquisition speed, relative to what can be practically achieved with spinning-disk confocal microscopy for imaging the ER, and a factor of ~2 improvement in resolution.
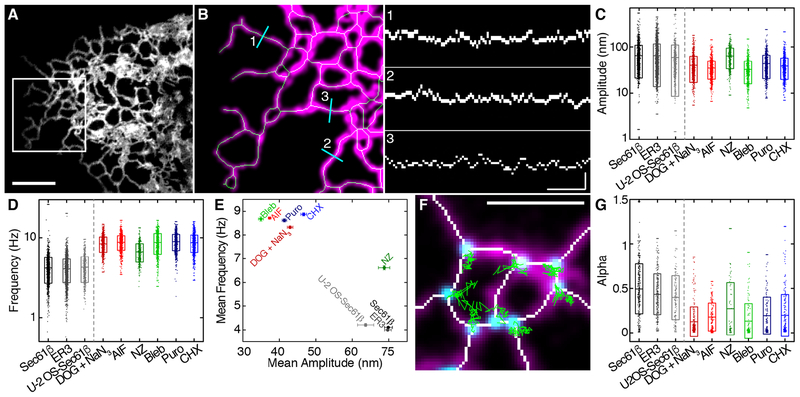
With GI-SIM, we imaged COS-7 cells expressing an ER membrane marker (mEmerald-Sec61β, henceforth Sec61β) to track ER tubules. Increased spatiotemporal resolution revealed a novel form of ER motion consisting of remarkably rapid tubular fluctuations (Fig. 1 and movie S1). Using a modified skeletonization algorithm (8) to track the movement of ER tubules (Fig. 1B), we identified oscillations with a mean peak-to-peak amplitude of 70 ± 50 nm, occurring an average of 4 ± 1 times per second (means ± SD; n = 1755 tubules from 8 cells) (Fig. 1, C and D). Traditional imaging modalities have the ability to localize precisely this tubular motion only if the tubules and junctions are sufficiently sparse. Also, a large proportion of this motion often occurs on too short a time scale to be effectively tracked using spinning-disk confocal microscopy at imaging speeds commonly reported in the literature (6, 9). This suggests that in dense regions, tubular ER motion and morphology are likely to be obscured when using traditional imaging modalities. These rapid fluctuations we observed in COS-7 cells were also found in an unrelated cell type (U-2 OS) as well as in COS-7 cells expressing a luminal ER marker (mEmerald-ER3, henceforth ER3; see materials and methods) instead of Sec61β (Fig. 1, C to E, and tables S1 and S2).
Fig. 1. The peripheral ER moves at high speeds.
(A) Tubular ER in the periphery of a COS-7 cell expressing Sec61β imaged live at 40 Hz using GI-SIM microscopy. Scale bar, 2 mm. (B) ER tubules within the boxed region in (A), identified using a skeletonization algorithm. Left: The midline of each tubule (green) is mapped onto the fluorescence (magenta). Right: Positions of the midlines are plotted as kymographs against time for each of the three locations shown in cyan at left; scale bars, 200 nm and 0.5 s. (C and D) Amplitudes (C) and frequencies (D) of tubular ER oscillations in COS-7 cells expressing Sec61β treated with deoxyglucose plus sodium azide (DOG + NaN3), AlF, nocodazole (NZ), blebbistatin (Bleb), puromycin (Puro), and cycloheximide (CHX). Untreated controls using a luminal ER marker (ER3) and results for a different cell line (U-2 OS–Sec61β) are also shown. (E) Plot of frequency versus amplitude for tubular oscillations in treated and untreated cells. Error bars represent SEM. (F) Locations of three-way junctions derived from skeletonized data (white). Original fluorescence is shown in magenta; example tracks of junctions (cyan) over 2.5 s are indicated in green. Scale bar, 2 mm. (G) MSD scaling exponent (α values) for treated and control cells. Box plots indicate the mean and SD in (C), (D), and (G); range is indicated by outer tick marks. See tables S1 and S2 for a detailed list of means and test statistics.
In addition to the tubules themselves, three-way junctions also exhibited appreciable motion over very short time scales (Fig. 1, F and G, and movie S1). Three-way junctions were identified from the skeletonization of fluorescent ER images. Skeletonized pixels with exactly two neighbors were considered to be part of a branch (Fig. 1F, white), and pixels with more than two neighbors were considered junctions (Fig. 1F, overlaid with cyan dots). Three-way junctions were then treated as single particles and tracked (Fig. 1F, green). The time-averaged mean square displacement (MSD) of three-way junctions can be described as MSD ~ tα, where a describes a particle’s motion as Brownian (α = 1), subdiffusive (α < 1), or super-diffusive (α > 1) (10). Junction dynamics exhibited a broad distribution of α (Fig. 1G and table S1). Again, these results were consistent among cells expressing membrane or luminal markers as well as in other cell types such as U-2 OS (Fig. 1G and tables S1 and S2).
Such rapid dynamics can often represent thermally derived motion in systems, but in this context thermal energy alone does not appear to be responsible for driving ER dynamics. Indeed, the motion of both tubules and three-way junctions characterized above was substantially altered by a variety of biological perturbations. Diverse treatments affecting access to cellular energy sources [deoxyglucose (DOG) + sodium azide (NaN3); aluminum fluoride (AlF)], cytoskeletal dynamics (blebbistatin), or protein translation (puromycin; cycloheximide) each reduced the amplitude and increased the frequency of oscillations to levels consistent with thermally derived Brownian motion (11) (Fig. 1, C to E). Additionally, the motion of three-way junctions was dampened (Fig. 1G). Although the broad susceptibility of rapid ER dynamics to pharmacological perturbation does not elucidate the direct source of the motion, it suggests a broader role for cellular dynamics in driving ER motion, as it is affected by a range of disparate processes. Of note, treatment with the microtubule-depolymerizing agent nocodazole increased the frequency of motion without any noticeable effect on amplitude (Fig. 1E and table S2), so it appears that at least in some situations, the amplitude and frequency of tubule oscillations can be uncoupled.
Peripheral “sheets” appear highly dynamic and are riddled with spaces
GI-SIM also permitted rapid imaging of the morphology and dynamics of structures that appeared to be flat peripheral sheets by diffraction-limited epifluorescence, leading to several highly unexpected observations. At the improved spatiotemporal resolution afforded by GI-SIM, most peripheral “sheets” do not appear continuous, but rather are riddled throughout with spaces devoid of Sec61β fluorescence (Fig. 2, A and B). These spaces are highly dynamic and densely distributed across the structure (Fig. 2B, kymo-graphs). To analyze these dynamics, we used a fluorescence inversion and image preparation protocol (see materials and methods) (12), transforming the dark areas into particle-like entities that are trackable using single-particle tracking (SPT) algorithms (13) (Fig. 2C and movie S2). The spaces were tracked and their lifetimes were extracted from the trajectories. For distances between tubules (spaces) larger than our ~100-nm limit of resolution, we quantified the average lifespan (250 ± 250 ms, n = 4292 tracks from 4 cells) and detectable separation between tubules (260 ± 350 nm, n = 1273 spaces from 4 cells) (Fig. 2D and tables S3 and S4). There was no significant difference in the lifespan or apparent size of voids in intensity in either U-2 OS or COS-7 cells expressing Sec61β (table S4). Measurable gaps between tubules did, however, appear significantly smaller in cells expressing Sec61β than in cells expressing ER3, presumably because of the relocation of the fluorescence tag from the lumen to the cytoplasmic surface of the ER membrane (Fig. 2D and tables S3 and S4; see also supplementary text). This apparent dilatation of the structure would be predicted to lead to an appearance of constriction in remaining nonfluorescent spaces. Certainly, the size and lifetime of the spaces observed with GISIM within these peripheral structures would render a substantial proportion of them undetectable by more traditional imaging modalities.
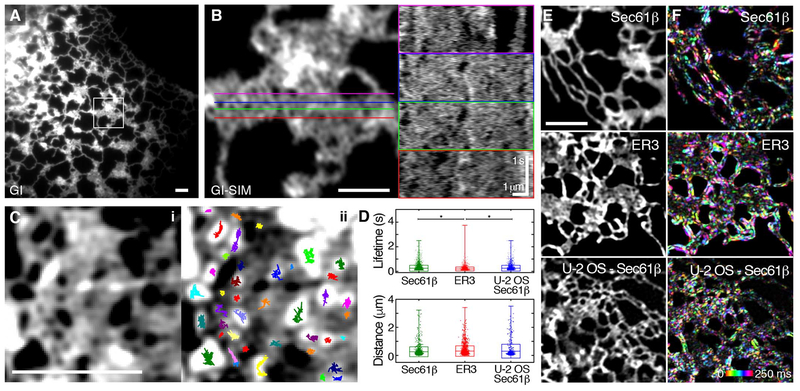
Fig. 2. Peripheral ER “sheets” are highly dynamic and riddled with spaces.
