Abstract
Melanoma is the most lethal cutaneous cancer with a highly aggressive and metastatic phenotype. While recent genetic and epigenetic studies have shed new insights into the mechanism of melanoma development, the involvement of regulatory non‐coding RNAs remain unclear. Long non‐coding RNAs (lncRNAs) are a group of endogenous non‐protein‐coding RNAs with the capacity to regulate gene expression at multiple levels. Recent evidences have shown that lncRNAs can regulate many cellular processes, such as cell proliferation, differentiation, migration and invasion. In the melanoma, deregulation of a number of lncRNAs, such as HOTAIR, MALAT1, BANCR, ANRIL, SPRY‐IT1 and SAMMSON, have been reported. Our review summarizes the functional role of lncRNAs in melanoma and their potential clinical application for diagnosis, prognostication and treatment.
Keywords: lncRNA, prognosis, SLNCR1, UCA1
1. INTRODUCTION
Melanoma is the leading cause of skin cancer‐related deaths and characterized by high metastatic potentials.1, 2, 3 The incidence of melanoma has been increasing in recent years, and is generally higher in fair‐skinned population.4, 5 For early‐stage melanoma, surgery remains the mainstay of treatment with a high cure rate.6 However, prognosis of advanced melanoma is dismal because of its resistance to conventional therapies, including chemotherapy and radiotherapy.7 Early melanoma detection is therefore the key to improving the survival. Nevertheless, the histopathologic diagnosis of melanoma is sometimes difficult for dermatopathologists in a subset of cases.8 Moreover, there is no sensitive and specific biomarker for melanoma owing to its unclear molecular pathogenesis.
It has been demonstrated that more than 90% of transcripts from the human genome are not translated into proteins.9 These non‐protein coding RNAs are an important class of regulatory molecules that play crucial roles in the regulation of gene expression 10 and their dysregulation has been implicated in the development of different types of cancer.11, 12 Non‐coding regulatory RNAs are classified into two categories depending on their length: small non‐coding RNA (≤200 bp) and long non‐coding RNA (lncRNA, >200 bp).13, 14 LncRNA can modulate gene expression through various mechanisms, including chromatin modification, transcriptional activation/repression, RNA editing/splicing/degradation, microRNA sequestration, and translational efficiency regulation.15, 16 Historically, lncRNAs were dismissed as junk or nonfunctional transcriptional noise.17 However, emerging evidence has demonstrated that lncRNAs play crucial functional roles in tumourigenesis, including melanoma.18
In this review, we summarize the published data on the deregulation and functions of lncRNAs in melanoma. We also discuss their potential diagnostic, prognostic and therapeutic applications.
2. DEREGULATED LNCRNAS IN MELANOMA
2.1. Hotair
The HOX transcript antisense intergenic RNA (HOTAIR), which was named for its location at the antisense strand of the HOXC gene cluster, was initially identified to be an overexpressed lncRNA in primary and metastatic breast cancer.19 By recruiting polycomb repressive complex 2 and histone demethylase complex, HOTAIR was found to mediate gene silencing via tri‐methylation of lysine 27 on histone H3 (H3K27me3) and H3K4me2.19 Numerous studies have shown that HOTAIR expression is upregulated in human cancers, including breast, gastric, hepatocellular, colorectal, pancreatic and nasopharyngeal carcinomas,14, 20, 21, 22, 23, 24 in which overexpression of HOTAIR plays an oncogenic role and is associated with cancer metastasis and poor prognosis. In melanoma, HOTAIR was consistently overexpressed in lymph‐node metastasis as compared with primary lesions.25 In addition, knockdown of HOTAIR suppressed the motility and invasion of melanoma cells in vitro, accompanied by decreased ability to degrade gelatin matrix, indicating that HOTAIR might increase melanoma cell invasiveness through promoting gelatinase activity. Another study showed that none of the benign melanocytic lesions showed the presence of HOTAIR whereas the staining of HOTAIR was very weak in primary non‐metastatic melanomas but very strong in all pairs of primary tissues and corresponding metastases. Interestingly, HOTAIR could be detected in intratumoural lymphocytes as well as in the serum of a subset of metastatic patients.26 Portoso et al. showed that HOTAIR RNA can repress transcription in the context, but that this effect is PRC2 independent.27 Taken together, these data suggested that HOTAIR is involved in the metastatic progression of melanoma and may serve as a diagnostic marker for metastatic melanoma.
