Abstract
Accumulating evidence from genome‐wide analysis and functional studies has begun to unveil the important role of long non‐coding RNAs (lncRNAs) in cancer development. The lncRNA SPRY4‐IT1 is derived from an intron of SPRY4 gene and was originally reported to be upregulated in melanoma in which it functioned as an oncogene. Since this discovery, an increasing number of studies have investigated the expression and function of SPRY4‐IT1 in human cancers. Aberrant expression of SPRY4‐IT1 has now been documented in different cancer types, including osteosarcoma, breast, renal, oesophageal and prostate cancers. However, its deregulation and function in lung and gastric cancers remain controversial. Pertinent to clinical practice, SPRY4‐IT1 expression has been shown to predict survival of cancer patients. In this review, we summarize recent evidence concerning SPRY4‐IT1 deregulation and the associated mechanisms in human cancers. We also discuss the potential clinical utilization of this lncRNA as a diagnostic and prognostic biomarker for cancer patients.
Keywords: cancer, lncRNA, osteosarcoma, prognosis, SPRY4‐IT1
1. INTRODUCTION
Only ~2% of the human genome is transcribed into RNAs that encode proteins, whereas the function of most transcribed non‐coding sequences remained unclear until recently.1, 2, 3 It is now known that non‐coding RNAs, including small (precursors <200 nucleotides in length) and long (>200 nucleotides in length) non‐coding RNAs (lncRNAs), represent an important group of RNAs that are transcribed from the so‐called genomic “dark matter”.4, 5, 6 Recent studies have demonstrated that lncRNAs play significant roles in diverse biological and pathological processes, such as transcriptional regulation, cell fate determination and tumorigenesis.7, 8, 9 The importance of lncRNAs in cancer development can be exemplified by the frequent upregulation of the HOX antisense intergenic RNA (HOTAIR),10 Colon Cancer Associated Transcript 1 (CCAT1)11 and BRAF‐regulated lncRNA 1 (BANCR)12 in human cancers and their association with disease progression. Delineating the expression and functions of other lncRNAs is essential for understanding the molecular pathogenesis of cancer as well as for identifying novel molecular biomarkers for cancer diagnosis, prognostication and prediction of treatment response.13, 14, 15, 16
SPRY4‐IT1 (GenBank accession ID AK024556), a 708 bp lncRNA, was first discovered by Khaitan and colleagues in 2011.17 SPRY4‐IT1 is derived from an intron of SPRY4 gene resided on chromosome 5q31.3, which encodes an endogenous inhibitor of the receptor‐transduced mitogen‐activated protein kinase pathway.17, 18 Computational prediction suggested its secondary structure of SPRY4‐IT1 might contain several long hairpins. This lncRNA was originally reported to be highly upregulated in melanoma where it played an oncogenic role.17 Since its discovery, a growing number of studies have reported the aberrant expression of SPRY4‐IT1 in other cancer types, such as lung, breast, gastric, oesophageal and prostate cancers.19, 20, 21, 22, 23 In this review, we examine current evidence on the deregulation and function of SPRY4‐IT1 in human cancers (Table 1 and Figure 1). We also discuss the potential clinical utilities of SPRY4‐IT1 as a biomarker.
Table 1.
SPRY4‐IT1 expressions in human cancers
| Cancer type | Expression | References |
|---|---|---|
| Lung cancer |
Downregulaed Upregulated (Serum) |
34, 35 |
| Osteosarcoma | Upregulated | 26 |
| Gastric cancer |
Upregulated Downregulaed |
21, 36 |
| Breast cancer | Upregulated | 20 |
| Oesophageal cancer | Upregulated | 28, 29 |
| Prostate cancer | Upregulated | 32 |
| Bladder cancer | Upregulated | 30 |
| Melanoma | Upregulated | 24, 25, 27 |
| Renal cancer | Upregulated | 31 |
| Glioma | Upregulated | 33 |
Figure 1.
