Abstract
Objectives
Accumulating evidence demonstrated that the long noncoding RNA (lncRNA) HOTAIR (Hox transcript antisense intergenic RNA) plays key role in renal cell carcinoma (RCC) malignancy, while microRNA‐124 (miR‐124) is a tumour suppressor in RCC. The aim of this work was to assess the biological function of HOTAIR and to explore underlying mechanism involved in HOTAIR/miR‐124/alpha‐2, 8‐sialyltransferase 4 (ST8SIA4) axis‐regulated progression in RCC.
Materials and methods
Real‐time PCR analyses and western blots were performed to the levels of HOTAIR, miR‐124 and ST8SIA4 expression in human RCC tissues and RCC cell lines (ACHN and 786‐O). Bioinformatics analysis and dual‐luciferase reporter assay were used to illustrate relationship between HOTAIR and miR‐124 in RCC. Colony formation assays, EdU assays, Ki67 assays and apoptosis assays were taken to evaluate cell proliferation. Tumour xenograft was created to explore the functions of HOTAIR and ST8SIA4 in tumorigenesis in vivo. Migration assays, invasion assays and cell adhesion assays and were also taken to analyse the carcinoma progression.
Results
In this study, HOTAIR level was confirmed to be significantly upregulated in RCC samples and RCC cell lines compared with those in the paired adjacent tissues and normal renal cell line. Overexpression of HOTAIR promoted the capability of proliferation, migration and invasion in RCC cell lines. HOTAIR directly bound to miR‐124, while miR‐124 mediated the expression of ST8SIA4 in RCC cell lines. ST8SIA4 was upregulated in RCC tissues and RCC cell lines. Ectopic expression of ST8SIA4 modulated the proliferation, migration and invasion of RCC cells. Further results indicated that HOTAIR promoted the proliferation and metastasis as a competing endogenous RNA to regulate ST8SIA4 expression by sponging miR‐124 in RCC.
Conclusions
Our results demonstrated that HOTAIR mediated RCC progression in part through miR‐124/ST8SIA4 axis, which functioned as a new prognostic biomarker in RCC.
1. INTRODUCTION
Renal cell carcinoma (RCC) is one of the 10 most common cancers.1, 2 About 25%‐30% RCC patients have distant metastases at initial diagnosis, and a third of the patients undergoing potentially curative nephrectomy will eventually develop metastases.3, 4 Although several clinical parameters and surgical operations have been developed, they lack accuracy and foreseeability in early RCC.5 Therefore, it is urgently needed to develop efficient and reliable molecular biomarkers to predict RCC progression.6
Noncoding RNAs (ncRNAs) are emerging classes of regulatory RNA that play key roles in various cellular and physiological processes such as in gene regulation, cell differentiation and development.7, 8 LncRNAs, a class of noncoding RNAs, exert critical effects on cancer progression. MicroRNAs (miRNAs) are a class of small noncoding RNA molecule and modulate gene transcription by inhibiting translation or inducing mRNA degradation.9 Kinds of relationship among lncRNAs, miRNAs and mRNA have been studying extensively. LncRNA HNF1A‐AS1 mediated repression of miR‐34a/SIRT1/p53 feedback loop promoted the metastatic progression of colon cancer by functioning as a ceRNA.10 HOTAIR regulates cyclin J expression via inhibition of miR‐205 expression in bladder cancer.11 Accumulating evidence has shown that HOTAIR is associated with the poor prognosis of many cancers and promotes tumour cell invasion, metastasis in breast, pancreatic, hepatocellular carcinomas and renal cell carcinoma.10, 11, 12 miR‐124 inhibits bone metastasis of breast cancer by repressing Interleukin‐11.13 miR‐124 inhibits aggressive behaviour of RCC.14 However, the miR‐124 sponge role of HOTAIR in RCC remains largely undefined.
Glycosylation, which can have a profound structural and functional effect on the conjugation, is the most abundant and complex protein modification.15 Sialic acids are a family of 9‐carbon containing acidic monosaccharides that are found in terminal positions of N‐ and O‐linked glycans of glycoproteins or glycolipids.16 Moreover, glycan sialylation is governed by sialidases and sialyltransferases (STs), which transfer sialic acids to the terminal position of glycans.17 Increasing studies reveal that STs take part in cancer progression and perform vital role.ST3Gal I is an independent adverse prognostic factor for recurrence and survival of patients in ccRCC.18 ST6GalNAcVI involved in the synthesis of DSGG is suppressed during the malignant transformation of the proximal tubules in the kidney.19 Although STs have an important role in RCC progression, investigating the regulation of sialylation by HOTAIR and miR‐124 remain unknown.
In the present study, we investigated HOTAIR expression and explored its biological functions in RCC cell lines. HOTAIR promoted the progression of RCC through ST8SIA4 by sponging miR‐124. Furthermore, the underlying mechanism involved in HOTAIR/miR‐124/ST8SIA4 axis‐regulated progression in RCC was explored.