(A) COS-7 cell expressing Sec61β imaged live by GI at 40 Hz exhibits many peripheral sheet-like structures. (B) GI-SIM of the boxed region in (A) shows many discrete spaces throughout the structure. Colored lines at left correspond to the locations of the kymographs shown at right. Voids in intensity within the structure can be seen appearing and disappearing over time. (C) Single-particle tracking (SPT) of dark spaces within the structure. The fluorescence image (i) was inverted and spaces were tracked using SPT algorithms. Tracks overlaid onto the inverted image are shown in (ii), with trajectories shown in different colors. (D) Each track length corresponds to the lifetime of the space; distance across the space (i.e., distance between tubules) is also quantified. The box plot indicates the SD and mean; range is indicated by outer tick marks. The asterisks denote significant difference between means, detailed in table S4. (E and F) Temporal intensity derivative analysis (see materials and methods) of representative peripheral sheet-like structures in a COS-7 cell expressing Sec61β, with a luminal ER marker (ER3) and another cell line (U-2 OS–Sec61β) as controls. (E) Original fluorescence images. (F) Each consecutive frame over a 250-ms time period is color-coded, with intensity corresponding to the magnitude of fluorescence change. Scale bars, 2 μm. See tables S3 and S4 for a detailed list of means and test statistics.
To accompany the SPT of these transient spaces, we used an analysis that does not require a best-fit process, termed the temporal intensity derivative (detailed in materials and methods; see also fig. S1). This technique maps locations in the structure where substantial changes in fluorescence intensity occur over defined time windows. We found that even across a very short time frame (250 ms), we were able to detect substantial motion in ER tubules, consistent with the tubular oscillations described in Fig. 1. Intriguingly, nearly the entire area of structures that appeared as peripheral “sheets” by conventional imaging also underwent fluctuations in fluorescence intensity similar to that of isolated tubules, suggesting major structural rearrangements within these structures over this time frame (Fig. 2, E and F). This phenomenon was consistently observed in COS-7 cells expressing either Sec61β or ER3, as well as in U-2 OS cells expressing Sec61β. Collectively, the rapid rearrangement of spaces and the magnitude of fluorescence changes across very short time scales within peripheral sheet-like structures imply that these regions are not likely to be continuous in nature.
Rapid assembly and disassembly of sheet-like structures into isolated tubules
In further support of this idea, longer time-lapse imaging with GI-SIM revealed rapid assembly and disassembly of sheet-like structures from clearly isolated tubules (movie S3). Given that this process occurs over relatively short time frames, it seems unlikely that energetically costly fusion or fission of the ER membrane would be required. Instead, our data suggest a possible mechanism whereby tubules could coalesce until the spaces between them become too small to observe, leading to the discontinuities described above. Consequently, dense networks could expand outward to isolated tubules by the reverse mechanism, without requiring membrane fusion or fission. This could be achieved by well-characterized motion through molecular motors or by the sliding of three-way junctions along tubules (3).
SR imaging reveals the existence of dense tubular matrices in the peripheral ER
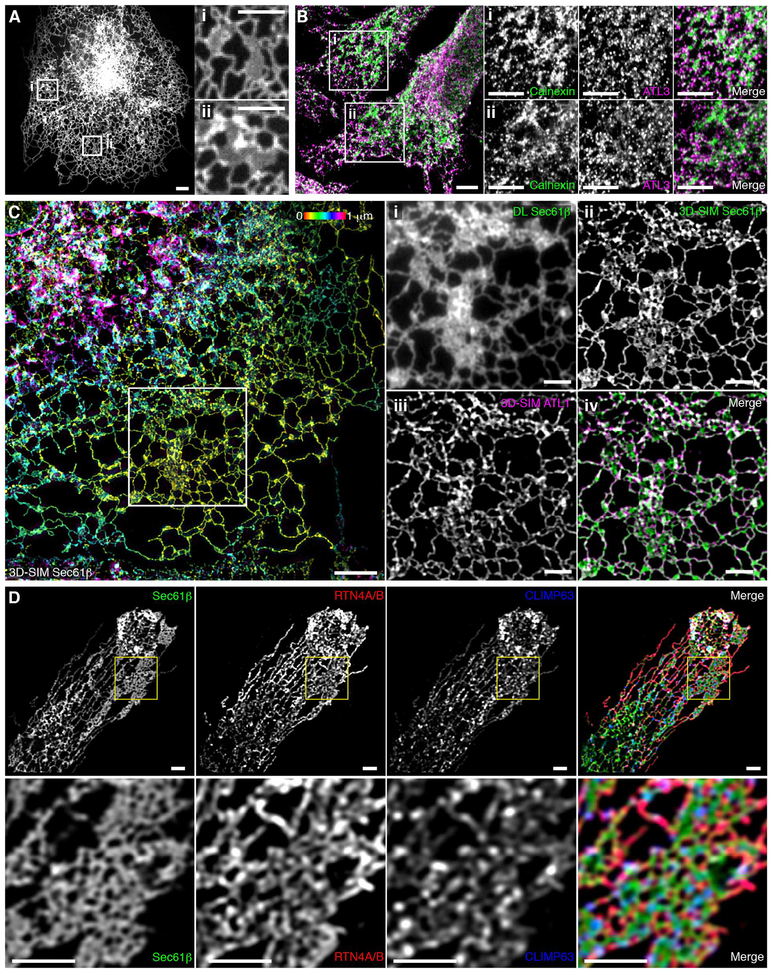
To gain a more comprehensive understanding of structures classically defined as peripheral sheets, we performed parallel experiments using SR imaging to reconstruct the protein and lipid distribution across the entire ER. COS-7 cells expressing Sec61β as an ER marker (thought to be expressed uniformly across the ER) were fixed and imaged by wide-field 3D-SIM, providing a representative map of ER transmembrane protein distribution throughout the cell (Fig. 3A). Remarkably, many structures that appeared to be intact sheets by diffraction-limited, wide-field imaging instead comprised a dense, cross-linked network of tubules enriched in three-way junctions (Fig. 3A). Notably, these structures were flat relative to their height but showed substantial variation in local topology (Fig. 3A, color coding).
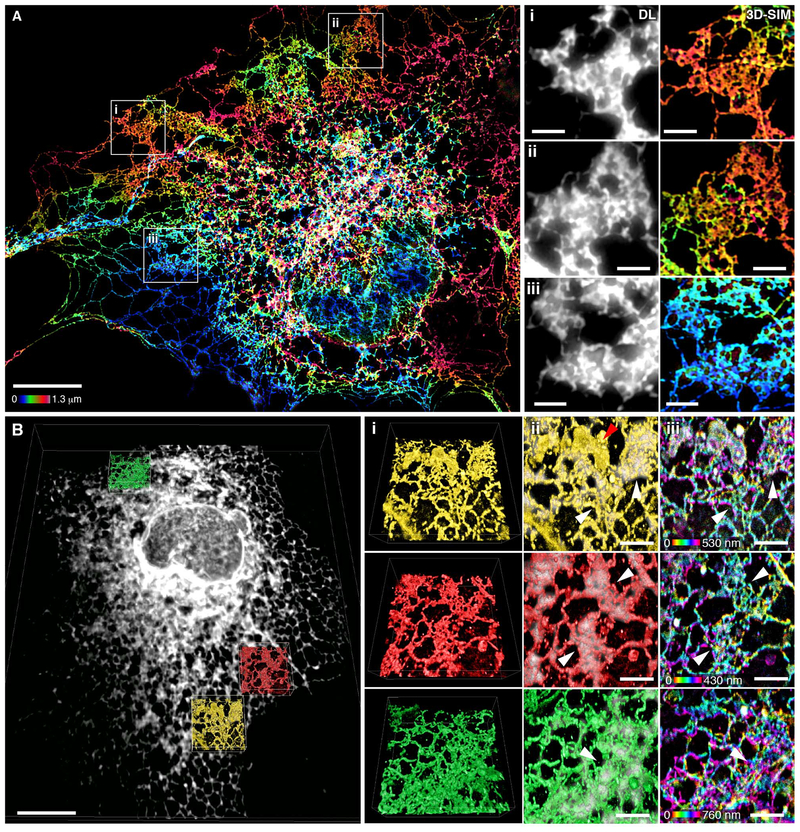
Fig. 3. Many peripheral structures classically identified as sheets are instead dense matrices of tubules.