2.2. Malat1
The metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear‐enriched transcript 2 (NEAT2),28 was initially identified as a prognostic marker for lung cancer metastasis.9 Accumulating studies have now demonstrated MALAT1 deregulation in different human malignancies.29 It has been shown that MALAT1 mainly plays an oncogenic role in tumourigenesis through promoting cancer‐cell proliferation, migration and invasion.30, 31 MALAT1 expression levels were higher in melanoma as compared with adjacent normal tissues.32 Moreover, knockdown of MALAT1 decreased the migration of melanoma cell line in vitro. A recent study also demonstrated that knockdown of MALAT1 promoted miR‐140 expression and suppressed Slug and ADAM10 expression in the uveal melanoma cell line MUM‐2C.33 These findings suggest that the aberrant upregulation of MALAT1 might contribute to melanoma metastasis through promoting cell migration via derepressing miR‐140‐mediated inhibition of Slug and ADAM10.
2.3. Bancr
BRAF‐activated non‐coding RNA (BANCR) is a 4‐exon transcript of 693 bp, whose encoding gene is located on chromosome 9.34, 35 BANCR is involved in a variety of human malignancies, including lung carcinoma, colorectal cancer, melanoma, gastric cancer and bladder cancer.36, 37, 38, 39, 40 Nevertheless, the direction of deregulation of BANCR was tissue‐specific in which this lncRNA could act as an oncogene or tumour‐suppressor gene.41 In melanoma, Flockhart and colleagues demonstrated that BANCR was recurrently overexpressed.35 Moreover, knockdown of BANCR decreased melanoma cell migration and this effect could be rescued by the chemokine CXCL11. Another study also showed that BANCR expression was upregulated in melanoma cell lines and tissues.42 In addition, its expression level was increased with advancing melanoma stages. Knockdown of BANCR expression significantly reduced the proliferation of melanoma cells through inactivating the extracellular signal–regulated kinases 1/2 (ERK1/2) and c‐Jun N‐terminal kinase (JNK) components of the mitogen‐activated protein kinase (MAPK) pathway. Moreover, BANCR knockdown suppressed melanoma growth in nude mice. Pertinent to clinical practice, patients with high BANCR expression in melanoma tissues showed a poorer prognosis and lower survival rate. These data indicated overexpression of BANCR contributes to the promotion and progression of melanoma.
2.4. Spry4‐it1
SPRY4‐IT1 is derived from an intron of the SPRY4 gene localized in the chromosomal region 5q31.3, which encodes an endogenous inhibitor of the receptor‐transduced mitogen‐activated protein kinase pathway.43 The secondary structure of SPRY4‐IT1 contains several long hair‐pins. It was initially identified to be upregulated in melanoma.43 In recent years, emerging studies have demonstrated the deregulation and pathogenic roles of SPRY4‐IT1 in human cancers, including lung cancer, gastric cancer, breast cancer and colorectal cancer.44, 45, 46, 47 In melanoma, SPRY4‐IT1 expression was predominantly localized in the cytoplasm.48 Knockdown of SPRY4‐IT1 impeded cell proliferation and differentiation but promoted apoptosis in melanoma cells.43 Differential expression of both SPRY4 and SPRY4‐IT1 was also detected in patient samples of primary in situ, regional metastatic, distant metastatic, and nodal metastatic melanoma. A subsequent mechanistic study identified lipin 2 as a major binding partner of SPRY4‐IT1. Lipin 2 is an enzyme that converts phosphatidate to diacylglycerol. Moreover, knockdown of SPRY4‐IT1 not only increased the protein expression of lipin 2, but also elevated the levels of diacylglycerol O‐acyltransferase 2 that converts diacylglycerol to triacylglycerol. Concordantly, SPRY4‐IT1 knockdown increased the levels of several lipid species, including fatty acyl chains, acyl carnitine and triacylglycerol.49 These findings indicated that aberrant upregulation of SPRY4‐IT1 plays a significant role in the pathogenesis of melanoma through promoting lipid synthesis.