2. MATERIALS AND METHODS
2.1. Cell culture
Human RCC cell lines ACHN and 786‐O were purchased from KeyGEN Company (Nanjing, China). ACHN cells was maintained in Modified Eagle's medium (MEM) supplemented with 10% foetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin‐streptomycin (HyClone, Logan, Utah, USA). 786‐O cells were cultured in RPMI‐1640 supplemented with 10% foetal bovine serum (FBS) and 1% penicillin‐streptomycin in a humidified atmosphere of 5% CO2 maintained at 37°C.
2.2. Tissue samples of patients
Thirty pairs of RCC tissues and their adjacent nontumour tissues were collected from patients who underwent surgical resections at the First Affiliated Hospital of Dalian Medical University from January 2016 to September 2017 after obtaining informed consent. The investigation project and the informed consent have been approved by the Ethics Committee of the First Affiliated Hospital of Dalian Medical University (Ethics References No: YJ‐KY‐FB‐2016‐37). The extracted specimens were confirmed to be ccRCC tissues with pathological diagnosis according to the International Union against Cancer (UICC). The specimens were immediately frozen in liquid nitrogen and stored at −80°C for analysis.
2.3. RNA isolation and quantitative real‐time PCR analysis
Total RNA from tissue specimen and from cell lines was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Real‐time PCR analyses were performed using SYBR‐Green‐quantitative real‐time PCR Master Mix kit (Toyobo Co., Osaka, Japan). The mirVanaTM qRT‐PCR microRNA Detection Kit (Ambion Inc., Austin, TX, USA) was used for miRNA detection according to manufacturer's instructions. LncRNA‐HOTAIR and mRNA expression data were normalized to GAPDH. The sequences of the upstream and downstream primers were as follows: 5′‐AGCCCTAGCCTTTGGAAGCT‐3′ (F) and 5′‐ACCCATGTGTCTCAAGATGCATT‐3′ (R) for HOTAIR, 5′‐TCCGTCATTGAGA CTTATTCAT‐3′(F); and 5′‐CACATTTAATGTTTTGAATTCT‐3′(R) for ST8SIA4, 5′‐ACCACAGTCCATGCCATCAC‐3′ (F) and 5′‐TCCACCACCCTGTTGCTGTA‐3′ (R) for GAPDH. The 2−△△Ct method was used for quantification and fold change for target genes was normalized by internal control. Each experiment was repeated three times.
2.4. Immunohistochemistry staining
The xenograft tumours were immediately fixed in 10% formaldehyde, dehydrated series of alcohol and then embedded in paraffin. Thick slices of 0.5 mm were cut, dried deparaffinized, rehydrated and then put on the IHC slides. Then tissue slices were deparaffinized and rehydrated. After washing with phosphate‐buffered saline (PBS), the tissue slices were incubated with Ki67 antibody (Abcam, Cambridge, UK) at a dilution of 1:1000 at 4°C overnight. The slices were counterstained with haematoxylin for 30 seconds and sealed with Neutral gum. Images were obtained from Olympus light microscope (20×10). The results of the experiments were analysed by software Image‐Pro plus 6.0 (Media Cybernetics, Bethesda, MD, USA).
2.5. Luciferase assay
About 5.0 × 104 human HEK293T cells in a 24‐well plate were cotransfected with 20 μm miR‐NC or miR‐124, 1 μg of luciferase reporter comprising 3′UTR of ST8SIA4, wild‐type or mutant HOTAIR fragment (Promega, Madison, WI, USA) using Lipofectamie 2000 (Invitrogen). HOTAIR wild sequence was CCAAAGAGUCUGAUGUUUACA and a mutant HOTAIR expression plasmid was changed to CCAAAGAGUCUGAUCUCUCCG constructed with the four mismatches of sequence in the putative target site of HOTAIR in pmirGLO plasmid (Promega). Luciferase activity was assayed 48 hours after transfection, using a Dual‐Luciferase reporter assay system (Promega). Transfection was repeated in triplicate.
2.6. Fluorescent in situ hybridization
The RCC and adjacent tissues were fixed in 10% formaldehyde, dehydrated series of alcohol and then embedded in paraffin. 0.5 mm thick slices were cut, dewaxed and rehydration. The samples were digested with proteinase K for 15 minutes in 37°C, denatured with formamide. The cells were incubated with the Cy3‐labelled HOTAIR probes and FITC‐labelled miR‐124 probes and incubated at 37°C overnight, washed 3 times with 2 × SSC for 3 minutes. DAPI (4′,6‐ diamidino‐2‐phenylindole) was used to counterstain the nuclei with 20 minutes in darkness. The sample was scanned and photographed under Olympus microscope.
