Abstract
Objectives
SNRPA is a protein component of U1 small nuclear ribonucleoprotein (U1 snRNP) complex, which takes part in the splicing of pre‐mRNAs. Its expression and function in tumour remain unknown. Herein, we elucidated the functional contribution of SNRPA to the progression of gastric cancer (GC).
Materials and methods
SNRPA expression was investigated in a GC tissue microarray by immunohistochemical staining. Cell proliferation was evaluated by CCK‐8, colony formation and EdU incorporation assays. A mouse xenograft model was used to detect the tumourigenicity. Gene expression profiling was performed and then the potential target genes were verified by quantitative real‐time PCR and western blot analyses. The functional relevance between SNRPA and its target gene was examined by cell growth assays.
Results
SNRPA expression was higher in tumour tissues than in matched normal gastric mucosa tissues, and it was positively correlated with the tumour size and progression. High SNRPA expression indicated poor prognosis of GC patients. Silencing SNRPA in GC cells markedly inhibited cell proliferation in vitro and tumour growth in a xenograft model, while overexpressing SNRPA exhibited opposite results. Moreover, we identified NGF (Nerve growth factor) as a downstream effector of SNRPA and further proved that NGF was crucial for SNRPA‐mediated GC cell growth.
Conclusions
These findings suggested that SNRPA may contribute to GC progression via NGF and could be a prognostic biomarker for GC.
1. INTRODUCTION
Gastric cancer (GC) is one of the most common types of digestive tumour in the world, with more than 70% of cases occurring in developing countries and remains one of the leading causes of cancer death worldwide.1, 2 Although advanced surgeries and chemotherapies have occurred in the past decades, there is currently no effective treatment strategy available to improve survival rates. Thus, identification of new biomarkers in GC progression is necessary to understand gastric cancer development and to design therapeutic targets.
Excision of introns from pre‐mRNA is an essential step for gene expression in eukaryotic cells.3 Most introns are removed by the major spliceosome composed of 5 fundamental RNA‐protein complexes: the U1, U2, U4, U5 and U6 snRNPs.4 Dysregulation of complex assembly or delocalization of snRNPs may initiate disease pathogenesis. The U1 snRNP is reported to initiate the assembly of the spliceosome by binding to the 5′‐splice site of pre‐mRNA.5 The U1 snRNP consists of the U1 snRNA molecule and several proteins: U1A (SNRPA), U1C, U1‐70K and a common set of proteins shared with other U‐type snRNPs.6, 7 Many studies have shown aberrant expression of genes encoding the spliceosomal members or mutations at splice sites of oncogenes and tumour suppressor genes may lead to cancer development, metastasis or drug resistance. These known splicing‐related genes include U2AF1,8 SRSF2,9 SF3B1,10 CD44,11 VEGF12 and so on.
SNRPA is a 282‐amino‐acid protein containing 2 RNA‐binding domains. The N‐terminal RNA‐binding domain, along with some flanking amino acids, is required for binding to U1 snRNA.13 SNRPA is important to form the spliceosome and promote the splicing process of mRNA. It is also involved in the SMN‐dependent snRNP biogenesis pathway known to regulate polyadenylation of mRNA.14, 15 SNRPA is moderately expressed in fat, weakly in muscle, and hardly expressed in small intestine, large intestine, spleen, liver and lung.16 SNRPA was found to bind the C‐terminal portion of importin α, by which SNRPA enters the nucleus independently of de novo snRNA synthesis.17 As for tumour development, one report has indicated that SNRPA is upregulated in hepatocellular carcinoma by cDNA microarray analysis;18 however, little is known about its function in human cancer to date.
In this study, we initially found an upregulation of SNRPA expression in GC tissues, which is closely associated with GC progression of patients. Overexpression or knockdown of SNRPA resulted in enhanced or inhibited phenotypes of GC cell growth in vitro and in vivo. Moreover, we demonstrated that NGF, the nerve growth factor, may act as a downstream effect of SNRPA on GC cell growth.
2. METHODS
2.1. Cell lines and culture conditions
Gastric cancer cell lines AGS, HGC27, SGC7901, BGC823 and MGC803 were obtained from Shanghai Cell Bank of Chinese Academy of Sciences. Cells were cultured in Modified Eagle's medium (MEM, Corning, US) supplemented with 10% FBS and penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA), in a humidified incubator under an atmosphere of 5% CO2 at 37°C.
2.2. Gastric cancer tissue samples
A human gastric cancer tissue microarray (Cat# HStmA180Su08), which contains 100 patients’ samples, was purchased from Outdo Biotech, Shanghai, China. Within the cohort, there are 80 paired of cancer samples and corresponding gastric mucosa specimens, and 20 cases of tumour tissues without adjacent normal tissues. Eighty‐nine of the 100 patients have detailed clinical information, among which are 34 women and 55 men, with an age range between 32 and 81 years old. The staining results were classified according to the product of percentage of positive cells (scored as 0%~100%) and staining intensity (scored as 0~3). 0~0.5, (−); 0.5~1.5, slight positive (+); 1.5~2.5, moderate positive (++); ≥2.5, strong positive (+++). To prevent bias from knowledge of clinical data, scoring of staining intensity was conducted by 2 pathologists in a blinded manner. Study protocol was approved by the ethics committee of Shanghai East Hospital, Tongji University School of Medicine.