(A) Left: Fixed COS-7 cell expressing Sec61β, imaged by 3D-SIM and color-coded by z position. Scale bar, 10 μm. Right: Magnified regions (i to iii) show that 3D-SIM reveals dense tubular matrices, which appear as sheets by diffraction-limited (DL) epifluorescence. Scale bars, 2 mm. (B) Left: Deconvolved, diffraction-limited LLS imaging of a fixed COS-7 cell overexpressing Sec61β (gray). All internal lipid membranes were reconstructed using LLS-PAINT microscopy. Data from three regions containing ER matrices are shown in colored insets. Scale bar, 10 μm. Right: The three boxed regions are enlarged, showing (i) 3D orientation of LLS-PAINT volume rendering, (ii) overlay of LLS-PAINT and diffraction-limited LLS imaging volume rendering (gray), and (iii) LLS-PAINT color-coded by z position. White arrowheads mark areas that appear as sheets by diffraction-limited imaging; the red arrowhead [top of (ii)] denotes a mitochondrion. Scale bars, 2 μm.
To verify that the ER membrane itself shared this structure, we turned to an even higher-resolution imaging technique that directly probes the locations of the membranes themselves. LLS-PAINT microscopy uses single-molecule localization of individual fluorescent lipid molecules as they stabilize on cellular membranes (14). COS-7 cells expressing Sec61β were fixed and the structure of the internal membranes was ascertained at the single-molecule level by LLS-PAINT microscopy (movie S4). In addition, for each cell, a single diffraction-limited 3D LLS image (15) was taken of the Sec61β signal to allow ER membranes to be distinguished from those of other organelles (Fig. 3B and movie S4). The resulting data set confirms that many ER structures that appear as continuous sheets with diffraction-limited imaging are shown to be dense tubular matrices when viewed using LLS-PAINT (Fig. 3B, insets). Many ER matrices had substantial topological variation across their structures, again supporting the notion that they are not strictly 2D (Fig. 3B, iii, and fig. S2). Thus, with improved spatial resolution in three dimensions, both protein and lipid components of the peripheral ER appear to comprise dense matrices of highly convoluted tubules.
Limitations in spatiotemporal resolution obscure dense tubular matrices
Given our observations that ER tubules undergo very rapid motion (Fig. 1), that many spaces in structures previously described as sheets are near or beneath the diffraction limit (Fig. 2), and that SR imaging of both ER membrane protein and lipid reveals most of these structures to be dense tubular networks (Fig. 3), we hypothesized that limitations in spatiotemporal resolution might obscure dense tubular matrices and lead to their frequent misinterpretation as sheets. To test this directly, we compared images of peripheral ER matrices collected by either diffraction-limited GI or GISIM using two different simulated exposure times (see materials and methods for details) (Fig. 4A). We found that the loss of either spatial or temporal resolution was sufficient to obscure the majority of gaps between tubules within the matrix.
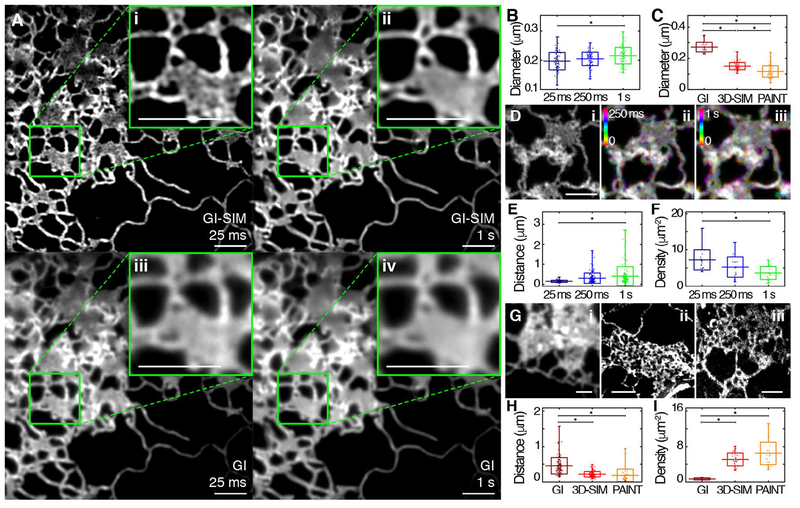
Fig. 4. Effects of spatiotemporal blurring on imaging ER tubular matrices.
(A) A COS-7 cell expressing Sec61β, imaged live using GI-SIM, showing both single 25-ms frames (i, iii) and 40 frames averaged over 1 s (ii, iv). The top row shows the GI-SIM images; the bottom row shows the corresponding diffraction-limited images with GI illumination, demonstrating the combined effects of spatial and temporal limitations in resolution. (B and C) The measurable diameter of isolated ER tubules found outside of matrices also decreases with increasing temporal (B) and spatial (C) resolution. (D) Structure of a representative tubular ER matrix imaged in a live cell by GI-SIM when integrating image frames for (i) 25 ms (1 frame), (ii) 250 ms (10 frames), or (iii) 1 s (40 frames) as in (A). (E and F) Quantification of the measurable distance between tubules (E) and density of these spaces (F) within tubular matrices for each functional exposure time. (G) Representative images of tubular matrices imaged in fixed cells by diffraction-limited GI (i), 3D-SIM (ii), and LLS-PAINT (iii). (H and I) Quantification of the measurable distance between tubules (H) and density of these spaces (I) in tubular matrices, as identified by imaging modalities of increasing spatial resolution. Scale bars, 2 μm. Box plots indicate the mean and SD in all panels; range is indicated by outer tick marks. The asterisks denote statistical significance between the means, as detailed in tables S5 and S6.
In the case of live-cell imaging, tubular motion can occur faster than the acquisition time of a single frame, creating a blurring artifact and thus increasing the apparent diameter of tubules as the temporal resolution decreases (Fig. 4B and tables S5 and S6). Likewise, the resolvable separation between tubules in a matrix is affected not only by the true distance across the space between tubules, but also by temporal blurring due to oscillations of the surrounding tubules and motion of their three-way junctions. We therefore expected gaps in tight tubular matrices to appear smaller in active, living cells than in fixed ones. Indeed, the average apparent distance between tubules in fixed 3D-SIM (220 nm) was greater than that observed using live GI-SIM (150 nm). Consequently, we predicted that decreasing temporal resolution would preferentially blur smaller spaces, as tubular oscillations and junction movement would mask them more readily. To quantify this, we summed GI-SIM images of tubular matrices over an increasing number of 25-ms frames to simulate longer exposure times. As predicted, longer simulated exposure times resulted in blurring of matrices until they ultimately resembled continuous sheets (Fig. 4D). On the other hand, improving temporal resolution decreased the minimum distance required between tubules before a space became detectable (Fig. 4E and tables S5 and S6), thereby increasing the density of measurable spaces within a dynamic matrix (Fig. 4F and tables S5 and S6).
To quantify the contribution of insufficient spatial resolution in obscuring the structure of ER tubular matrices, we compared cells in the absence of motion (i.e., in fixed cells) using three different imaging modalities of varying spatial resolution (diffraction-limited GI, 3D-SIM, or LLS-PAINT) (Fig. 4G). Predictably, improvements in spatial resolution decreased the measurable diameter of ER tubules (Fig. 4C and tables S5 and S6). Moreover, imaging with diminishing spatial resolution limited the detectable degree of separation of tubules in a matrix (Fig. 4H and tables S5 and S6), decreasing their detectable density (Fig. 4I and tables S5 and S6). Thus, sufficient spatial and temporal resolution are both required to resolve the fine structure of tubular matrices in living cells, and with any imaging modality an apparently continuous structure may conceal spaces if they are beneath the resolvable power of the technique.