2.7. Western blot
Whole RCC cell proteins were electrophoresed under the conditions in SDS‐PAGE polyacrylamide gels. The separated proteins were transferred to apolyvinylidenedifluoride membrane. Nonspecific binding was blocked with 5% skimmed milk for 2 hours at 37°C. The membrane was then incubated with PARP (1:1000 diluted; ThermoFisher Scientific, MA, USA), caspase 3 (1:1000 diluted; ThermoFisher Scientific), ST8SIA4 (1:500 diluted; ThermoFisher Scientific), at 4°C overnight. A GAPDH antibody (1:2500 diluted; Bioworld, Minneapolis, MN, USA) was used as a control. Next, the membrane was incubated with peroxidase‐conjugated anti‐rabbit IgG (1:5000 diluted; Thermo, Boston, MA, USA). Images were obtained from multifunctional gel imaging system (ImageQuant LAS 500, General Electric, Fairfield, CT, USA).
2.8. Transwell assays
The invasion assays were performed using 24‐well transwell chambers (8 μm; Corning, Shanghai, China) and the upper chambers were covered with 1 mg/mL Matrigel at first. Tumour cells were resuspended in serum‐free RPMI‐1640 medium and 1.0 × 104 cells were seeded into the upper chambers. About 0.5 mL RPMI‐1640 containing 10% FBS was added to the bottom chambers. After 24‐hour incubation, cells on the upper surface of the membrane were scrubbed off, and the migrated cells were fixed in 75% methanol, stained with 0.1% crystal violet with 25 minutes, and counted under a light microscope (40×10).
2.9. Xenografted tumour model
Four‐week‐old nude mice were purchased from the Model Animal Research Institute of Nanjing University. About 5.0 × 106 cells were inoculated into the right flank of every nude mouse by subcutaneous injection. When bearing palpable tumours, mice were randomly divided into six groups, each group containing six mice. Subsequently, the nude mice were injected intratumorally with HOTAIR, shHOTAIR, ST8SIA4, shST8SIA4 and their negative controls thrice a week for five weeks. The size of tumour was determined every 7 days. The nude mice were sacrificed, and tumours were isolated and weighed. Tumour volume was calculated as the following formula: (length × width2)/2.
2.10. Cell viability
The cell viability was performed by using the Cell Counting Kit‐8 (CCK‐8; KeyGEN, Nanjing, China). About 5.0 × 103/well cells were seeded into 96‐well plates with 100 μL of RPMI‐1640 medium containing 10% FBS. About 10 μL CCK8 solution was added to the plate after incubation. The absorbance was measured in a microplate reader at 450 nm (168‐1000 Model 680, Bio‐Rad). Each experiment was performed at least three times.
2.11. 5‐Ethynyl‐2′‐deoxyuridine (EdU) assay
Cell proliferation was determined by 5‐Ethynyl‐2′‐deoxyuridine assay using an KeyFluor488 Click‐iTEdU kit (KeyGEN, Nanjing, China) following the manufacturer's instructions. About 5.0 × 103 cells/well was seeded in a 96‐well plate, then transfection with vector, miR‐NC, HOTAIR, siHOTAIR, NC inhibitor, miR‐124, miR‐124 inhibitor, ST8SIA4 or siST8SIA4. After transfection, cells were incubated with EdU for 2 hours at 37°C, and the cells were fixed with 4% polyformaldehyde containing PBS. Cells were incubated with Hoechst 33342 solution. The cells were then visualized under a fluorescence microscopy (20 × 10). Experiments were repeated three times.
2.12. Colony formation assays
786‐O and ACHN transfected cells were seeded into six‐well plates at a density of 1000 cells/well and cultured in RPMI‐1640 medium containing 10% FBS for 10 days. Cell colonies were fixed with 10% formaldehyde for 40 minutes, stained with 0.1% crystal violet at room temperature for 20 minutes, and then photographed. Experiments were repeated three times.
2.13. Wound healing assay
About 4.0 × 105 ACHN or 786‐O cells were seeded in a 12‐well plate. One night after inoculation, the cell layer was scratched using a pipette tip when adherent cells were observed. Next, images of cell morphology were captured at initiation time and 24 hours under the Olympus microscope (10 × 10). The migratory ability was quantified and normalized by relative gap distance. The results of the experiments were analysed by software IPP6.0.
2.14. Apoptosis analysis
Apoptosis was assayed through dual staining with AnnexinV‐FITC/propidium iodide (PI; KeyGen BioTECH, Nanjing, China). Annexin V‐FITC and PI were added to the cellular suspension according to the manufacturer's instructions. Measurements were repeated independently three times.
2.15. Statistical analysis
Each experiment was performed at least in triplicate. Data are displayed as mean standard deviation (SD) and analysed by using spss 17.0 (SPSS Inc., Chicago, IL, USA). The significance of differences in multiple comparisons was determined using Student's t test. *P < 0.05 was considered to be statistically significant.