2.3. RNA extraction and quantitative real‐time PCR
The relative expression of SNRPA was determined with qRT‐PCR with the commercial PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, Japan) and SYBR green reagent (TaKaRa, Japan). The total RNA was extracted with TRIzol (Sigma, USA), and reverse transcription was performed with the RNAiso Plus reagent (TaKaRa, Japan). The relative expression was calculated with the double delta comparative CT (2−∆∆Ct) method and β‐actin was used as an endogenous control. Each measurement was performed in triplicate independently. The following primer pairs were used to amplify and measure the amount of SNRPA and β‐actin: SNRPA‐qF: 5‐CAAACCTATGCGTATCCAGT‐3, SNRPA‐qR: 5‐GGATTCTCAGAAAGAGGCTG‐3 and β‐actin‐qF: 5‐CCTGGCACCCAGCACAATG‐3, β‐actin‐qR: 5‐GGGCCGGACTCGTCATACT‐3.
2.4. Establishment of stable cell lines
The lentivirus mediating overexpression of SNRPA (Gene ID: 6626) was purchased from Tuzhu Biotech, Shanghai, China. The C‐terminal of SNRPA was fused a FLAG tag. The lentivirus knocking down SNRPA was packaged and purchased from GenePharma, Shanghai, China. The targeting sequence for SNRPA mRNA is 5‐CCAAGACCGACUCAGAUAU‐3. Before the infection, the exponentially growing cells were seeded into six‐well plates, then the lentivirus with appropriate dilution was applied to each well for 48 hours and stably transfected cell lines were isolated by puromycin selection. The efficacy of knockdown or overexpression of SNRPA was verified by western blot assay.
2.5. Cell transfection and RNA interference
GC cells were plated in six‐well or other plates and transfected using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For knockdown of NGF expression, NGF‐specific small interference RNAs (siRNAs) were chemically synthesized (GenePharma, Shanghai, China). The sequence of si‐NGF was designed as follows: sense 5‐CCACAGACAUCAAGGGCAA‐3 and a non‐specific control (si‐NC) sequence is sense 5‐UUCUCCGAACGUGUCACGUdTdT‐3. Knockdown of NGF was verified by quantitative real‐time PCR.
2.6. Western blot analysis
Protein extracts of cells were electrophoresed by 10% sodium dodecyl sulphatepolyacrylamide gel electrophoresis (SDS‐PAGE), then transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore, USA). Membranes were blocked with 5% non‐fat milk in PBST (0.1% Tween 20 in PBS) for 2 h and incubated with primary antibodies against SNRPA (ProteinTech, 10212‐1‐AP), NGF (Abcam, ab6199), and β‐actin (Santa Cruz Biotechnology, #81171) overnight at 4°C. After washing for 15 minutes by PBST, the membranes were incubated with secondary antibodies for 1 hour. Detection of proteins was achieved by using the Odyssey Infrared Imaging System (Li‐COR, USA) according to the manufacturer's instructions.
2.7. Cell proliferation assay
Cell proliferation was determined with a commercial Cell Counting Kit‐8 (CCK‐8) kit following the manufacturer's instructions. Briefly, the appropriate number of cells was seeded in a 96‐well plate and subjected to the indicated treatment. A volume of 10 microlitres of CCK‐8 solution was added to each well and cultured for 1.5 hour at 37°C. Absorbance at 450 nm was measured with a microplate reader, and the relative cell viability was calculated. Each experiment was performed in triplicate independently.
2.8. Animal experiments
2 × 106 MGC803/LV‐SNRPA and MGC803/LV‐VEC cells or BGC823/LV‐shSNRPA and BGC823/LV‐shNC cells were collected and inoculated subcutaneously into the right flank regions of 4‐week‐old male BALB/c nude mice (Sippr‐BK laboratory animal corporation, Shanghai, China). Mice were euthanized after 30 days. The developed tumours were removed, photographed and weighted. All animal handling and experimental procedures were approved by the Ethics Committee of Shanghai East Hospital.
2.9. Colony formation assay
Stable cells expressing or silencing SNRPA were seeded, respectively, at a density of 3000 per well in 5 cm dish. Cells were allowed to culture for 2‐3 weeks; then, the colonies were stained with crystal violet for 30 minutes, photographed and counted. For soft agar colony formation assay, cells were seeded at density of 1000 per well in 24‐well plate in 0.4% low melting agarose over a 1% bottom agarose layer. The culture medium was changed every 3‐4 days for up to 2 weeks. The colonies were photographed and counted. All the experiments above were performed as least 3 times in triplicates.
2.10. EdU labelling and immunofluorescence
GC cells were seeded in 24‐well culture plates. After 24 hours, cells were incubated with 50 mM 5‐ethynyl‐2′‐deoxyuridine (EdU, RIBOBIO, China) for 2 hours and stained with Apollo®567 according to the manufacturer's instruction. The stained cells were observed with microscope and counted. All experiments were independently repeated at least 3 times.