FIB-SEM reveals tubular matrices with heterogeneous topology
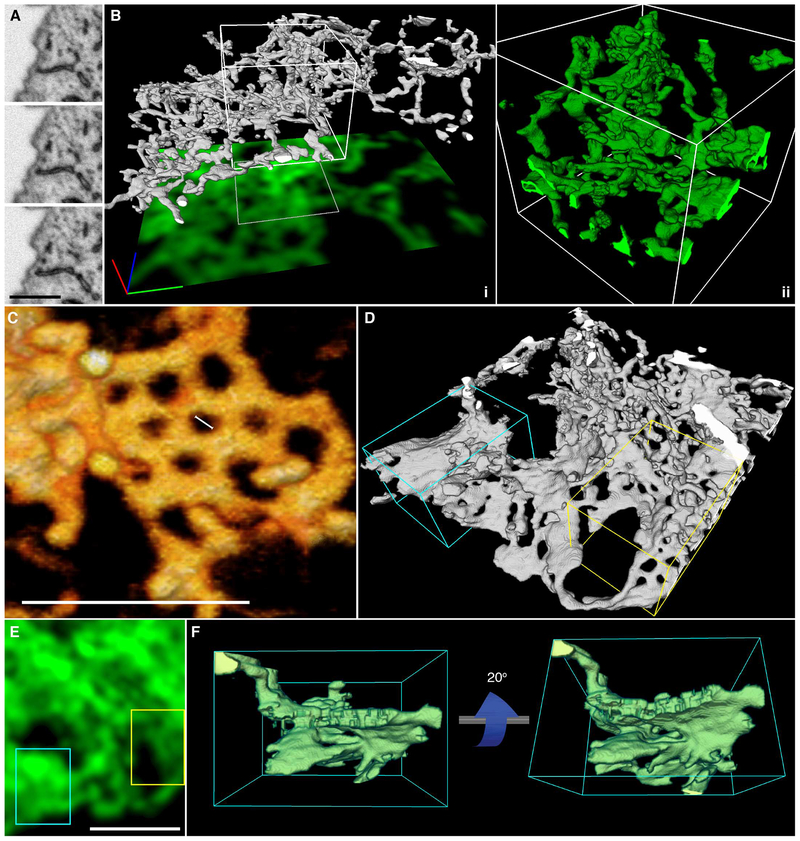
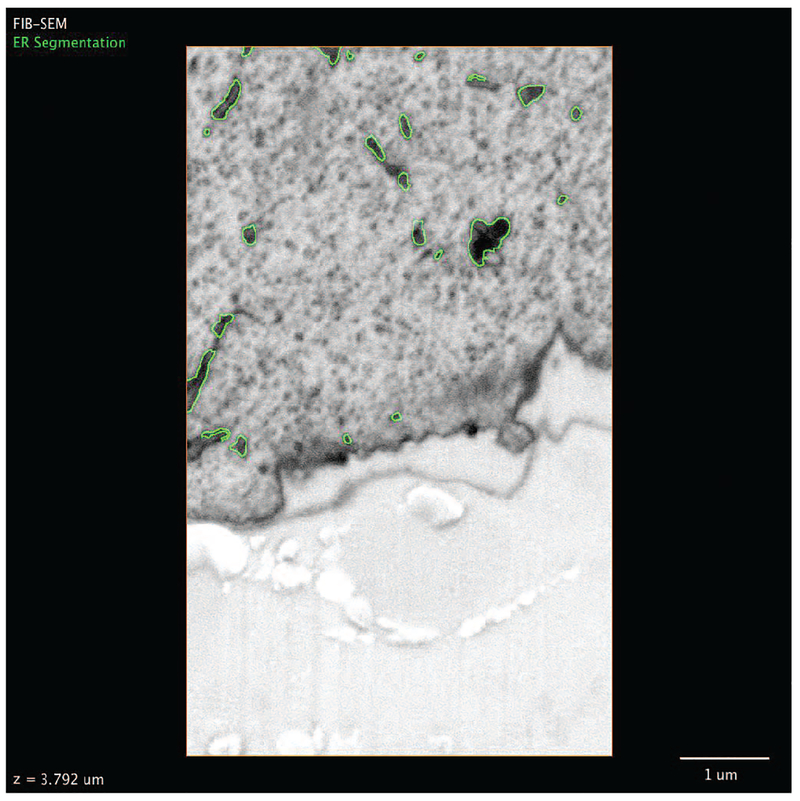
To overcome the limitations of optical microscopy in resolving the very fine structure of tubular matrices, we studied their morphology by electron microscopy (EM). FIB-SEM was performed in native, untransfected cells to provide an additional control against the possibility that over-expression of both Sec61β and ER3 coincidentally induces matrices. The conditions used for FIBSEM were selected to result in 8-nm steps in the z position (see materials and methods), which provides extremely high resolution in the z dimension and ensures that even thin structures such as tubular matrices can be captured (Fig. 5A). This fine z resolution allowed the reconstruction of remarkably intricate 3D tubular structures even within very thin sections of the cell (Fig. 5B and Movie 1). In contrast, we found that the topological complexity of matrices in the z dimension would be lost with diffraction-limited confocal imaging, as nearly the entire structure shown in Fig. 5B falls within the focal plane of a single confocal slice. Indeed, a projection of a theoretical confocal image derived from the EM data in Fig. 5B (see materials and methods) results in an image indistinguishable from that of an intact ER sheet (Fig. 5B, green footprint). In agreement with the LLS-PAINT data described above, we found that these structures can contain substantial vertical topology even within a thin space, and a single slice through the structure often revealed only a few isolated tubules (Fig. 5A). Inspection of the FIB-SEM data also revealed heterogeneity in matrix structures, from highly convoluted 3D structures to nearly planar arrays of tubes (e.g., Fig. 5B versus Fig. 5C). Additionally, some matrices were incredibly tightly clustered, with spaces less than 50 nm between tubules (Fig. 5C). As such, it would be impossible to resolve these structures as distinct from sheets by SIM, and it would be difficult to do so even by LLS-PAINT.
Fig. 5. ER tubules form highly convoluted and intricate structures within the resolvable volume limits of any optical imaging technique.
(A) Three consecutive FIB-SEM slices through an ER matrix spaced by 8 nm. Scale bar, 1 mm. (B) Three-dimensional reconstruction of a tubular matrix in a thin section (~600 to 1200 nm between the plasma membranes) of the cell. (i) The footprint shows the theoretically highest resolution that could be achieved with a single confocal slice through the structure directly shown above. Scale bar, 1 mm in each direction. (ii) Close-up of the reconstruction of the boxed region in (i). (C) 3D rendering of raw EM data showing an example of an approximately planar ER matrix with subresolution spaces (large scale bar, 500 nm; small scale bar, 50 nm). (D) 3D rendering of the ER at the border of perinuclear and peripheral regions of the cell, showing stacked helicoidal sheets (cyan box) and ER matrices (yellow box). (E) A theoretical confocal image of the structure, showing the difficulty in distinguishing these structures by diffraction-limited imaging. (F) Views from the side of a stacked helicoidal sheet in the perinuclear region of the ER [from cyan box in (D)], showing the pitch of the intact membranes.
Movie 1. Three-dimensional reconstruction of FIB-SEM data reveals a convoluted ER matrix.
Raw 2D FIB-SEM data of ER tubules are shown in a series of sequential planes. The ER is segmented in green; the 3D reconstruction is shown, revealing an ER matrix. When confocal resolution is simulated, the convoluted nature of the structure is masked.
Generally, areas of flat continuous membrane are described as having net zero curvature, whereas highly curved regions of membrane have either net positive or negative curvature (1). In addition to the aforementioned tubular matrices, we also observed a number of structures in the ER that show local regions of near-zero curvature in the ER membrane. The largest of these regions are in true stacked ER sheets near the nucleus (Fig. 5D, cyan box), which have been described extensively (16). When imaged with diffraction-limited techniques, these structures are nearly impossible to differentiate from tubular ER matrices (Fig. 5, D and E). We also observed that whenever stacked sheets are seen, they appear to be connected by membrane “ramps” [as described in (16)] and generally are closeto the nucleus, where the cell height is much greater than in the periphery (Fig. 5F and movie S5).
Localizing ER-shaping proteins to tubular matrices
Given the highly variable and complex nature of ER matrices identified in the FIB-SEM data, we speculated that examining the distribution of well-characterized ER-shaping proteins may provide insights into the diversity of these structures. ATL GTPases have been well characterized as three-way junction-forming proteins localized within ER tubules, where they also bind ER-shaping RTNs (17, 18). Thus, although they would not be expected to be enriched in sheets, they would be expected in matrices composed of densely packed tubules and three-way junctions. We transfected COS-7 cells with HaloTag-ATL1 (one of three human ATL paralogs) and assessed the localization of ATL1 in live cells by means of diffraction-limited confocal microscopy (Fig. 6A). ATL1 was present in all structures classically considered as peripheral sheets. This was verified by immunostaining of endogenous ATL3 in HeLa cells (Fig. 6B).