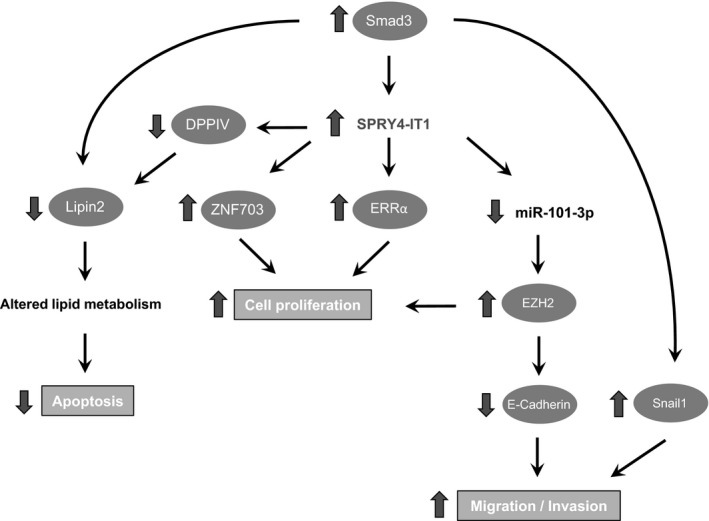
Upstream regulatory and downstream molecular mechanisms underlying SPRY4‐IT1 upregulation in human cancers
2. OSTEOSARCOMA
Xu et al.24 measured the role of SPRY4‐IT1 in the development of osteosarcoma. They firstly measured the expression of SPRY4‐IT1 in the osteosarcoma cell lines and tissues. They found that the SPRY4‐IT1 expression was upregulated in osteosarcoma cell lines and tissues. In addition, they showed that knockdown expression of SPRY4‐IT1 suppressed the osteosarcoma cell growth through inducing G1 arrest and increasing cell apoptosis. Furthermore, SPRY4‐IT1 inhibition expression suppressed osteosarcoma cell invasion and migration. These findings proved that SPRY4‐IT1 acted as an oncogene in development of osteosarcoma.
3. MELANOMA
SPRY4‐IT1 was upregulated in human melanomas as compared with the normal melanocytes.25, 26 There was also a strong correlation between SPRY4‐IT1 and SPRY4 expression.17 RNA‐fluorescent in situ hybridization revealed that SPRY4‐IT1 was predominantly expressed in the cytoplasm of melanoma cells, in which knockdown of SPRY4‐IT1 decreased cell survival, proliferation, migration and invasion.17 A subsequent study identified lipin 2, an enzyme that converts phosphatidate to diacylglycerol, as a major binding partner of SPRY4‐IT1.27 Knockdown of SPRY4‐IT1 not only elevated the protein expression of lipin 2 but also increased the levels of diacylglycerol O‐acyltransferase 2, an enzyme involved in the conversion of diacylglycerol to triacylglycerol. Consistently, SPRY4‐IT1 knockdown increased the levels of several lipid species, including fatty acyl chains, acyl carnitine and triacylglycerol. These findings suggested that SPRY4‐IT1 might alter lipid metabolism to induce lipotoxicity in melanoma cells through lipin 2.
4. BREAST CANCER
Shi and colleagues found that SPRY4‐IT1 expression was significantly higher in breast cancer tissues as compared with normal tissues.20 Increased SPRY4‐IT1 expression was associated with larger tumour size and more advanced pathological stage. Further experiments demonstrated that the knockdown of SPRY4‐IT1 significantly inhibited cell proliferation and induced apoptosis in breast cancer cell lines.