2.11. Global cDNA microarray analysis and target gene verification
The RNA was extracted and reverse transcribed into cDNA as described above, and then sent to KangChen Biotech, Shanghai, China for microarray hybridization. The processed slides were scanned with an Agilent DNA microarray scanner. Differentially expressed genes were identified through Fold‐change screening. The resulting text files extracted from Agilent Feature Extraction Software (version 9.5.3) were imported into the Agilent GeneSpring GX software (version 7.3) for further analysis. Differentially expressed genes were identified through Fold‐change screening. For target gene verification, we used quantitative real‐time PCR and western blot analyses. The primers for the target genes are listed in Table S1.
2.12. Statistical analysis
Quantitative values are represented as mean ± standard deviation (±SD), statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc., San Diego, CA, USA) and Student's t test was used to compare values for paired data. The Kaplan‐Meier and log‐rank tests were used for the overall survival analysis. P < .05 was considered to be statistically significant.
3. RESULTS
3.1. Elevated SNRPA expression in GC and its association with GC development
SNRPA expression was initially examined by immunohistochemical staining in a GC tissue microarray with specific antibody against SNRPA. As shown in Figure 1A,B, SNRPA staining was stronger in cancer tissue cohort than that of normal gastric mucosa tissue cohort. In a total of 80 paired of clinical samples, 51/80 (63.8%) of cancer tissues exhibited an upregulated SNRPA expression compared with corresponding adjacent non‐cancerous tissues. Additionally, according to the staining intensity of cancer tissues, we divided the 89 patients with available clinical information into high SNRPA (n = 52) and low SNRPA (n = 37) groups, after which a series of clinicopathological features were statistically compared between the 2 groups (Table 1). The results revealed that SNRPA expression was positively correlated with tumour size, T classification and JACC classification. Importantly, Kaplan‐Meier overall survival analysis demonstrated that the patients with higher SNRPA expression have a significantly shorter overall survival time than those with low SNRPA expression (Figure 1C), indicating that high expression of SNRPA was closely associated with poor outcomes of GC patients, and these findings were consistent with the Kaplan‐Meier overall survival and disease‐free survival analysis obtained from Kaplan‐Meier Plotter Dataset (Figure S1). Taken together, these data suggested that SNRPA expression was increased in GC and was associated with larger tumour size, more advanced pathological stages and worse prognosis.
Figure 1.
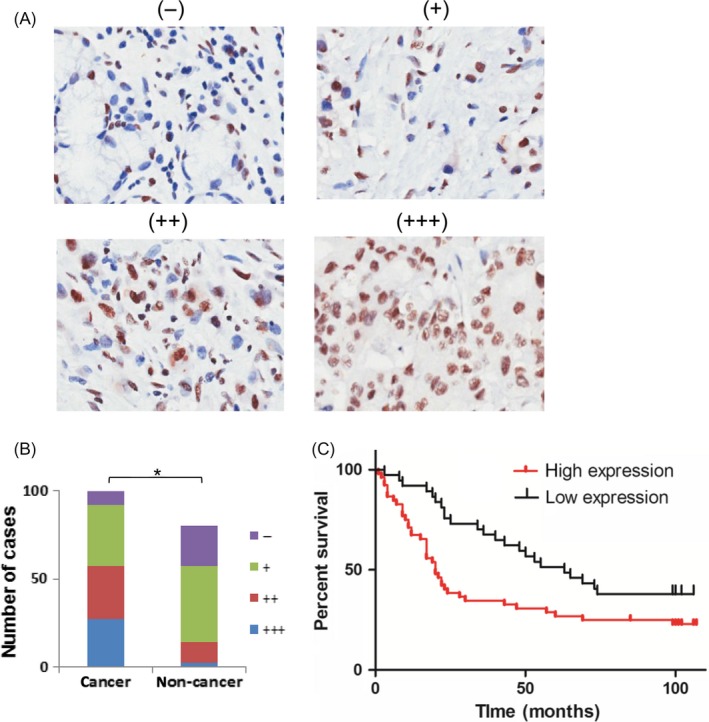
SNRPA expression is correlated with GC prognosis. A, The protein expression of SNRPA was performed with immunohistochemical staining on a GC tissue microarray. (−), representative sections of negative staining; (+), slight positive; (++), moderate positive; (+++), strong positive. Magnification: x200. B, The positive ratio of SNRPA staining in cancer and non‐cancer tissues. *P < .01. C, Overall survival plot comparing patients demonstrating high SNRPA expression (red line; n = 37) and low SNRPA expression in tumours (black line; n = 52; P = .0136, Log‐rank test)
Table 1.