Fig. 6. Localization of ER-shaping proteins within dense tubular matrices.
(A) COS-7 cell expressing low levels of HaloTag-ATL1 imaged live with conventional spinning disk microscopy. Boxed regions (i) and (ii) demonstrate ATL1 localization throughout structures that appear to be peripheral sheets by spatiotemporally limited imaging techniques. (B) Left: A fixed HeLa cell acquired with conventional scanning point confocal microscopy, stained for the endogenous ER marker calnexin and endogenous ATL3 (merged image); boxed regions (i) and (ii) show endogenous ATL3 localization to structures that appear as sheets. Right: Regions shown in (i) and (ii) were stained for the endogenous ER marker calnexin and endogenous ATL3. (C) A fixed COS-7 cell expressing Sec61β and HaloTag-ATL1 imaged by wide-field SIM. Sec61β fluorescence is color-coded by z position. Boxed region is enlarged in panels at right. Structures that appear as sheets by diffraction-limited imaging (i) are revealed to be dense tubular matrices (ii) that are positive for ATL1 (iii); the merged image of (ii) and (iii) is shown in (iv). (D) U-2 OS cell expressing Sec61β stained for endogenous RTN4A/B and CLIMP63 with the boxed region magnified (bottom row), illustrating localizations of both proteins within a tubular matrix. Merged images are at the far right. Scale bars, 2 μm.
Overexpression of ATLs was previously shown to induce massive proliferation of “aberrant sheet-like structures” in a GTPase-dependent manner (17). This finding has been difficult to explain in light of the known role of ATL GTPases as mediators of three-way junction formation between tubules. We hypothesized that ATL overexpression might drive the formation of increasingly dense tubular matrices, which would appear as peripheral sheets under standard confocal imaging because of insufficient spatiotemporal resolution. COS-7 cells coexpressing Sec61β and HaloTag-ATL1 were fixed and imaged using both diffraction-limited epifluorescence and 3D-SIM (Fig. 6C). Epifluorescence revealed the presence of peripheral sheet-like structures (Fig. 6C, i), as previously reported (17). However, the improved spatial resolution offered by 3DSIM showed these “aberrant sheet-like structures” to be dense tubular matrices (Fig. 6C, ii). These matrices contained ATL1 throughout (Fig. 6C, iii). Collectively, these data suggest that over-expression of ATLs does not drive formation of aberrant sheets, but rather induces the formation of dense tubular matrices, consistent with the known cellular functions of ATLs.
We also looked at the distributions of other proteins associated with driving or stabilizing particular ER shapes. RTNs are a highly conserved family of ER proteins sharing substantial sequence homology; multiple isoforms are present in most cell types and are typically associated with inducing and maintaining the curvature of the ER membrane in tubules (3). Classic work in the field has demonstrated localization of RTNs to tubular structures and their exclusion from sheet-like structures (19, 20). Conversely, CLIMP63 is a traditional marker used to identify ER sheets, as it is believed to be involved in stabilizing the diameter of the ER lumen through interactions between long, dimeric coiled-coil domains (2). U-2 OS cells expressing Sec61β were stained with antibodies to endogenous CLIMP63 and RTN4A/B and imaged using Airyscan, a technique capable of achieving subdiffraction-limited imaging independent of SIM. In concordance with their tubular nature, many peripheral ER matrices were found to be positive for RTN4A/B, and some of these were also enriched in CLIMP63 (Fig. 6D). There are also a smaller number of ER matrices that exclude RTN4A/B staining, as has been previously demon strated in some peripheral sheet-like structures (2) (fig. S3). This complex heterogeneity in protein localization across matrices is unlikely to be a staining artifact, as overexpression of CLIMP63 or a variety of RTN isoforms also resulted in the presence of these proteins in some, but not all, ER matrices (fig. S3 and supplementary text). It is possible that heterogeneity in the distribution of these proteins is linked to the highly variable topologies shown in Fig. 5, which may correspond to functionally distinct classes of structures that appear identical when imaged with insufficient resolution.
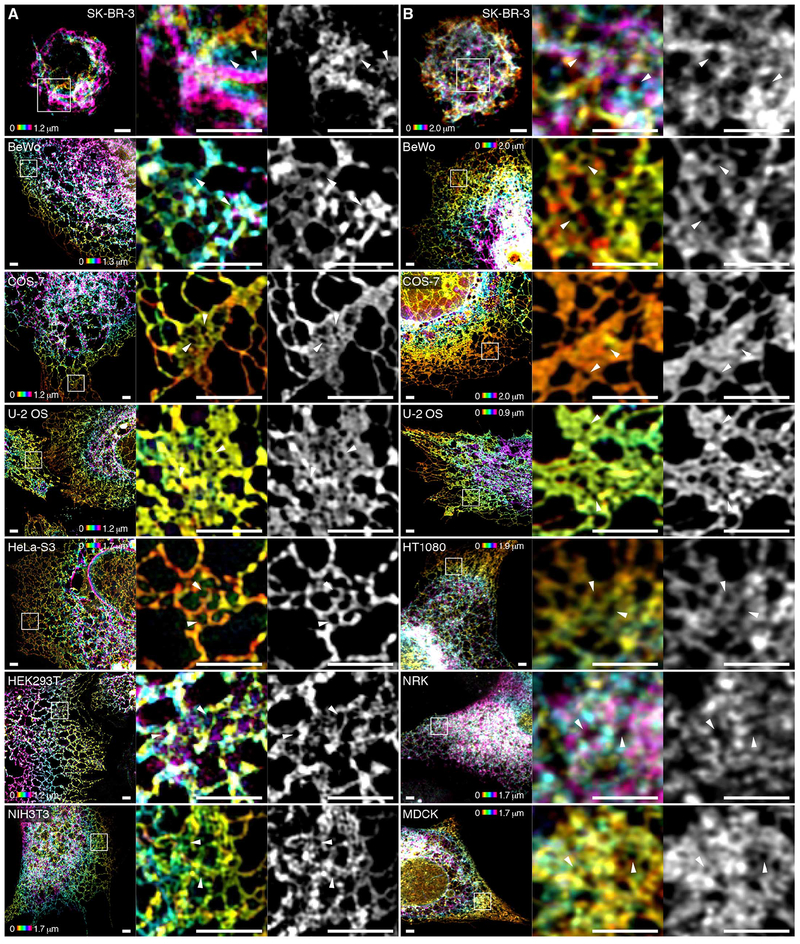
To explore whether our observations hold true for multiple cell types, we examined the structure of the ER in three dimensions using either 3D-SIM or Airyscan in a variety of cell lines with highly variant morphology and diverse organisms and tissues of origin. In all 10 cell lines examined, peripheral ER matrices were visible (Fig. 7). Although across cell types there seems to be substantial variation in the topology, density, and cellular location of these structures, they are uniformly present and clearly visible using either of the two independent imaging techniques.
Fig. 7. Tubular ER matrices are present in different cell types.
(A and B) Various cell lines expressing Sec61β were imaged using 3D-SIM (A) or, where fluorescence intensity was insufficient, Airyscan (B).The first four cell lines were imaged using both modalities, demonstrating that the dense matrix structures are not artifacts of any given imaging modality. Boxed regions highlighting representative tubular ER matrices in each cell type are magnified at the right. White arrowheads indicate subdiffraction-limited spaces between tubules. Scale bars, 2 μm. The signal of Sec61β fluorescence is color-coded by z position in the left and center panels corresponding to each cell line and technique.
Discussion
Taken together, our data indicate that most previously described “sheet-like” ER structures within the thin periphery of cells are actually dense tubular matrices. Limitations in spatiotemporal resolution using conventional microscopy result in their appearance as continuous or fenestrated sheets. Additionally, we show a previously uncharacterized, rapidly dynamic state in the peripheral ER that is broadly dependent on cellular energy sources and that contributes to the misidentification of ER matrices in living cells.
Why would the peripheral ER be organized in this way? The structural conformation of tubular matrices is likely to be imperative to multiple features of ER biology, such as the ability of the ER to rapidly alter its conformation in response to changing cellular needs. Interconversions among loose polygonal networks and dense matrices could be accomplished by simply sliding tubules along one another, rather than requiring energetically costly fusion or fission of ER membranes. This rapid intercon-version between loose and tight polygonal arrays of tubules (e.g., movie S3) is likely to be important in enabling the ER to rapidly reconfigure its spatial footprint in response to intracellular structural rearrangements or cell shape changes, or during cell migration. Indeed, the ER and cytoskeleton coexert a driving force for cytoplasmic streaming during cell expansion in Arabidopsis, and this is altered in mutants in which ER morphology is affected (21).