2.5. Anril
ANRIL (antisense non‐coding RNA in the INK4 locus) is named since it is expressed in the opposite direction from the INK4A‐ARF‐INK4B gene cluster.50 ANRIL gene has been reported to be a genetic susceptibility locus shared by coronary heart disease, type 2 diabetes and also cancers.51 ANRIL was significantly upregulated in many cancers, including gastric cancer, non‐small cell lung carcinoma, ovarian cancer and gallbladder cancer.52, 53, 54, 55 Chromosome 9p21, which harbours ANRIL gene, is frequently inactivated in multiple human cancers.56 Moreover, recurrent fusion transcripts of MTAP and ANRIL can be detected in ~25% of melanoma cell lines and tumour tissues.6 In another study, ANRIL was shown to be upregulated whereas INK4A and INK4B were downregulated in uveal and cutaneous melanoma tissues and melanoma cell lines. Interestingly, knockdown of ANRIL restored INK4A and INK4B expression and inhibited colony formation and migration in vitro and growth of melanoma xenograft in vivo.57 These findings indicate that ANRIL exerts its oncogenic action in melanoma probably through regulation of its encoding locus that also harbours the tumour suppressors INK4A and INK4B.
2.6. Llme23
Wu and colleagues identified a previously unreported lncRNA known as Llme23 in a human melanoma cell line. This lncRNA was found to interact with polypyrimidine tract‐binding protein‐associated splicing factor (PSF). In addition, Llme23 expression was exclusively detected in human melanoma lines.58 Knockdown of Llme23 remarkably inhibited the malignant phenotypes of melanoma cells and deceased expression of the proto‐oncogene Rab23. These findings suggest that Llme23 might play an oncogenic role in human melanoma through direct binding to PSF.
2.7. Uca1
Urothelial carcinoma associated 1 (UCA1) was initially identified to be upregulated in bladder cancer cells.59 UCA1 expression was significantly upregulated in melanomas compared with paired adjacent normal tissues.32 Moreover, the expression level of UCA1 was significantly higher in more advanced stages (stages 3‐4) melanoma than those at early stages (stages 1‐2). Knockdown of UCA1 inhibited the migration of melanoma cells. A subsequent mechanistic study demonstrated that UCA1 could sponge miR‐507 and derepress miR‐507‐mediated inhibition of FOXM1 expression in melanoma, leading to increased cell proliferation and invasion.60 These findings indicate that increased UCA1 expression might contribute to melanoma growth and metastasis through the miR‐507‐FOXM1 axis.
2.8. Slncr1
SRA‐like non‐coding RNA1 (SLNCR1) is a novel lncRNA with significant sequence similarity to the lncRNA steroid receptor RNA activator 1. Schmidt and colleagues reported that SLNCR1 is an abundantly‐expressed lncRNA associated with worse overall survival in melanoma patients. Further functional and mechanistic characterization demonstrated that SLNCR1 increases melanoma invasion by transcriptionally upregulating matrix metalloproteinase 9 (MMP9) in cooperation with the brain‐specific homeobox protein 3a (Brn3a) and the androgen receptor (AR).61 This study may partially why males have higher incidence of melanoma metastases and exhibit an overall lower survival.62
2.9. Sammson
SAMMSON is a recently annotated lncRNA with its encoding gene located on chromosome 3p13–3p14, which also harbours the melanoma‐specific oncogene MITF.63 Leucci and colleagues demonstrated that SAMMSON was frequently co‐amplified with MITF and its expression is lineage‐specific. Functional assays showed that exogenous SAMMSON increased the clonogenic potential of melanoma cells whereas SAMMSON knockdown drastically decreased melanoma cell viability and sensitized melanoma to MAPK‐targeting therapeutics. Mechanistically, SAMMSON interacts with p32 to increase its mitochondrial localization for regulating mitochondrial homeostasis and metabolism. Concordantly, targeting SAMMSON decreased oxidative phosphorylation, mitochondrial ribosome biogenesis, and respiratory chain complex activity in a cancer‐cell‐specific manner.63 These results suggest that SAMMON silencing may deliver effective anti‐melanoma therapeutic responses (Table 1 and Figure 1).
Table 1.