The correlation of SNRPA expression with clinicopathological features of gastric cancer
| Number of cases | P value | ||
|---|---|---|---|
| Strong staining | Weak staining | ||
| Age (y) | |||
| >60 | 39 | 22 | .120 |
| ≤60 | 13 | 15 | |
| Gender | |||
| Male | 31 | 24 | .615 |
| Female | 21 | 13 | |
| Tumour size | |||
| >5 | 30 | 11 | .009 |
| ≤5 | 22 | 26 | |
| Differentiation | |||
| Well‐differentiated | 21 | 13 | .615 |
| Poorly differentiated | 31 | 24 | |
| T classification | |||
| T1, T2 | 6 | 13 | .007 |
| T3, T4 | 46 | 24 | |
| N classification | |||
| N0 | 12 | 13 | .212 |
| N1‐3 | 40 | 24 | |
| M classification | |||
| M0 | 46 | 36 | .127 |
| M1 | 6 | 1 | |
| JACC classification | |||
| I, II | 16 | 24 | .001 |
| III, IV | 36 | 13 | |
3.2. Knockdown of SNRPA inhibits cell proliferation of GC cells in vitro
To investigate the role of SNRPA in GC, we first detected the expression levels of SNRPA in 5 kinds of GC cell lines. The western blot data showed that SNRPA was highly expressed in BGC823 and AGS cells (Figure 2A). Therefore, the 2 cell lines were employed for knockdown experiments whereas SGC7901 and MGC803 cells were employed for overexpression experiments due to the relative low expression of SNRPA. Two stable cell lines with lentivirus‐mediated SNRPA silencing were generated in BGC823 and AGS cells. The efficacy of SNRPA knockdown was confirmed by western blot analysis (Figure 2B). CCK‐8 assay was used to determine the effect of SNRPA knockdown on GC cell proliferation. The results indicated that SNRPA knockdown significantly slowed cell growth relative to control cells (Figure 2B), Consistently, SNRPA silenced cells formed fewer and smaller colonies in culture plate and soft agar (Figure 2C,D), revealing that SNRPA knockdown also inhibited anchorage‐dependent and ‐independent cell growth. These findings collectively suggested that SNRPA knockdown significantly inhibits cell proliferation of GC cells in vitro.
Figure 2.
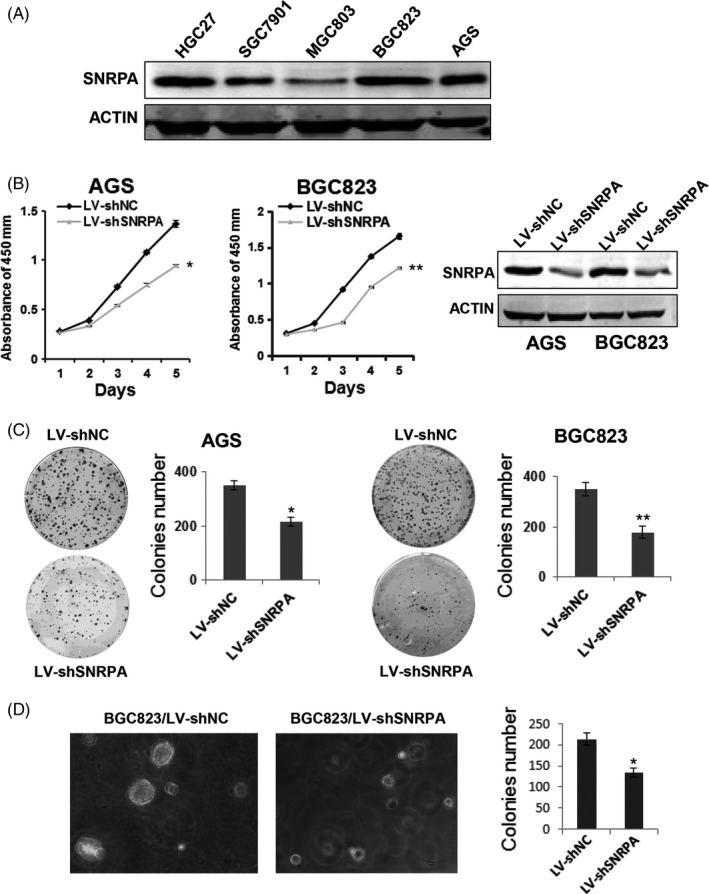
SNRPA depletion inhibits GC cell growth. A, The expression of SNRPA was evaluated by western blot in 5 gastric cancer cell lines. B, The effects of SNRPA knockdown on cell proliferation in AGS and BGC823 cells. The knockdown efficiency of SNRPA was determined by western blotting. C, The effects of SNRPA knockdown on colony formation capacity of AGS and BGC823 cells. D, Soft agar assays was performed in BGC823 cells. Cell colonies were photographed and counted after 2 wk. All the above experiments were repeated at least 3 times. The data represent mean ± SD of 3 independent experiments. *P < .05, **P < .01
3.3. Overexpression of SNRPA enhances cell growth in GC
To determine whether SNRPA overexpression promotes cell growth in GC, we generated 2 stable cell lines by infecting MGC803 and SGC7901 cells with SNRPA overexpressed lentivirus. The effect of SNRPA overexpression was verified by western blot analysis (Figure 3A). CCK‐8 assay and colony formation assay data demonstrated SNRPA accelerated the cell growth rates of MGC803 and SGC7901 cells (Figure 3A,B). Moreover, the cell growth was also elucidated by EdU incorporation assay. As expected, overexpression of SNRPA in MGC803 and SGC7901 cells resulted in a higher percentage of EdU‐positive cells (Figure 3C), suggesting that cells with overexpression of SNRPA grew faster than control groups.
Figure 3.