3. RESULTS
3.1. HOTAIR level was upregulated in RCC tissues and RCC cell lines
To evaluate correlation between HOTAIR expression and clinical characteristics, the HOTAIR expression was examined in 30 pairs of RCC tissues and their corresponding normal renal tissues by using qRT‐PCR. We found that the expression of HOTAIR was significantly higher in RCC tissues than that in normal renal tissues (Figure 1A,C). HOTAIR expression was also higher in 786‐O and ACHN cells compared within normal kidney cell HK‐2 cells (Figure 1B). In addition, upregulated HOTAIR was correlated with distant metastasis (Figure 1D). The survival data based on the expression of HOTAIR in the Cancer Genome Atlas (TCGA) showed that HOTAIR expression level affected the patient survival (FigureS1A,B). These results indicated that HOTAIR was associated with RCC progression and overexpression of HOTAIR might be useful for diagnosis and estimate of RCC.
Figure 1.
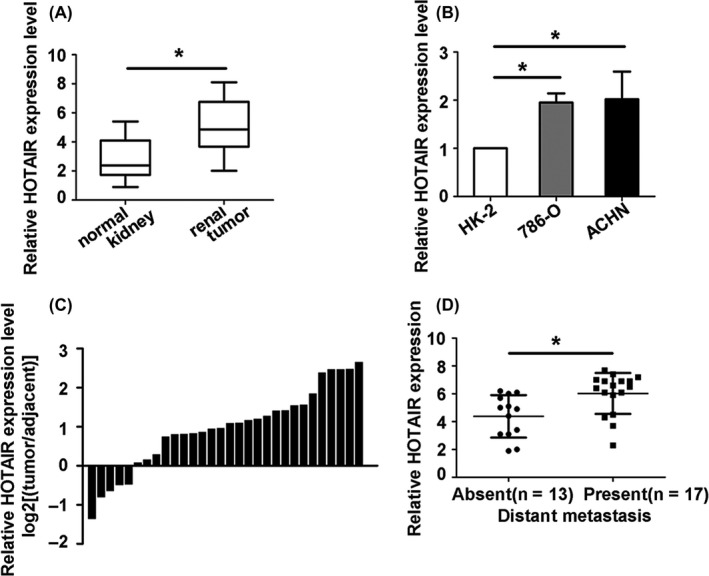
Hox transcript antisense intergenic RNA (HOTAIR) level was upregulated in RCC tissues and RCC cell lines. (A and C) Relative expression of HOATIR in RCC tissues was compared with corresponding nontumour tissues. B, HOTAIR level was examined in normal kidney cell HK2 and RCC cell lines ACHN and 786‐O. D, HOTAIR upregulated was correlated with distant metastasis. Each experiment was independently repeated at least three times. Data were presented as mean ± SD (n = 3). *P < 0.05
3.2. HOTAIR mediated cell proliferation, apoptosis, migration and invasion in RCC cell lines
To study the effect of HOTAIR on the proliferation and invasion of RCC cells, we performed proliferation‐related assays in 786‐O and ACHN cells transfected with HOTAIR and siHOTAIR. Transfection of HOTAIR affected the HOTAIR levels in RCC cells (Figure 2A). CCK‐8 assay and cell colony formation assay showed that HOTAIR significantly promoted cell proliferation (Figure 2B,C). The cells transfected with HOTAIR showed incremental level of Ki67 and EdU, whereas knockdown of HOTAIR had opposite effect on the expression (Figure 2D). Apoptosis rate was decreased after transfecting with HOTAIR, and apoptosis rate was increased after transfecting with siHOTAIR in ACHN and 786‐O cells (Figure 2E). The cell adhesion per cent was increased after transfecting with HOTAIR, while the result was opposite in siHOTAIR cells (Figure 2F,G). Wound healing assay and cell invasion assay showed that the ability of metastasis was strongly increased in RCC cells after transfecting with HOTAIR, while the ability was obviously decreased in cells transfected with siHOTAIR (Figure 3A,B).
Figure 2.