5. OESOPHAGEAL CANCER
SPRY4‐IT1 expression was significantly upregulated in oesophageal squamous cell carcinoma (ESCC) tissues and cell lines as compared with their normal counterparts.28 In addition, higher SPRY4‐IT1 expression predicted poorer prognosis and more advanced clinical stages. Multivariate analysis revealed that SPRY4‐IT1 was an independent prognostic factor in patients with ESCC. Knockdown of SPRY4‐IT1 not only reduced ESCC cell proliferation, invasiveness and migration in vitro but also ESCC xenograft growth in vivo. Another study demonstrated that the plasma levels SPRY4‐IT1 were significantly higher in ESCC patients than with normal controls.29 These findings indicated that SPRY4‐IT1 is involved in ESCC pathogenesis and may serve as a potential diagnostic and prognostic biomarker as well as a target for therapeutic intervention.
6. BLADDER CANCER
SPRY4‐IT1 expression was significantly increased in urothelial carcinoma of the bladder (UCB) as compared with adjacent non‐tumour tissues.30 Moreover, high SPRY4‐IT1 expression levels were correlated with advanced histological grade, tumour stage, lymph node metastasis as well as shortened overall survival. The prognostic significance of SPRY4‐IT1 for overall survival of UCB patients was independent of other clinicopathological parameters. Knockdown of SPRY4‐IT1 reduced bladder cancer cell proliferation, migration and invasion. These results suggested that SPRY4‐IT1 was implicated in the development and progression of UCB and might serve as a novel prognostic biomarker.
7. RENAL CANCER
SPRY4‐IT1 expression was significantly higher in clear cell renal cell carcinoma (ccRCC) tissues and renal cancer cell lines as compared with their normal counterparts. Higher SPRY4‐IT1 expression was also associated with more advanced clinical stages and poorer survival in ccRCC patients. In vitro assays indicated that knockdown of SPRY4‐IT1 could inhibit renal cancer cell proliferation, migration and invasion.31 SPRY4‐IT1 might be a potential prognostic indicator and therapeutic target in renal cancer.
8. PROSTATE CANCER
SPRY4‐IT1 expression was higher in primary human prostatic adenocarcinomas and prostate cancer cell line PC3 as compared with matched normal tissues and normal prostate epithelial cells. Functional investigation revealed that knockdown of SPRY4‐IT1 reduced PC3 cell proliferation and invasion and promoted apoptosis.32
9. GLIOMA
SPRY4‐IT1 expression was significantly elevated in human glioma tissues as compared with the adjacent normal brain tissues. The functional role of SPRY4‐IT1 in glioma was also investigated. SPRY4‐IT1 knockdown inhibited glioma cell proliferation and migration as well as epithelial‐mesenchymal transition,33 suggesting SPRY4‐IT1 might be a new therapeutic target for patients with glioma.
10. LUNG CANCER
The expression of SPRY4‐IT1 was decreased in ~95% of cancer tissues compared with corresponding adjacent normal tissues in non‐small‐cell lung cancer (NSCLC). In addition, low SPRY4‐IT1 expression was correlated with a poor prognosis in NSCLC patients independent of other clinicopathological parameters. Functional characterization revealed that SPRY4‐IT1 inhibited lung cancer cell proliferation, migration and invasion, and promoted apoptosis. More importantly, ectopic overexpression of SPRY4‐IT1 inhibited tumorigenesis and metastasis of NSCLC in vivo.34 Contrary to the reported downregulation of SPRY4‐IT1 in NSCLC tissues, another study demonstrated that the levels of circulating SPRY4‐IT1 was significantly higher in NSCLC plasma samples, indicating that this lncRNA might serve as a potential predictive biomarker for the early diagnosis of NSCLC.35
11. GASTRIC CANCER
Similar to lung cancer, the expression of SPRY4‐IT1 in gastric cancer is controversial. One study reported that SPRY4‐IT1 expression was downregulated in gastric cancer tissues as compared with normal tissues. Lower SPRY4‐IT1 expression was also associated with larger tumour size and more advanced clinical and pathological stages and predicted shorter survival of gastric cancer patients. Further investigation showed that SPRY4‐IT1 might suppress metastasis through the regulation of epithelial‐mesenchymal transition (EMT) in gastric cancer.21 However, another study showed that SPRY4‐IT1 expression was upregulated in both gastric cancer tissues and cell lines.36 Additionally, SPRY4‐IT1 expression levels were positively correlated with advanced TNM stage and served as an independent prognostic indicator of overall survival and disease‐free survival in gastric cancer patients. In vitro assays showed that the inhibition of SPRY4‐IT1 decreased gastric cancer cell proliferation and migration, and invasion (Table 1).