Functional characterization of the lncRNAs in melanoma
| lncRNAs | Expression | Functional role | Related gene | Role | References |
|---|---|---|---|---|---|
| HOTAIR | Up | Motility invasion gelatin matrix | Oncogene | 25, 26 | |
| MALAT1 | Up | Migration metastasis |
miR‐140 Slug ADAM10 |
Oncogene | 32, 33 |
| BANCR | Up | Migration proliferation |
ERK1/2 JNK MAPK |
Oncogene | 35, 42 |
| SPRY‐IT1 | Up | Proliferation differentiation apoptosis | lipin 2 | Oncogene | 43, 48, 49 |
| ANRIL | Up | Colony formation migration | INK4A INK4B | Oncogene | 6, 57 |
| Llme23 | Up | Malignant phenotypes |
PSF Rab23 |
Oncogene | 58 |
| UCA1 | Up | Migration proliferation |
miR‐507 FOXM1 |
Oncogene | 32, 60 |
| SLNCR1 | Up | Invasion |
MMP9 Brn3a AR |
Oncogene | 60 |
| SAMMSON | Up | Viability oxidative |
MITF
MAPK |
Oncogene | 63 |
Figure 1.
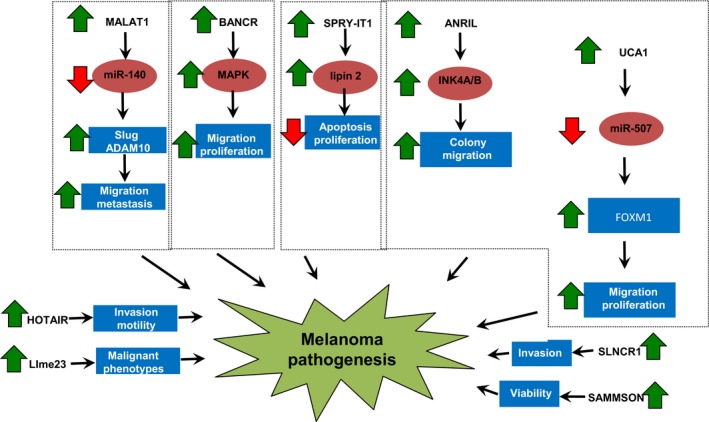
Functional roles of specific deregulated lncRNAs in melanoma
3. CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Melanoma is the most aggressive type of skin cancer with rapid metastatic progression. Early diagnosis is crucial for melanoma management as advanced melanomas are refractory to conventional treatment and associated with dismal survival outcomes. LncRNAs were initially considered to be functionless and therefore termed “genomic dark matter”. However, emerging studies have revealed their important functions. Although thousands of lncRNAs have been identified, only few have been functionally characterized. Current research has revealed the importance of lncRNA in tumourigenesis. In melanoma, several lncRNAs have been demonstrated to be differentially expressed in melanoma and function as potent regulators of melanoma progression and metastasis. These lncRNAs include HOTAIR, MALAT1, BANCR, ANRIL, SPRY‐IT1, Llme23, UCA1, SLNCR1 and SAMMSON. However, the current knowledge of lncRNAs in terms of their deregulation and mechanisms in melanoma is far from complete. Moreover, the clinical utilities of lncRNAs remain not fully established. Future investigations are therefore needed to clarify the upstream and downstream mechanisms as well as clinical implications of lncRNA deregulation in melanoma.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
Yu X, Zheng H, Tse G, Chan MTV, Wu WKK. Long non‐coding RNAs in melanoma. Cell Prolif. 2018;51:e12457 10.1111/cpr.12457
References
- 1. Miller AJ, Mihm MC Jr. Melanoma. N Engl J Med. 2006;355:51‐65. [DOI] [PubMed] [Google Scholar]
- 2. Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: a new player enters the game. Oncotarget. 2015;6:4562‐4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun L, Wang Q, Gao X, Shi D, Mi S, Han Q. MicroRNA‐454 functions as an oncogene by regulating PTEN in uveal melanoma. FEBS Lett. 2015;589:2791‐2796. [DOI] [PubMed] [Google Scholar]
- 4. MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(Suppl 6):vi1‐vi7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gangemi R, Amaro A, Gino A, Barisione G, Fabbi M, Pfeffer U, et al. ADAM10 correlates with uveal melanoma metastasis and promotes in vitro invasion. Pigment Cell Melanoma Res. 2014;27:1138‐1148. [DOI] [PubMed] [Google Scholar]
- 6. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ‐line deletion, including the entire INK4/ARF locus, in a melanoma‐neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Can Res. 2007;67:3963‐3969. [DOI] [PubMed] [Google Scholar]
- 7. Singh BP, Salama AK. Updates in therapy for advanced melanoma. Cancers. 2016;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonazzi VF, Stark MS, Hayward NK. MicroRNA regulation of melanoma progression. Melanoma Res. 2012;22:101‐113. [DOI] [PubMed] [Google Scholar]
- 9. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559‐1563. [DOI] [PubMed] [Google Scholar]
- 10. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5‐nucleotide resolution. Science. 2005;308:1149‐1154. [DOI] [PubMed] [Google Scholar]
- 11. Xin Y, Li Z, Chan MT, Wu WK. Circulating epigenetic biomarkers in melanoma. Tumour Biol. 2015;37:1487‐1492. [DOI] [PubMed] [Google Scholar]