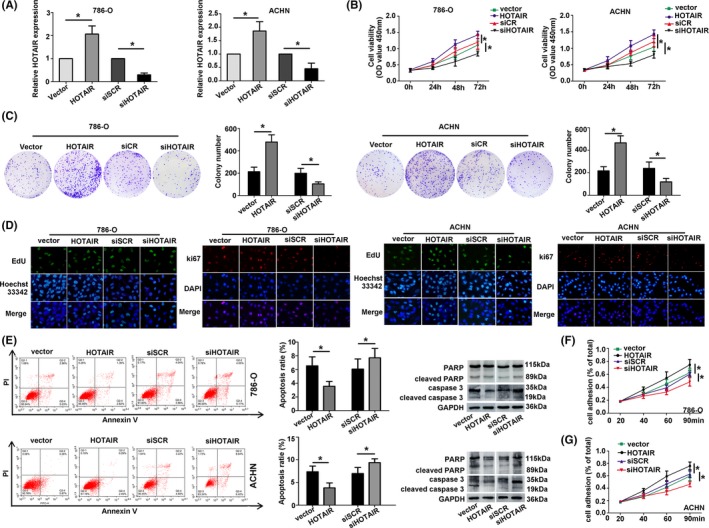
Hox transcript antisense intergenic RNA (HOTAIR) mediated RCC cell proliferation, apoptosis. A, Related HOTAIR expression was measured to verify the transfection. (B and C) Overexpressing HOTAIR significantly enhanced cell proliferation by colony formation assay and CCK‐8 assay. Immunofluorescence analysis with EdU and Ki67 D, and inhibited the apoptosis rate (E). (F and G) Cell adhesion ability was increased after transfected with HOTAIR. Data were presented as mean ± SD (n = 3). *P < 0.05
Figure 3.
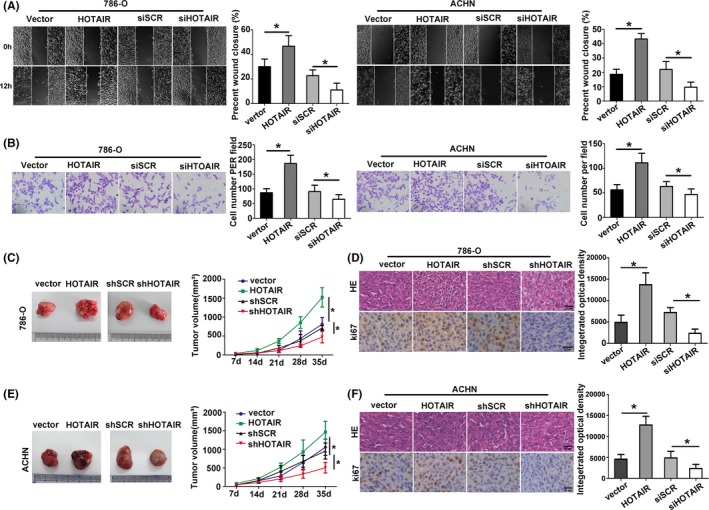
Hox transcript antisense intergenic RNA (HOTAIR) mediated RCC migration, invasion in vitro and tumorigenesis in vivo. A, Wound healing assays were performed to investigate changes in cell migration. B, Transwell assays were carried out in ACHN and 786‐O cells. C, Xenografted tumours injected with 786‐O cells were measured to evaluate the cell proliferation in vitro. D, Above: HE staining of xenografted tumours. Blow: IHC staining with ki67 antibody. E, Xenografted tumours injected with ACHN cells were measured to evaluate the cell proliferation in vitro. F, Above: HE staining of xenografted tumours. Blow: IHC staining with ki67 antibody. Each experiment was independently repeated at least three times. Data were presented as mean ± SD (A‐B n = 3, C‐F n = 6). *P < 0.05
To further confirm whether HOTAIR affected the tumorigenesis, transfected 786‐O and ACHN cell lines were inoculated into nude mice. As shown in Figure 3C,E, the tumour growth in HOTAIR group was faster than that in vector group. Furthermore, the tumour growth of group transfected with shHOTAIR was slower than that in the shSCR group. The expression of Ki67 in tumour tissues was consistent with the results in vitro (Figure 3D,F). These data indicated that overexpression of HOTAIR promoted RCC cell growth and metastasis.
3.3. miR‐124 directly bound to HOTAIR and negatively modulated the expression of ST8SIA4 in RCC cells
A bioinformation analysis was performed using miRcode algorithm (http://www.mircode.org/) and the result showed that miR‐124 contained a binding site for HOTAIR (Figure 4A). To investigate the molecular mechanism, we first determined the subcellular location of HOTAIR and miR‐124 through FISH in RCC tissues and adjacent tissues (Figure S1C). The luciferase activity was decreased when miR‐124 and wild‐type 3′‐UTR HOTAIR cotransfected into 293T cells (Figure 4B). The luciferase activity was unchangeable in mutation‐type group. To determine whether HOTAIR and miR‐124 were in the same RNA‐induced silencing complex (RISC), RNA‐binding protein immunoprecipitation (RIP) assay was performed. The levels of HOTAIR and miR‐124 were higher in anti‐Ago2 group than those in anti‐normal IgG group (Figure 4C). 786‐O cells were chosen for further investigated. The expression of miR‐124 was significantly decreased in 786‐O cells transfected with HOTAIR and increased in cells transfected with siHOTAIR (Figure 4D). In addition, a negative correlation was exhibited between HOTAIR and miR‐124 in 30 pairs of clinical RCC tissues (Figure 4E). We also found that ST8SIA4 as potential miR‐124 targeted gene by bioinformatics analysis miRcode algorithm (http://www.mircode.org/) and Target Scan Human 7.1 (http://www.targetscan.org/) (Figure 4F). Luciferase expression in cells transfected with ST8SIA4‐WT and the miR‐124 mimic was decreased than that of control cells (Figure 4G), confirming that ST8SIA4 was a direct target of miR‐124. The expression level of miR‐124 was higher in normal kidney samples and kidney normal cells than in RCC tissues and RCC cells (Figure 4H,I). Furthermore, negative correlation was observed between the expression of miR‐124 and ST8SIA4 in RCC tissues (Figure 4J). The ST8SIA4 mRNA and protein levels were mediated by miR‐124 (Figure 4K). The results indicated miR‐124 regulated HOTAIR and ST8SIA4 expression.