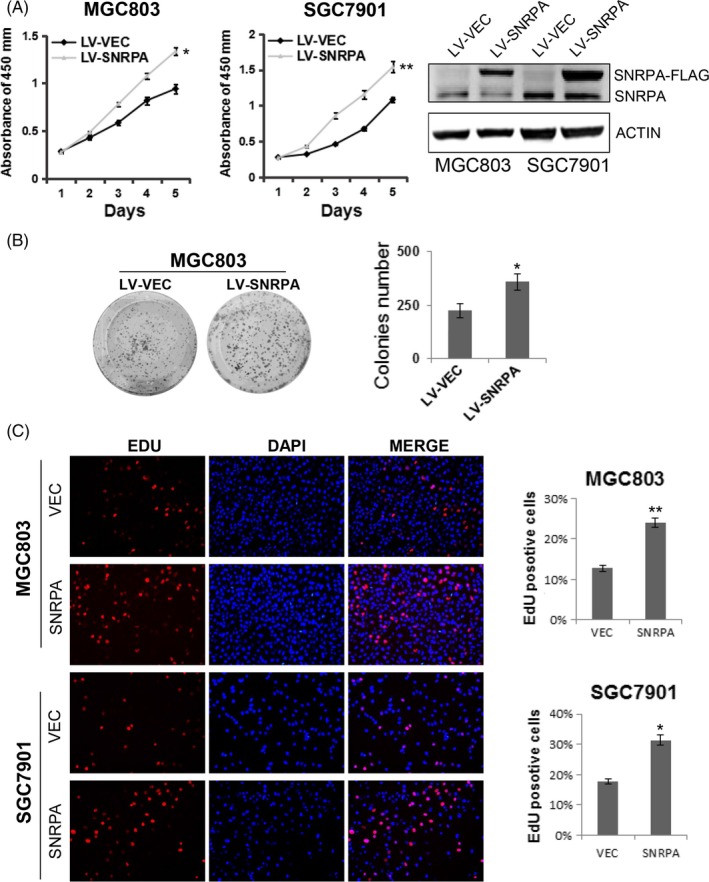
SNRPA enhances GC cell proliferation. A, Cell growth was examined by CCK8 method in SNRPA overexpressed SGC7901 and MGC803 cells. Western blot analysis was used to detect SNRPA expression in the 2 cell lines. B, The colony formation capacity of MGC803 cells was evaluated upon SNRPA overexpression. The above experiments were repeated at least 3 times. C, EdU incorporation assay was used to measure cell proliferation in MGC803/LV‐shSNRPA, SGC7901/LV‐shSNRPA cells and their corresponding control cells. Data represented as mean ± SD deviation of 3 independent experiments. *P < .05, **P < .01
3.4. SNRPA promotes tumour growth in a xenograft model
To investigate the role of SNRPA on tumourigenicity, the nude mice xenograft model was employed. BGC823‐LV‐shSNRPA and control cells were injected subcutaneously into 2 separate groups of nude mice (n = 7), respectively. The mice were euthanized and tumour weights were measured after 1 month. As shown in Figure 4A, SNRPA knockdown significantly suppressed the tumourigenicity of BGC823 cells whether in tumour size or tumour weight. On the contrary, SNRPA overexpression promoted the tumourigenicity of MGC803 cells in another tumour xenograft assay (Figure 4B, n = 5), implying that SNRPA is strongly associated with the proliferation capacity of GC cells in vivo.
Figure 4.
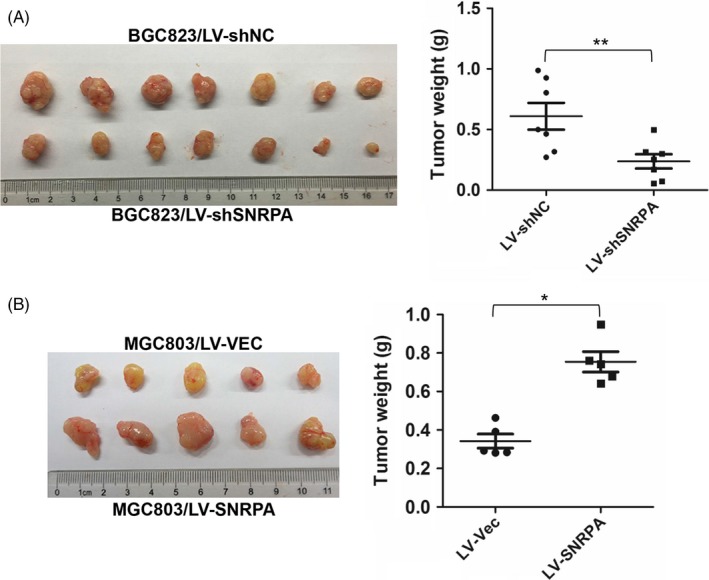
SNRPA facilitates the tumourigenicity of GC cell in nude mice. A, The impact of SNRPA knockdown on tumourigenicity of BGC823 cells. B, The impact of SNRPA overexpression on tumourigenicity of MGC803 cells. Animals were sacrificed and tumour tissues were collected and photographed, tumour weight was measured at the end of the experiment. Data are expressed as mean ± SD. *P < .05, **P < .01
3.5. NGF acts as a downstream target of SNRPA in GC cell growth
To figure out the molecular mechanism of how SNRPA promotes GC cell growth, we performed the genome‐wide transcriptome profile in SNRPA knockdown cells by Agilent Xenopus 4 × 44K Gene Expression Microarray. Compared to control cells, 414 genes were upregulated and 522 genes were downregulated in BGC823‐LV‐shSNRPA cells. Forty‐five genes whose Fold‐change was over 2.5 were shown in Figure 5A. Three upregulated genes (STAB 2, FAP and RAB17) and 5 downregulated genes (HSPG2, LEF1, NGF, NOTCH3 and RREB1) were chosen for further verification by quantitative real‐time PCR. The PCR results were consistent with the microarray data except STAB 1 gene (Figure 5B). However, when these genes were examined in MGC803 cells, only NGF, the nerve growth factor, was upregulated with SNRPA overexpression (Figure 5B), suggesting NGF might be a potential downstream effector of SNRPA. Further studies revealed that SNRPA had a positive effect on NGF expression in mRNA level (Figure 5C) and in protein level (Figure 5D), demonstrating that NGF was regulated by SNRPA.