- 12. Yu X, Li Z. Long non‐coding RNA growth arrest‐specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953‐1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li PF, Chen SC, Xia T, Jiang XM, Shao YF, Xiao BX, et al. Non‐coding RNAs and gastric cancer. World J Gastroenterol. 2014;20:5411‐5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu X, Li Z. Long non‐coding RNA HOTAIR: a novel oncogene (Review). Mol Med Rep. 2015;12:5611‐5618. [DOI] [PubMed] [Google Scholar]
- 15. Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155‐159. [DOI] [PubMed] [Google Scholar]
- 16. Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non‐coding RNAs. Nucleic Acids Res. 2012;40:6391‐6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark MB, Amaral PP, Schlesinger FJ, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253‐1261. [DOI] [PubMed] [Google Scholar]
- 19. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non‐coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119‐2128. [DOI] [PubMed] [Google Scholar]
- 21. Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875‐883. [DOI] [PubMed] [Google Scholar]
- 22. Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, et al. Upregulation of the long non‐coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908‐915. [DOI] [PubMed] [Google Scholar]
- 23. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro‐oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cantile M, Scognamiglio G, Marra L, Aquino G, Botti C, Falcone M, et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J Cell Physiol. 2017;232:3422‐3432. [DOI] [PubMed] [Google Scholar]
- 27. Portoso M, Ragazzini R, Brenčič Ž, Moiani A, Michaud A, Vassilev I, et al. PRC2 is dispensable for HOTAIR‐mediated transcriptional repression. EMBO J. 2017;36:981‐994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22:8031‐8041. [DOI] [PubMed] [Google Scholar]
- 29. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non‐coding RNA in cancer. Biochem Biophys Acta. 2015;1859:192‐199. [DOI] [PubMed] [Google Scholar]
- 30. Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91:791‐801. [DOI] [PubMed] [Google Scholar]
- 31. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, et al. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial‐mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5:15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian Y, Zhang X, Hao Y, Fang Z, He Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat‐1 in metastasis of melanoma. Melanoma Res. 2014;24:335‐341. [DOI] [PubMed] [Google Scholar]
- 33. Sun L, Sun P, Zhou Q, Gao X, Han Q. Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and invasion by silencing of miR‐140. Am J Transl Res. 2016;8:3939‐3946. [PMC free article] [PubMed] [Google Scholar]
- 34. Epigenetics McCarthy N. Going places with BANCR. Nat Rev Cancer. 2012;12:451. [DOI] [PubMed] [Google Scholar]
- 35. Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109‐112. [DOI] [PubMed] [Google Scholar]
- 37. Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, et al. Downregulated long noncoding RNA BANCR promotes the proliferation of colorectal cancer cells via downregualtion of p21 expression. PLoS ONE. 2015;10:e0122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li R, Zhang L, Jia L, et al. Correction: long non‐coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE. 2015;10:e0118728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang W, Zhang D, Xu B, et al. Long non‐coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomed Pharmacother. 2015;69:90‐95. [DOI] [PubMed] [Google Scholar]
- 40. He A, Liu Y, Chen Z, Li J, Chen M, Liu L, et al. Over‐expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Cancer Res. 2016;35:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT, et al. BRAF activated non‐coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF‐kappaB1. Biochem Biophys Res Comm. 2015;465:225‐231. [DOI] [PubMed] [Google Scholar]