Figure 4.
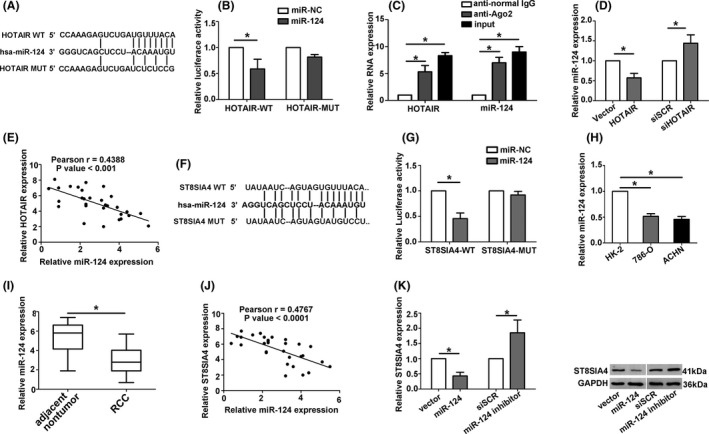
miR‐124 directly bound to HOTAIR and negatively modulated the expression of ST8SIA4 in RCC cells. A, Bioinformatics analysis predicted that HOTAIR bound to miR‐124. B, Dual‐luciferase assays indicated a significant reduction in luciferase activities after cotransfection of miR‐124 and the wild‐type HOTAIR reporter vector, but not the mutant‐type HOTAIR. C, A RIP assay was performed using input from cell lysates, anti‐normal mouse IgG or anti‐Ago2. The relative expression levels of HOTAIR and miR‐124 were detected by quantitative real‐time PCR. D, Relative miR‐124 expression was measured by qRT‐PCR in 786‐O cells transfected with ST8SIA4 or siHOTAIR. E, Pearson's correlation curve revealed negative correlation between HOTAIR and miR‐124 was found in 30 pairs of clinical RCC tissues. F, Bioinformatics analysis predicted that ST8SIA4 bound to miR‐124. G, Dual‐luciferase assays indicated a significant reduction in luciferase activities after cotransfection of miR‐124 and wild‐type ST8SIA4 reporter vector. (H and I) The relative miR‐124 expression was detected by qRT‐PCR in RCC tissues and RCC cell lines. J, A negative relationship between miR‐124 and ST8SIA4 was observed in 30 pairs of clinical RCC tissues. K, The ST8SIA4 mRNA and protein expression level were changed after cells transfected with miR‐124 or miR‐124 inhibitor. Each experiment was independently repeated at least three times. Data were presented as mean ± SD (n = 3). *P < 0.05
3.4. ST8SIA4 promoted the RCC cell proliferation and metastasis
The level of ST8SIA4 mRNA was upregulated in RCC tissues and RCC cell lines than those in adjacent kidney tissues and kidney normal cell (Figure 5A). To research the function of ST8SIA4 in RCC cell lines, the colony formation, EdU assay, wound healing and invasion assay were performed. As shown in Figure 5B, the higher level ST8SIA4 was observed in RCC tissues than that in adjacent tissues by IHC. The mRNA and protein levels of ST8SIA4 were assessed in 786‐O cells transfected with ST8SIA4 or siST8SIA4 (Figure 5C,D). The colony formation number was higher in ST8SIA4 overexpression cells, while decreased in ST8SIA4 knockdown cells (Figure 5E). EdU assay indicated that the fluorescence intensity elevated in 786‐O cells after transfecting with ST8SIA4. Cells transfected with siST8SIA4 showed the inverse results, suggesting that ST8SIA4 influenced the proliferative ability of RCC cell lines (Figure 5F).
Figure 5.