12. CONCLUSION AND FUTURE PROSPECTIVE
LncRNAs are important regulators of cellular processes. The lncRNA SPRY4‐IT1 is upregulated in many cancer types, including breast, oesophageal, prostate, bladder and renal cancers as well as melanoma and glioma. However, the expression of SPRY4‐IT1 in NSCLC and gastric cancer remain controversial. Although the discrepancy in the literature might reflect the tissue‐ and context‐specific function of SPRY4‐IT1, replication with larger sample size in independent cohorts may help to clarify the role of this lncRNA. Validation of the prognostic significance of SPRY4‐IT1 is also necessary before it can be established as a clinically utilizable biomarker for cancer diagnosis or prognostication. SPRY4‐IT1 plays significant roles in the regulation of cancer cell proliferation, migration and invasion. These findings not only contribute to the current understanding of the molecular pathogenesis of cancers but may also help to identify novel strategies for cancer treatment. Further investigation should focus on the molecular mechanisms upstream and downstream of SPRY4‐IT1 deregulation in different cancer types.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (Grant Numbers: 81401847, 81272053 and 81330044) and Beijing Natural Science Foundation (Grant Number: 15G10025).
Li Z, Shen J, Chan MTV, Wu WKK. The long non‐coding RNA SPRY4‐IT1: An emerging player in tumorigenesis. Cell Prolif. 2018;51:e12446 10.1111/cpr.12446
REFERENCES
- 1. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non‐coding RNA in cancer. Biochim Biophys Acta. 2015;1859:192‐199. [DOI] [PubMed] [Google Scholar]
- 2. Tano K, Akimitsu N. Long non‐coding RNAs in cancer progression. Front Genet. 2012;3:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non‐coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai). 2014;46:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sorensen KP, Thomassen M, Tan Q, et al. Long non‐coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor‐positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529‐536. [DOI] [PubMed] [Google Scholar]
- 5. He X, Bao W, Li X, et al. The long non‐coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. 2014;33:325‐332. [DOI] [PubMed] [Google Scholar]
- 6. Zhao HX, Hou WG, Tao JG, et al. Upregulation of lncRNA HNF1A‐AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta‐catenin signaling pathway. Am J Transl Res. 2016;8:3503‐3512. [PMC free article] [PubMed] [Google Scholar]
- 7. Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: a pivotal oncogenic long non‐coding RNA in human cancers. Cell Prolif. 2017;50:e12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xin Y, Li Z, Shen J, Chan M, Wu W. CCAT1: a pivotal oncogenic long non‐coding RNA in human cancers. Cell Prolif. 2016;49:255‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li P, Xue WJ, Feng Y, Mao QS. Long non‐coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8:3522‐3529. [PMC free article] [PubMed] [Google Scholar]
- 10. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang F, Xue X, Bi J, et al. Long noncoding RNA CCAT1, which could be activated by c‐Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li R, Zhang L, Jia L, et al. Long non‐coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE. 2014;9:e100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109‐112. [DOI] [PubMed] [Google Scholar]
- 14. Sun Y, Zheng ZP, Li H, Zhang HQ, Ma FQ. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Mol Med Rep. 2016;14:1714‐1720. [DOI] [PubMed] [Google Scholar]
- 15. Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36:4851‐4859. [DOI] [PubMed] [Google Scholar]
- 16. Tian T, Zhang T, Zhou T, Lin S, Shi S, Lin Y. Synthesis of an ethyleneimine/tetrahedral DNA nanostructure complex and its potential application as a multi‐functional delivery vehicle. Nanoscale. 2017;9:18402‐18412. [DOI] [PubMed] [Google Scholar]