Clustering of tubules into tight arrays of three-way junctions might also function to decrease curvature stress across the ER, because the negative curvature of three-way junctions could help to neutralize the positive curvature associated with tubules (1). Thus, a relatively planar network could be formed (e.g., Fig. 5C) in the thin periphery of cells, where a lack of vertical space might prevent the formation of stacked helicoidal sheets (16). Dense ER matrices are also predicted to have greater membrane surface area than a flat sheet of similar dimensions, so they may allow storage of excess membrane proteins and lipids or provide increased surface for modulating lipid synthesis or protein folding. Such a tubular membrane reservoir may also be needed to facilitate the availability of ER membrane for modulating interactions with other organelles, such as mitochondria, lipid droplets, or endocytic compartments.
Our data do not conflict with the impressive literature describing the structures of flattened regions of the ER associated with specialized functions, such as the nuclear envelope (22), helicoidal stacks in the perinuclear region (16), or flattened cisternal structures close to the plasma membrane (23). Rather, these structures represent one end of a spectrum of curvatures across the ER membrane, with the other end of the spectrum dominated by more prevalent ER tubules and the dense tubular matrices we have described.
The heterogeneity observed in fine ultrastructure and ER protein content also suggests that there may be several distinct types of ER matrices. It is therefore possible that these different structures may carry out distinct functions. For instance, our FIB-SEM data support previous 3D EM reconstructions in suggesting that tubular morphology is far more heterogeneous than the cylindrical structures often depicted in models based on optical microscopy studies (24, 25). Tubules can also take on flattened or highly irregular structures along their lengths, as has been described in ER contact sites with the plasma membrane (23). There is no reason to think that tubules within ER matrices could not also undergo these sorts of deformations; hence, ER matrices close to the plasma membrane or other organelles (e.g., Fig. 3B) could potentially play important roles in rapidly facilitating calcium signaling or lipid transfer. These altered morphologies may also explain some of the variability in distribution of ER-shaping proteins across matrices, as structures may become too dense or too deformed to stably hold certain classes of ER proteins. These data may thus suggest another mechanism for modulating peripheral ER function. It seems clear, in any event, that given the pathogenic role of impaired ER shaping and protein distribution in disorders such as the hereditary spastic paraplegias (4), the ER structure and dynamics described here will have important implications for understanding both basic cell biology and disease pathogenesis.
Materials and methods
Plasmids and antibodies
Constructs expressing Myc- and HA-tagged atlastins have been previously described (26), with HaloTag-ATL1 purchased from Promega. Constructs expressing mEmerald-Sec61β and mApple-Sec61β were generated by replacing the GFP cassette of pAcGFP1-Sec61β (2) with corresponding fluorescent proteins, using flanking AgeI and BsrG1 restriction sites. Reticulon constructs were cloned from whole brain cDNA and inserted into pmCherry-C1 using the BglII and SalI restriction sites. Lifeact-mApple has been previously described (27), and mEmerald-ER-3 (ER3) was a gift from Michael Davidson (Addgene plasmid # 54082). ER3 consists of an ER lumen-targeting motif fused to mEmerald with a C-terminal KDEL tag.
Commercially available mouse monoclonal anti-Myc epitope (1:200; IgG1, clone 9E10; Santa Cruz Biotechnology) and rabbit polyclonal anti-HA epitope (1:200; IgG, clone Y-11; Santa Cruz Biotechnology) antibodies were used for Immunocytochemistry. In addition, a custom affinity-purified mouse monoclonal anti-ATL3 (No. 6115, IgG, clone 9H2B12; residues 561–578; acetyl-CATVRDAVVGRPSMDKKAQ-OH) antibody was used at 1:100. The anti-RTN4A/B antibody was a kind gift from Riqiang Yan, and was used as described previously (28). CLIMP63 was stained using a commercial antibody purchased from Enzo Lifesciences (1:250; IgG2a, clone G1/296).
Cell culture, transfection, and plating
COS-7, U-2 OS and HeLa cells (ATCC) were grown in phenol red-free Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) FBS (Corning), 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C and 5% CO2. MDCK cells were a line stably expressing ER-RFP (29, 30) generously provided by Erik Snapp. All other cells (SK-BR-3, BeWo, HeLa-S3, HEK293T, HT1080, NRK and NIH3T3) were grown according to manufacturer’s specifications (ATCC).
Coverslips and chambers were pre-coated with 400–600 μg/ml Matrigel (Corning), and cells were seeded to achieve ~60% confluency at the time of imaging. Transfections were executed using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s specifications. Fluorescently-tagged Sec61β alone was transfected at 1 μg/ 35 mm chamber, or else cotransfected at a ~3–4:1 ratio with the additional plasmid. Imaging was performed between 14–22 hours post-transfection. Where indicated, HaloTag-ATL1 was labeled with JF549 as previously described (31), and cells were imaged immediately post-labeling.
Drug treatments
All drugs used in the paper were purchased from Sigma Aldrich and used as has been described (32). Drugs were diluted from high concentration stocks reconstituted in DMSO, and diluted to the appropriate concentration in complete medium unless otherwise indicated. Cycloheximide (CHX) and Puromycin (Puro) were used at a final concentration of 100 μg/ml, Nocodazole (NZ) was used at 30 μM, and Blebbistatin (Bleb) was used at a final concentration of 50 μM. ATP depletion was accomplished by incubating the cells for one hour in 10 mM 2-deoxyglucose (DOG) and 2 mM sodium azide (NaN3) in PBS at 37°C, and AlF treatment was performed as previously described (33) in HBSS. HBSS-only controls were also run for all experiments using AlF, and no difference in any phenotype studied was seen compared to untreated controls (data not shown).
Immunocytochemistry
For immunocytochemistry staining, cells were seeded into No. 1.5 imaging chambers (Lab-Tek) coated with Matrigel, and then fixed with 4% (w/v) paraformaldehyde (PFA) for 20 min at room temperature (RT). Cells were then permeabilized with 0.2% (w/v) saponin (Sigma-Aldrich) for 30 min, and blocked in 5% (v/v) donkey serum (Sigma-Aldrich) doped with 0.05% (w/v) saponin for 1 hour at RT. Primary antibodies were diluted in block to the aforementioned concentrations, added to samples, and incubated overnight at 4°C. Secondary anti-mouse and anti-rabbit antibodies conjugated to Alexa 488, Alexa 555 (1:1000; Life Technologies) were made in block and incubated with samples for 30 min at RT. Imaging was performed in fresh PBS.
Confocal microscopy
Live-cell confocal imaging was performed using a customized Nikon TiE inverted scope outfitted with a Yokogawa spinning-disk scan head (#CSUX1, Yokogawa) and a Photometrics EM-CCD camera (Evolve 512) with 500 ms exposure time. Fluorescence was collected using a 60× Plan-Apochromat 1.40 NA oil objective (Nikon) with the additional use of a 1.5× optovar to create a final pixel size of 130 nm. Cells were imaged in DMEM and incubated with a LiveCell Imaging Chamber (Nikon) at 37°C and 5% CO2.
Fixed-cell confocal microscopy was performed using a Zeiss 780 laser scanning confocal microscope equipped with a 32-channel multi-anode spectral detector. Excitations were performed sequentially using 405, 488, 561, or 633 nm lines as needed, and imaging conditions were experimentally selected to minimize crosstalk. The resulting fluorescence was collected using a 100× Plan-Apochromat 1.4 NA oil objective (Carl Zeiss) and images were prepared using the commercial Zen software package (Carl Zeiss).
Airyscan imaging was performed in fixed cells using a Zeiss 880 outfitted with an Airyscan module. Cells were seeded onto matrigel-coated cover-slips, fixed in 4% (w/v) PFA supplemented with 0.2% (w/v) glutaraldehyde at RT for 20 min. After fixation, cells were washed in RT PBS and imaged in clean PBS. Data was collected using a 63× 1.4 NA objective and immersion oil optimized for 30°C (Carl Zeiss). Colors were collected sequentially to minimize crosstalk, and Airyscan processing was performed using the Airyscan module in the commercial ZEN software package (Carl Zeiss).