Figure 5.
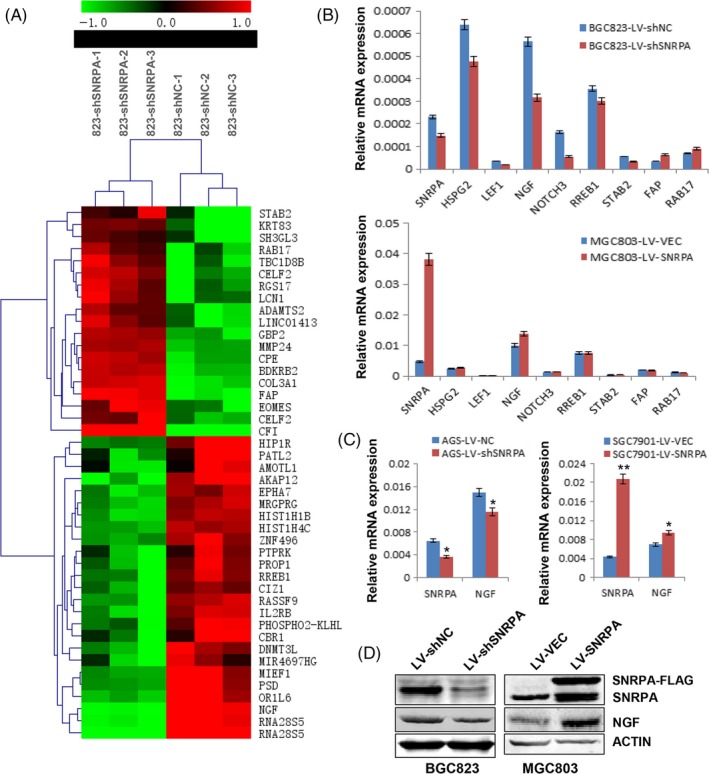
Target gene identification by global microarray analysis and effects of SNRPA on the expression of NGF in GC cells. A, The heatmap of differentially expressed genes which Fold‐change >2.5. Row represents gene, column represents experimental cells as indicated. Upregulated genes are shown in red and downregulated genes in green. B, Verifications of SNRPA target genes in SNRPA silenced BGC823 cells and SNRPA overexpressed MGC803 cells. C, The expression of NGF was evaluated in AGS, SGC7901 and their corresponding control cells by real‐time PCR in which SNRPA was silenced (AGS) or overexpressed (SGC7901). D, The protein expression of NGF was detected by western blot in BGC823 or MGC803 cells. SNRPA has a positive effect on NGF expression. The PCR data are expressed as mean ± SD. *P < .05, **P < .01
To explore the functional relevance of SNRPA and NGF, the si‐NGF was transiently transfected into SGC7901 and MGC803 cells, respectively. The results of quantitative real‐time PCR and western blot analysis indicated that NGF was successfully downregulated in these cells (Figure 6A,B). Importantly, we found Cyclin D1 was also decreased with NGF knockdown (Figure 6C). Cell growth curves by CCK‐8 method showed that cells transfected with si‐NGF grew much slower than those cells transfected with si‐NC (Figure 6D), consistent with previous report that NGF acts as an oncogene in tumour.19 Meanwhile, cell viability assays indicated that NGF depletion attenuated the promotion of SNRPA overexpression on GC cell growth (Figure 6E), suggesting that NGF may serve as a downstream effect of SNRPA on tumour cell growth in GC.
Figure 6.
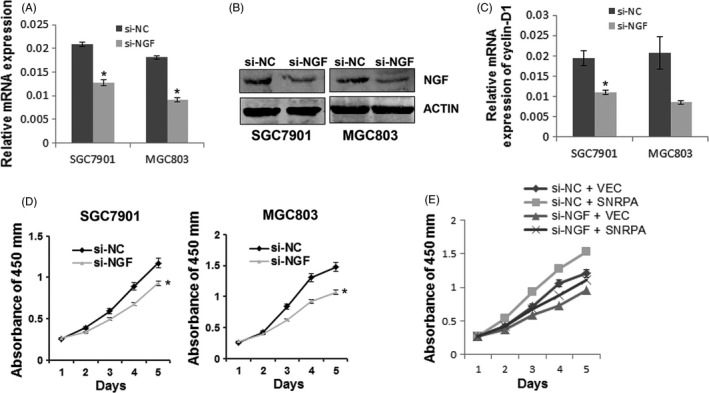
NGF depletion reduces the effect of SNRPA overexpression on cell growth. The knockdown efficiency of NGF was examined by quantitative RT‐PCR (A) and Western blotting (B). C, Cyclin D1 expression was examined by quantitative RT‐PCR in NGF knocked down cells as indicated. D, The effects of NGF knockdown on cell growth in SGC7901 and MGC803 cells. E, The effects of SNRPA overexpression on cell growth in NGF‐silenced MGC803 cells. All above experiments were repeated at least 3 times. *P < .05, **P < .01
4. DISCUSSION
Previous studies have reported that the U1 snRNP complex is involved in a lot of essential activities in eukaryotic cells, such as pre‐mRNA splicing and apoptosis.20, 21 Among its components, in addition to their splicing roles, U1 snRNA molecule is a major target of autoimmunity in SLE‐overlap syndromes,22 U1‐70K was shown to be highly associated with apoptosis.23 However, the roles of U1 snRNP complex and its components in tumourigenesis remain unclear. Our study is the first demonstrating that SNRPA, as a member of U1 snRNP complex, contributes to gastric cancer progression and indicates poor prognosis. We found that SNRPA protein is highly expressed in GC tumours and its high expression is strongly associated with larger tumour size, more advanced tumour stage and worse prognosis of patients with GC. Importantly, the results of cell viability assays, colony formation assays, EdU incorporation assays and the nude mice xenograft model support the oncogenic role of SNRPA in GC by promoting cell proliferation.