- 17. Khaitan D, Dinger ME, Mazar J, et al. The melanoma‐upregulated long noncoding RNA SPRY4‐IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852‐3862. [DOI] [PubMed] [Google Scholar]
- 18. Leeksma OC, Van Achterberg TA, Tsumura Y, et al. Human sprouty 4, a new ras antagonist on 5q31, interacts with the dual specificity kinase TESK1. Eur J Biochem. 2002;269:2546‐2556. [DOI] [PubMed] [Google Scholar]
- 19. Zhang X, Chi Q, Zhao Z. Up‐regulation of long non‐coding RNA SPRY4‐IT1 promotes tumor cell migration and invasion in lung adenocarcinoma. Oncotarget. 2017;8:51058‐51065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4‐IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie M, Nie F, Sun M, et al. Decreased long noncoding RNA SPRY4‐IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial‐mesenchymal transition. J Transl Med. 2015;13:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui F, Wu D, He X, Wang W, Xi J, Wang M. Long noncoding RNA SPRY4‐IT1 promotes esophageal squamous cell carcinoma cell proliferation, invasion, and epithelial‐mesenchymal transition. Tumour Biol. 2016;37:10871‐10876. [DOI] [PubMed] [Google Scholar]
- 23. Mouraviev V, Lee B, Patel V, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:14‐20. [DOI] [PubMed] [Google Scholar]
- 24. Xu J, Ding R, Xu Y. Effects of long non‐coding RNA SPRY4‐IT1 on osteosarcoma cell biological behavior. Am J Transl Res. 2016;8:5330‐5337. [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao W, Mazar J, Lee B, et al. The long noncoding RNA SPRIGHTLY regulates cell proliferation in primary human melanocytes. J Invest Dermatol. 2016;136:819‐828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee B, Sahoo A, Marchica J, et al. The long noncoding RNA SPRIGHTLY acts as an intranuclear organizing hub for pre‐mRNA molecules. Sci Adv. 2017;3:e1602505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazar J, Zhao W, Khalil AM, et al. The functional characterization of long noncoding RNA SPRY4‐IT1 in human melanoma cells. Oncotarget. 2014;5:8959‐8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4‐IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743‐7754. [DOI] [PubMed] [Google Scholar]
- 29. Tong YS, Wang XW, Zhou XL, et al. Identification of the long non‐coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao XL, Zhao ZH, Xu WC, Hou JQ, Du XY. Increased expression of SPRY4‐IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Int J Clin Exp Pathol. 2015;8:1954‐1960. [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non‐coding RNA SPRY4‐IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801‐5809. [PMC free article] [PubMed] [Google Scholar]
- 32. Lee B, Mazar J, Aftab MN, et al. Long noncoding RNAs as putative biomarkers for prostate cancer detection. J Mol Diagn. 2014;16:615‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu H, Lv Z, Guo E. Knockdown of long noncoding RNA SPRY4‐IT1 suppresses glioma cell proliferation, metastasis and epithelial‐mesenchymal transition. Int J Clin Exp Pathol. 2015;8:9140‐9146. [PMC free article] [PubMed] [Google Scholar]
- 34. Sun M, Liu XH, Lu KH, et al. EZH2‐mediated epigenetic suppression of long noncoding RNA SPRY4‐IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial‐mesenchymal transition. Cell Death Dis. 2014;5:e1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu X, Bao J, Wang Z, et al. The plasma lncRNA acting as fingerprint in non‐small‐cell lung cancer. Tumour Biol. 2015;37:3497‐3504. [DOI] [PubMed] [Google Scholar]
- 36. Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4‐IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36:6751‐6758. [DOI] [PubMed] [Google Scholar]