Structured illumination microscopy (SIM)
Fixed cell, 3D-SIM was performed using a commercial Zeiss microscope (ELYRA SR-SIM, Carl Zeiss Microimaging) outfitted with a Plan-Aprochromat 63× 1.4 NA objective lens. Samples were fixed in 4% PFA supplemented with 0.1% glutaraldehyde at RT for 20 min. Cells were imaged in PBS at RT with a final pixel size of 40 nm and 110 nm z plane spacing using three rotations of the SIM grating. SIM processing was performed using the SIM module in the Zen software package (Carl Zeiss Microimaging), and multicolor images were channel aligned using a matrix generated with Tetraspeck beads (LifeTechnologies) imaged on the same day as the cells.
Grazing incidence-structured illumination microscopy (GI-SIM)
GI-SIM was performed in a high speed SIM microscope previously adapted for total internal reflection fluorescence (TIRF)-SIM (34). GI-SIM shared the same beam path configuration as the high-speed TIRF-SIM system. In TIRF illumination mode, because the intensity of evanescent wave of excitation light exponentially decayed from the interface between cover slip and cell sample, the characteristic penetration depth of the evanes-cent wave limited the TIRF-SIM imaging depth to around 100 nm (34), in which much of the ER network lies outside. In order to increase the imaging depth, it is straightforward to tune down the incident angle that is inversely proportional to the penetration depth of the evanescent wave (34). We realized that when the incident angle was tuned to slightly smaller than the critical angle, where the refraction angle at the interface was near 90 degrees, and the refracted light grazed the surface of the cover slip, the grazing incident light actually formed a thin light sheet parallel to the cover slip surface. The thin light sheet intensity remained constant in both lateral and axial directions, and its thickness could be adjusted by tuning the incident angle of excitation light. The optimum thickness of grazing incident light sheet should match the depth-of-focus of the high NA objective (Zeiss alpha Plan-Apochromat 100x/1.57 Oil-HI), which is around 700 nm. To implement GI-SIM, we used the grating pattern generation algorithm previously developed for patterned activation nonlinear SIM. It permitted us to finely tune the incident angle of excitation light, i.e., the period of illumination pattern (34). We identified the optimum incident angle by keeping the outof-focus background of the TIRF image as little as possible, meanwhile observing as much ER structure as possible. After we identified the critical angle for grazing incidence, the raw image acquisition and SIM image reconstruction procedure is the same as TIRF-SIM (34). Time-lapse images were also subject to a traditional bleach correction algorithm by histogram matching in ImageJ (NIH). Cells were plated and transfected on Matrigel-coated high-NA coverslips (Zeiss) and imaging was performed the following day.
Lattice light sheet-point accumulation for imaging in nanoscale topography (LLS-PAINT)
LLS-PAINT was performed as described elsewhere (14) using a custom built Lattice Light Sheet microscope (15). Membrane labeling was performed sequentially with BODIPY-TR (LifeTechnologies) followed by AZEP-Rh (31) to label intracellular membranes and carried out over 14 days total. The final image was reconstructed from 548,792,627 individual molecular localizations with a median precision of 7.2 nm laterally and 41.0 nm axially. Immediately prior to PAINT imaging, a diffraction limited dithered LLS image of mEmerald-Sec61β was taken for comparison in the same cell.
Electron microscopy (EM)
In preparation for EM, cells were grown in 100 mm culture dishes (Corning) in standard cell culture conditions. Cells were fixed in 2% (w/v) glutaraldehyde in 0.08 M cacodylate buffer for one hour. Cells were then post-fixed with osmium according to a modified ROTO (reduced osmium thiocarbohydrazide-osmium) protocol (35). Briefly, fixation was performed in 1% (w/v) OsO4 in 0.1 M cacodylate buffer for 30 min on ice, followed by a wash in cacodylate buffer. The cells were then incubated with 1% (w/v) thiocarbohydrazide in water for 10 min at room temperature, followed by immersion in 1% (w/v) OsO4 in 0.1 M cacodylate for 30 min at 4°C. Cells were contrasted en bloc with 1% (v/v) uranyl acetate, dehydrated in ethanol, and embedded in Durcupan ACM (Fluka). FIB-SEM was performed using a Zeiss NVision40 Focused Ion Beam Scanning Electron Microscope. SEM and FIB milling steps were optimized to produce isotropic 8 nm voxels. The SEM image stack was acquired at 300 kHz/voxel using a 3-nA electron beam at 1.5 kV landing energy for imaging and a 30-kV gallium ion beam for FIB milling.
Structured illumination microscopy reconstruction and Fourier filtering
SIM reconstruction was performed utilizing a modified reconstruction algorithm based on the previously described Gustafsson algorithm (36). During reconstruction, data was filtered in Fourier space using a variety of filters to minimize the appearance of reconstruction artifacts. This included at least a one log scan of the Wiener filter and a variety of suppression radii around the peaks at Abbe’s limit (fig. S4), in addition to a variety of apodization functions designed to roll off the noise at the limit of resolution. While the first two filters were selected individually for each image, the apodization function was decided collectively for the data and applied to every image in the paper. The apodization function was a single Gaussian blur using a radius that is smaller than the resolution limit of the technique, σ = 45 nm in real space. This allows most SIM reconstruction artifacts to be filtered out, maximizing the potential signal to noise with only a small price in functional resolution (fig. S4).
Reconstruction of three-dimensional EM data
Three dimensional FIB-SEM data was reconstructed and the ER segmented using a pseudo-automated approach. First, images were prepped, cropped, and inverted using ImageJ, so thatosmium signal appeared as fluorescence for subsequent analysis. Images were then loaded into Ilastik (37) for pixel classification. The pixel classification algorithm was used to generate a probability map for cellular membranes, based upon the osmium signal. A carving algorithm within Ilastik was then utilized. The resulting segmentation was overlaid onto the raw EM data using Amira (FEI) and quality checked by eye throughout each slice of the reconstruction. Simulation of serial section data was performed by simply summing the requisite number of FIB-SEM slices that would have been present in a single slice acquired by serial section.
Data visualization
Two dimensional image preparation and analysis was generally performed using ImageJ (NIH), and three dimensional image preparation was performed using Amira (FEI).
Skeletonization
Skeletonization of images was performed using ImageJ (NIH). First, images were pre-processed using enhanced local contrast (CLAHE) to help flatten the intensity of the ER. The images were then manually thresholded, made binary, and skeletonized. Using the AnalyzeSkeleton (8) plug-in in ImageJ, branches and junctions were determined from the skeletonized images. In short, skeletonized pixels with exactly two neighbors are considered branches and pixels with more than two neighbors, junctions.
Analysis of tubule motion
After obtaining the skeletonized image, lines were drawn perpendicular to the skeletonized structure over a number of tubules that were to be analyzed. In order to avoid the confounding effects of junctions crossing the line, lines were placed on sections of tubule that were spatially separated from any junctions. Kymographs of the skeletonized data were then generated along the lines over a total time lapse of 100 frames (2.5 s) (see Fig. 1B, for example). Amplitude was extracted from the kymograph by using a custom written peak finding algorithm in Labview, then measuring the distance between the maximum and minimum of peaks of the skeleton during the kymograph’s time window. The frequency was defined as the inverse of the period, which was measured by dividing the length of the time course by the number of paired maxima and minima within the data.
Junction tracking
Junctions were determined directly from the AnalyzeSkeleton plugin in ImageJ. Junctions from the tagged skeleton output of the AnalyzeSkeleton plugin typically occupy 1–5 pixels. The binary junction images were smoothed with a Gaussian kernel having a standard deviation of 1.0 pixel, resulting in single-particle-like images. The resulting images were then fed directly into the u-track SPT software (13).
Mean square displacement analysis
Trajectories, with a lifetime of at least 10 frames (0.25 s) were obtained. The trajectories were characterized through their mean square displacement (MSD). MSD =(1/T) where T the total movie time and r the displacement. The MSD can be described as MSD ~ tα where α can be used to describe a particle’s motion as Brownian (α = 1), subdiffusive (α < 1), or super-diffusive (α > 1) (10).
Kymographs
Kymographs were prepared from time lapse images by manually drawing lines across the image and using the standard reslice package in ImageJ. The resulting figure represents the intensity by pixel along the line graphed against time. Axes are labeled to indicate the respective x (μm) and t (s).
Tracking spaces in sheets
GI-SIM images of the ER were cropped such that the ROI was an individual sheet. The area outside of the sheet was then subtracted and the intensity of the images inverted. The transformation resulted in local intensity minima (spaces) becoming local intensity maxima. These maxima were then directly entered into the u-track SPT algorithm.