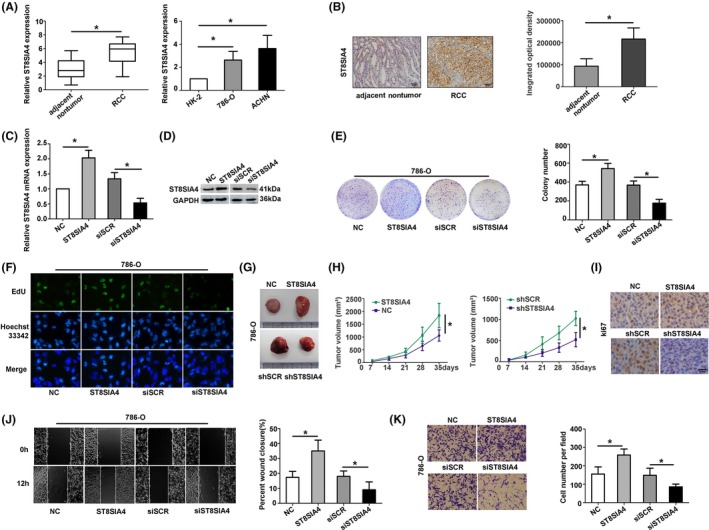
Alpha‐2,8‐sialyltransferase 4 (ST8SIA4) promotes the RCC cell proliferation and metastasis. A, The expression of ST8SIA4 was detected in RCC tissues, corresponding nontumour tissues, and RCC cell lines. B, Immunohistochemistry (IHC) staining was performed to analysis the ST8SIA4 expression in RCC tissues and corresponding nontumour tissues. (C and D) The relative levels of ST8SIA4 mRNA and protein were detected by qRT‐PCR and Western blot in 786‐O cells transfected with ST8SIA4 or siST8SIA4. (E and F) Upregulated ST8SIA4 significantly increased colony formation and immunofluorescence intensity with EdU. (G, H and I) The nude mice were injected with 786‐O cells transfected with ST8SIA4 or shST8SIA4, the xenografted tumours were measured and IHC staining was performed. (J and K) The wound healing and cell invasion assays stimulated by ectopic expression of ST8SIA4 could be repressed by knockdown of siST8SSIA4. Each experiment was independently repeated at least three times. Data were presented as mean ± SD (A‐F, J, K n = 3 and G‐I n = 6). *P < 0.05
To further study the ST8SIA4 function in vivo, 786‐O cells transfected with ST8SIA4 or shST8SIA4 were injected into nude mice. After 35 days, the mice were sacrificed. The tumours were measured (Figure 5G), and the tumours volume were recorded (Figure 5H). The group injected with ST8SIA4 presented larger tumour volume than the control group. The group transfected with ST8SIA4 showed higher integrated optical density by IHC staining (Figure 5I). The migratory and invasive ability was increased in 786‐O cell transfected with ST8SIA4, while the inverse result presented in cell transfected with siST8SIA4 (Figure 5J,K). The results revealed that ST8SIA4 functioned as an oncogene in RCC.
3.5. HOTAIR and miR‐124 mediated RCC progression through regulating ST8SIA4 expression
The level of ST8SIA4 mRNA and protein was decreased in 786‐O cells cotransfected with miR‐124 and vector, while the level was increased in cells transfected with miR‐NC and HOTAIR. The results were reversed when cells were cotransfected with miR‐124 and HOTAIR (Figure 6A,B). Knockdown of miR‐124 increased the expression of ST8SIA4. The level of ST8SIA4 was decreased in cells transfected with siHOTAIR, and the effect was reversed by transfected with siHOTAIR and miR‐124 inhibitor (Figure 6E,F).
Figure 6.
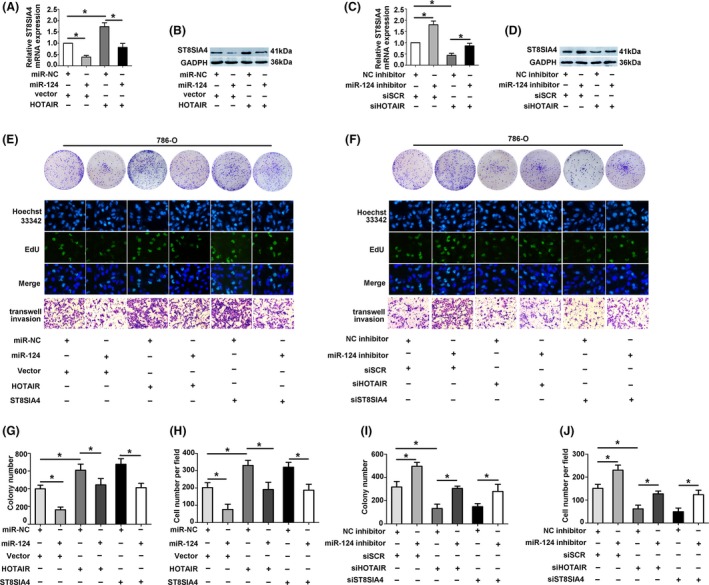
Hox transcript antisense intergenic RNA (HOTAIR) and miR‐124 mediated RCC progression through regulating ST8SIA4 expression. (A and B) The protein and mRNA expression level were measured in 786‐O cells after transfected with HOTAIR and miR‐124 by western blotting and qRT‐PCR. (C and D) The protein and mRNA expression level were measured in 786‐O cells after transfected with siHOTAIR or miR‐124 inhibitor. (E and J) Functional assays were performed to verify HOTAIR mediated the ST8SIA4 expression by competing for miR‐124. Each experiment was independently repeated at least three times. Data were presented as mean ± SD (n = 3). *P < 0.05
To figure out whether HOTAIR function in RCC cells through miR‐124, the proliferation and metastasis‐related assays were carried out. The proliferative and metastatic ability were decreased when 786‐O cells were cotransfected with vector and miR‐124. Overexpression of HOTAIR or ST8SIA4 promoted the proliferative and invasive ability, while the abilities were recovered when cells were cotransfected with HOTAIR or ST8SIA4 and miR‐124. On the contrary, suppression of miR‐124 boosted the proliferation and metastasis in 786‐O cells. Knockdown of HOTAIR or ST8SIA4 significantly inhibited the ability of proliferation and metastasis; however, the suppression could be restored by cotransfected with siHOTAIR or siST8SIA4 and miR‐124 inhibitor (Figure 6G‐J). These data suggested that HOTAIR promoted the carcinogenesis to regulate ST8SIA4 expression by sponging miR‐124 in RCC.