- 42. Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, et al. Long non‐coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE. 2014;9:e100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma‐upregulated long noncoding RNA SPRY4‐IT1 modulates apoptosis and invasion. Can Res. 2011;71:3852‐3862. [DOI] [PubMed] [Google Scholar]
- 44. Zhang X, Chi Q, Zhao Z. Up‐regulation of long non‐coding RNA SPRY4‐IT1 promotes tumor cell migration and invasion in lung adenocarcinoma. Oncotarget. 2017;8:51058‐51065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan W, Song Z, Xu Q, Qu X, Li Z, Wang Y, et al. Up‐regulated expression of SPRY4‐IT1 predicts poor prognosis in colorectal cancer. Med Sci Monit. 2017;23:309‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, et al. The long noncoding RNA SPRY4‐IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie M, Nie F, Sun M, Xia R, Liu Y, Zhou P, et al. Decreased long noncoding RNA SPRY4‐IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial‐mesenchymal transition. J Transl Med. 2015;13:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, et al. The functional characterization of long noncoding RNA SPRY4‐IT1 in human melanoma cells. Oncotarget. 2014;5:8959‐8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mazar J, Zhao W, Khalil A, Lee B, Shelley J, Govindarajan S, et al. The functional characterization of long noncoding RNA SPRY4‐IT1 in human melanoma cells. Oncotarget. 2014;5:8959‐8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non‐coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956‐1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Popov N, Gil J. Epigenetic regulation of the INK4b‐ARF‐INK4a locus: in sickness and in health. Epigenetics. 2010;5:685‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deng W, Wang J, Zhang J, Cai J, Bai ZG, Zhang ZT. TET2 regulates LncRNA‐ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life. 2016;68:355‐364. [DOI] [PubMed] [Google Scholar]
- 53. Lin L, Gu ZT, Chen WH, Cao KJ. Increased expression of the long non‐coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol. 2015;10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX, Hua KQ. The long non‐coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget. 2016;7:32478‐32492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu B, Shen ED, Liao MM, Hu YB, Wu K, Yang P, et al. Expression and mechanisms of long non‐coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumor Biol. 2016;37:9875‐9886. [DOI] [PubMed] [Google Scholar]
- 56. Sasaki S, Kitagawa Y, Sekido Y, Minna JD, Kuwano H, Yokota J, et al. Molecular processes of chromosome 9p21 deletions in human cancers. Oncogene. 2003;22:3792‐3798. [DOI] [PubMed] [Google Scholar]
- 57. Xu S, Wang H, Pan H, Shi Y, Li T, Ge S, et al. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4‐hub in melanoma. Cancer Lett. 2016;381:41‐48. [DOI] [PubMed] [Google Scholar]
- 58. Wu CF, Tan GH, Ma CC, Li L. The non‐coding RNA llme23 drives the malignant property of human melanoma cells. J Genet Genomics. 2013;40:179‐188. [DOI] [PubMed] [Google Scholar]
- 59. Wang X, Zhang Z, Wang H, Cai J, Xu Q, Li M, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851‐4858. [DOI] [PubMed] [Google Scholar]
- 60. Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, et al. LncRNA UCA1‐miR‐507‐FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. [DOI] [PubMed] [Google Scholar]
- 61. Schmidt K, Joyce C, Buquicchio F, Brown A, Ritz J, Distel R, et al. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1‐like region. Cell Rep. 2016;15:2025‐2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aubuchon M, Bolt L, Janssen‐Heijnen M, Verleisdonk‐Bolhaar S, van Marion A, van Berlo C. Epidemiology, management and survival outcomes of primary cutaneous melanoma: a ten‐year overview. Acta Chir Belg. 2017;117:29‐35. [DOI] [PubMed] [Google Scholar]
- 63. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non‐coding RNA SAMMSON. Nature. 2016;531:518‐522. [DOI] [PubMed] [Google Scholar]