Space lifetime and density
Track length corresponds to the lifetime of the spaces. To account for clipping of lifetimes at either the start or end of the movie, the distribution of lifetimes was corrected following Loerke and colleagues (38, 39). To measure the density of spaces, the area of each sheet was measured by drawing a freehand ROI around the sheet and then measuring the area of the ROI in ImageJ. The density of spaces was then calculated as the number of spaces within the sheet divided by the area of that particular sheet.
Temporal intensity derivative
The derivative was calculated by choosing a defined region and time series of interest and processing the data as described for GI-SIM (fig. S1A). Each frame of the time-lapse image was then subtracted pixel by pixel from the following frame using a floating 32-bit depth pixel to ensure negative signals were not lost (fig. S1B). The resulting image was squared on a per-pixel basis, to make all changes positive integers (fig. S1C). The upper limit of the dynamic range was reset to the theoretical maximum in order to normalize the derivative between samples. The resulting time-lapse image was temporally color coded, yielding a spatial map of the change in fluorescence intensity over time (fig. S1D).
Measuring the diameter of tubules and spaces in matrices
The size of apparent spaces within tubular matrices was measured by fitting the intensity cross-section profile of each minima to a Gaussian curve. The full width at half-maximum of the Gaussian curves provides a good estimate of the distance across the space. The diameter of tubules was measured in a similar fashion: intensity cross-sections along several locations of tubules were fit to a Gaussian curve and the full width at half-maximum was reported as the diameter of the tubule.
Temporal blurring
To simulate the effects of longer exposure times in GI-SIM, the appropriate number of SIM frames collected at 40 Hz were merged using a simple sum projection in ImageJ. Thus, 250 ms images are the sum of 10 individual 25 ms frames, and 1 s images are the sum of 40 separate 25 ms frames. When color-coding by frame is shown, the temporal color code projection tool was used in place of the simple sum projection.
Supplementary Material
ACKNOWLEDGMENTS
Supported by a NIGMS Postdoctoral Research Associate Program fellowship (A.V.W.), an intramural AIDS research fellowship from the NIH Office of AIDS Research (C.J.O.), a grant from the NIH Intramural AIDS Targeted Antiviral Program (J.L.-S. and C.J.O.), National Key Research Program of China grant 2016YFA0500200 (D.L.), and HHMI and the Intramural Research Programs of NINDS and NICHD. J.N.-A. is supported by a UCL School of Pharmacy PhD studentship (to K.H.). We thank H. White for assistance with cell culture, L. Shao for assistance with imaging and data processing, P.-P. Zhu for advice on immunocytochemistry, R. Yan for a generous gift of an RTN4A/B antibody, and E. Snapp for provision of an MDCK ER-RFP stable cell line and critical reading of the manuscript. The authors declare that there are no conflicts of interest. All data to support the conclusions are provided either in the manuscript or in the supplementary materials.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Shemesh T et al. , A model for the generation and interconversion of ER morphologies. Proc. Natl. Acad. Sci. U.S.A. 111, E5243–E5251 (2014). doi: 10.1073/pnas.1419997111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibata Y et al. , Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788 (2010). doi: 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goyal U, Blackstone C, Untangling the web: Mechanisms underlying ER network formation. Biochim. Biophys. Acta 1833, 2492–2498 (2013). doi: 10.1016/j.bbamcr.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackstone C, Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci 35, 25–47 (2012). doi: 10.1146/annurev-neuro-062111-150400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Chen LB, Dynamic behavior of endoplasmic reticulum in living cells. Cell 54, 37–46 (1988). doi: 10.1016/0092-8674(88)90177-8 [DOI] [PubMed] [Google Scholar]
- 6.English AR, Voeltz GK, Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol 15, 169–178 (2013). doi: 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D et al. , Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015). doi: 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C, 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc. Res. Tech 73, 1019–1029 (2010). doi: 10.1002/jemt.20829 [DOI] [PubMed] [Google Scholar]
- 9.Chen S et al. , Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A 112, 418–423 (2015). doi: 10.1073/pnas.1423026112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzler R, Klafter J, The random walk’s guide to anomalous diffusion: A fractional dynamics approach. Phys. Rep 339, 1–77 (2000). doi: 10.1016/S0370-1573(00)00070-3 [DOI] [Google Scholar]
- 11.Guo M et al. , Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell 158, 822–832 (2014). doi: 10.1016/j.cell.2014.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kural C et al. , Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc. Natl. Acad. Sci. U.S.A 104, 5378–5382 (2007). doi: 10.1073/pnas.0700145104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaqaman K et al. , Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). doi: 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legant WR et al. , High-density three-dimensional localization microscopy across large volumes. Nat. Methods 13, 359–365 (2016). doi: 10.1038/nmeth.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B-C et al. , Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). doi: 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terasaki M et al. , Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell 154, 285–296 (2013). doi: 10.1016/j.cell.2013.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J et al. , A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561 (2009). doi: 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orso G et al. , Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 (2009). doi: 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- 19.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA, A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 (2006). doi: 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- 20.Shibata Y et al. , The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem 283, 18892–18904 (2008). doi: 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefano G, Renna L, Brandizzi F, The endoplasmic reticulum exerts control over organelle streaming during cell expansion. J. Cell Sci 127, 947–953 (2014). doi: 10.1242/jcs.139907 [DOI] [PubMed] [Google Scholar]
- 22.Hetzer MW, The nuclear envelope. Cold Spring Harb. Perspect. Biol 2, a000539 (2010). doi: 10.1101/cshperspecta000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Busnadiego R, Saheki Y, De Camilli P, Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. U.S.A 112, E2004–E2013 (2015). doi: 10.1073/pnas.1503191112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E, Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell 23, 2424–2432 (2012). doi: 10.1091/mbc.E10-12-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puhka M, Vihinen H, Joensuu M, Jokitalo E, Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol 179, 895–909 (2007). doi: 10.1083/jcb.200705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu P-P, Soderblom C, Tao-Cheng J-H, Stadler J, Blackstone C, SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet 15, 1343–1353 (2006). doi: 10.1093/hmg/ddl054 [DOI] [PubMed] [Google Scholar]
- 27.Riedl J et al. , Lifeact: A versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008). doi: 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He W et al. , Reticulon family members modulate BACE1 activity and amyloid-b peptide generation. Nat. Med 10, 959–965 (2004). doi: 10.1038/nm1088 [DOI] [PubMed] [Google Scholar]
- 29.Lai CW, Aronson DE, Snapp EL, BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Mol. Biol. Cell 21, 1909–1921 (2010). doi: 10.1091/mbc.E09-12-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS, Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. U.S.A 103, 6536–6541 (2006). doi: 10.1073/pnas.0510657103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm JB et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). doi: 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole NB, Compendium of drugs commonly used in cell biology research. Curr. Protoc. Cell Biol 00:1B:1.B.1–1.B.26 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Donaldson JG, Kahn RA, Lippincott-Schwartz J, Klausner RD, Binding of ARF and beta-COP to Golgi membranes: Possible regulation by a trimeric G protein. Science 254, 1197–1199 (1991). doi: 10.1126/science.1957170 [DOI] [PubMed] [Google Scholar]
- 34.Steyer JA, Almers W, A real-time view of life within 100 nm of the plasma membrane. Nat. Rev. Mol. Cell Biol 2, 268–275 (2001). doi: 10.1038/35067069 [DOI] [PubMed] [Google Scholar]
- 35.Deerinck TJ, Bushong EA, Thor A, Ellisman MH, NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy (National Center for Microscopy and Imaging Research, 2010); www.ncmir.ucsd.edu/sbem-protocol.
- 36.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL, Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6, 339–342 (2009). doi: 10.1038/nmeth.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer C, Straehle C, Kothe U, Hamprecht FA, Ilastik: Interactive learning and segmentation toolkit. In 2011 8th IEEE International Symposium on Biomedical Imaging (ISBI 2011), pp. 230–233 (2011). doi: 10.1109/ISBI.2011.5872394 [DOI] [Google Scholar]
- 38.Loerke D et al. , Cargo and dynamin regulate clathrin-coated pit maturation. PLOS Biol 7, e1000057 (2009). doi: 10.1371/journal.pbio.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigel AV, Tamkun MM, Krapf D, Quantifying the dynamic interactions between a clathrin-coated pit and cargo molecules. Proc. Natl. Acad. Sci. U.S.A 110, E4591–E4600 (2013). doi: 10.1073/pnas.1315202110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.