Furthermore, through gene expressing profiles we compared changes in gene expression after silencing of SNRPA in GC cells and finally identified NGF as a potential target gene of SNRPA. The nerve growth factor (NGF) is one of the members of neurotrophin families. NGF has 2 different binding receptors: the p75 neurotrophin receptor (p75NTR) and tropomyosin‐related kinase A (TrkA). When binding to TrkA, NGF can promote cell proliferation and metastasis.24, 25, 26 On the other hand, signalling via p75NTR can result in an opposite effect. Studies have revealed that NGF activates c‐Jun N‐terminal kinase (JNK) and NF‐κB pathways to promote apoptosis and cell survival.27, 28 NGF is not only discovered in nervous system, but also detected in a variety of normal and neoplastic human tissues.29 Recently, there have been many researches about the role of NGF in tumour progression. It has been reported that early, sustained sequestration of NGF reduces tumour‐induced bone destruction and reduces weight loss in animals with bone cancer.30 Qing‐li LU et al demonstrated that NGF is highly expressed in NSCLC tissues compared with para‐cancerous lung tissues.31 Another recent study indicated that NGF promotes the development of new blood vessels via interacting with α9β1 integrin.32 NGF has also been shown to promote the EMT process of tumour cells, migration and invasion in pancreatic cancer through the activation of the ERK/CD133 signal cascade and promote gastric cancer tumourigenesis via aberrant cholinergic signalling.33
In our case, we found that overexpression of SNRPA increased NGF level and knockdown of SNRPA decreased NGF level. Functionally, NGF knockdown inhibited GC cell growth, in accordance with previous report. More importantly, NGF knockdown attenuated the effect of SNRPA overexpression on the proliferation ability of GC cells. These results may imply that NGF serves as a potential downstream target of SNRPA in human GC. Further study is needed to be done in future in order to reveal the molecular mechanism by which SNRPA regulates the NGF expression in GC cells.
In conclusion, we identified SNRPA as a potential oncogene in GC, which is frequently elevated in tumour tissues and promotes the proliferation ability of GC cells in vitro and in vivo. Our work provided new insights into the treatment strategies against GC and a valuable biomarker for GC diagnosis and prognosis in the future.
CONFLICT OF INTEREST
The authors disclose no potential conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by grants from National Natural Science Foundation of China (81772568, 81772567 and 81472576) and Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2017‐13).
Dou N, Yang D, Yu S, Wu B, Gao Y, Li Y. SNRPA enhances tumour cell growth in gastric cancer through modulating NGF expression. Cell Prolif. 2018;51:e12484 10.1111/cpr.12484
Ning Dou and Dong Yang are the authors who contributed equally to this work.
Contributor Information
Yong Gao, Email: gaoyon@hotmail.com.
Yandong Li, Email: yandongli2009@gmail.com.
REFERENCES
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115‐132. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 3. Krausova M, Stanek D. snRNP proteins in health and disease. Semin Cell Dev Biol. 2017;79:92‐102. [DOI] [PubMed] [Google Scholar]
- 4. Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805‐815. [DOI] [PubMed] [Google Scholar]
- 6. Guiro J, O'Reilly D. Insights into the U1 small nuclear ribonucleoprotein complex superfamily. Wiley Interdiscip Rev RNA. 2015;6:79‐92. [DOI] [PubMed] [Google Scholar]
- 7. Scherly D, Kambach C, Boelens W, van Venrooij WJ, Mattaj IW. Conserved amino acid residues within and outside of the N‐terminal ribonucleoprotein motif of U1A small nuclear ribonucleoprotein involved in U1 RNA binding. J Mol Biol. 1991;219:577‐584. [DOI] [PubMed] [Google Scholar]
- 8. Kim SY, Kim K, Hwang B, et al. The high frequency of the U2AF1 S34Y mutation and its association with isolated trisomy 8 in myelodysplastic syndrome in Asians, but not in Caucasians. Leuk Res. 2017;61:96‐103. [DOI] [PubMed] [Google Scholar]
- 9. Zhang J, Lieu YK, Ali AM, et al. Disease‐associated mutation in SRSF2 misregulates splicing by altering RNA‐binding affinities. Proc Natl Acad Sci USA. 2015;112:E4726‐E4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin S, Su H, Tran NT, et al. Splicing factor SF3B1K700E mutant dysregulates erythroid differentiation via aberrant alternative splicing of transcription factor TAL1. PLoS ONE. 2017;12:e0175523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prochazka L, Tesarik R, Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014;26:2234‐2239. [DOI] [PubMed] [Google Scholar]
- 12. Pentheroudakis G, Kotoula V, Kouvatseas G, et al. Association of VEGF‐A splice variant mRNA expression with outcome in bevacizumab‐treated patients with metastatic breast cancer. Clin Breast Cancer. 2014;14:330‐338. [DOI] [PubMed] [Google Scholar]
- 13. Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”‐U2A’ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645‐650. [DOI] [PubMed] [Google Scholar]
- 14. Gunderson SI, Polycarpou‐Schwarz M, Mattaj IW. U1 snRNP inhibits pre‐mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255‐264. [DOI] [PubMed] [Google Scholar]
- 15. Vagner S, Ruegsegger U, Gunderson SI, Keller W, Mattaj IW. Position‐dependent inhibition of the cleavage step of pre‐mRNA 3′‐end processing by U1 snRNP. RNA. 2000;6:178‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. An QC, Liu GY. Molecular cloning, sequence identification, and tissue expression profile analysis of three novel porcine genes: SDHB, SNRPA and CRYBB1. Mol Biol Rep. 2009;36:683‐690. [DOI] [PubMed] [Google Scholar]
- 17. Hieda M, Tachibana T, Fukumoto M, Yoneda Y. Nuclear import of the U1A splicesome protein is mediated by importin alpha/beta and Ran in living mammalian cells. J Biol Chem. 2015;290:9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y, Zhu X, Zhu J, et al. Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol Rep. 2007;18:943‐951. [PubMed] [Google Scholar]
- 19. Urzua U, Tapia V, Geraldo MP, Selman A, Vega M, Romero C. Nerve growth factor stimulates cellular proliferation of human epithelial ovarian cancer. Hormone Metab Res. 2012;44:656‐661. [DOI] [PubMed] [Google Scholar]
- 20. Degen WG, Aarssen Y, Pruijn GJ, Utz PJ, van Venrooij WJ. The fate of U1 snRNP during anti‐Fas induced apoptosis: specific cleavage of the U1 snRNA molecule. Cell Death Differ. 2000;7:70‐79. [DOI] [PubMed] [Google Scholar]
- 21. Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539‐542. [DOI] [PubMed] [Google Scholar]
- 22. Satoh M, Vazquez‐Del Mercado M, Krzyszczak ME, et al. Coexistence of anti‐RNA polymerase III and anti‐U1RNP antibodies in patients with systemic lupus erythematosus: two cases without features of scleroderma. Lupus. 2012;21:68‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dieker J, Cisterna B, Monneaux F, et al. Apoptosis‐linked changes in the phosphorylation status and subcellular localization of the spliceosomal autoantigen U1‐70K. Cell Death Differ. 2008;15:793‐804. [DOI] [PubMed] [Google Scholar]
- 24. Johnson TV, Bull ND, Martin KR. Neurotrophic factor delivery as a protective treatment for glaucoma. Exp Eye Res. 2011;93:196‐203. [DOI] [PubMed] [Google Scholar]
- 25. Masoudi R, Ioannou MS, Coughlin MD, et al. Biological activity of nerve growth factor precursor is dependent upon relative levels of its receptors. J Biol Chem. 2009;284:18424‐18433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hellebrand EE, Varbiro G. Development of mitochondrial permeability transition inhibitory agents: a novel drug target. Drug Discov Ther. 2010;4:54‐61. [PubMed] [Google Scholar]
- 27. Nykjaer A, Willnow TE, Petersen CM. p75NTR–live or let die. Curr Opin Neurobiol. 2005;15:49‐57. [DOI] [PubMed] [Google Scholar]
- 28. Nykjaer A, Willnow TE. Sortilin: a receptor to regulate neuronal viability and function. Trends Neurosci. 2012;35:261‐270. [DOI] [PubMed] [Google Scholar]
- 29. MacGrogan D, Saint‐Andre JP, Dicou E. Expression of nerve growth factor and nerve growth factor receptor genes in human tissues and in prostatic adenocarcinoma cell lines. J Neurochem. 1992;59:1381‐1391. [DOI] [PubMed] [Google Scholar]
- 30. McCaffrey G, Thompson ML, Majuta L, et al. NGF blockade at early times during bone cancer development attenuates bone destruction and increases limb use. Can Res. 2014;74:7014‐7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu QL, Liu J, Zhu XL, Xu WJ. Expression of nerve growth factor and hypoxia inducible factor‐1alpha and its correlation with angiogenesis in non‐small cell lung cancer. J Huazhong Univ Sci Technolog Med Sci. 2014;34:359‐362. [DOI] [PubMed] [Google Scholar]
- 32. Walsh EM, Kim R, Del Valle L, et al. Importance of interaction between nerve growth factor and alpha9beta1 integrin in glial tumor angiogenesis. Neuro‐oncology. 2012;14:890‐901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayakawa Y, Sakitani K, Konishi M, et al. Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell. 2017;31:21‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