4. DISCUSSION
LncRNAs, which are more than 200 nucleotides in length with limited protein coding potential, are special expressed in differentiated tissues or specific cancer.20, 21, 22 Recent reports revealed that HOTAIR was regarded as an oncogene in renal cell carcinoma.23 It has been reported that high HOTAIR expression indicated a poorer prognosis in RCC; HOTAIR modulated cells proliferation and migration in vitro.24, 25 Hiromichi Katayama et al also verified that HOTAIR enhanced the metastasis of RCC cells in vivo.26 In this study, we verified that the level of HOTAIR was markedly increased in RCC tissues and RCC cell lines compared to adjacent normal kidney tissues and normal kidney cell HK‐2. The results of distant metastasis showed that HOTAIR positively associated with the aggressiveness in RCC tissues. We further demonstrated that alteration of HOTAIR mediated the proliferation, invasion and metastasis of RCC cells by series of experiments. The xenografted tumour model and IHC staining were performed to verify the proliferation ability of HOTAIR for the first time. Moreover, the location of HOTAIR and miR‐124 in RCC tissues, adjacent tissues and cell lines were first detected by dual‐FISH staining. These data suggested that HOTAIR acted as a tumour oncogene in RCC progression.
It has been reported that lncRNAs containing miRNA‐binding sites could regulate and communicate with mRNAs by competing specifically for shared miRNAs, thus acting as ceRNAs.27, 28 The ceRNAs functioned as natural miRNA sponges that reduced the binding of endogenous miRNAs to their target genes, thereby regulating gene expression. HOTAIR played essential role in cancer progression as a ceRNA.29 HOTAIR functioned as a ceRNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer.20 The level of miR‐124 was negatively related to HOTAIR expression in gastric cancers.22 In this study, we firstly found that miR‐124 directly targeted HOTAIR, and HOTAIR and miR‐124 were in the same RISC by RIP assay. MiR‐124 level was decreased when cells were transfected with HOTAIR, while the opposite result occurred in HOTAIR knockdown cells. Additionally, miR‐124 expression was inversely correlated with HOTAIR in RCC tissues. miR‐124 has been identified as a tumour suppressor gene in many malignant tumours, such as colorectal cancer, breast cancer, ovarian cancer and renal cell cancer.13, 14, 30 In this study, we revealed that ST8SIA4 was a potential target gene of miR‐124. miR‐124, as a tumour suppressor, showed lower expression in RCC tissues and RCC cell lines compared to adjacent normal kidney tissues and normal kidney cell HK‐2. Moreover, a negative correlation existed between miR‐124 and ST8SIA4 in RCC tissues. Overexpression of miR‐124 decreased ST8SIA4 mRNA and protein levels, which was upregulated by miR‐124 inhibitor. Taken together, the results indicated that HOTAIR sponged miR‐124 to direct target ST8SIA4.
Sialic acids are commonly found as the terminal monosaccharides of the glycans and play pivotal roles in many physiologically and pathologically important processes, including cancer metastasis.31, 32 STs are key enzymes in the biosynthesis of sialic acid‐containing oligosaccharides and glycoconjugates.33 It was demonstrated that increased level of ST3GalII mRNA might be involved in carcinogenesis of kidney and malignant progression of human RCC.34 ST8SIA4 mediated tumour cell metastasis by modifying the sialylation profile in breast cancer cells.35 In this study, ST8SIA4 level was markedly increased in RCC tissues and RCC cell lines compared to adjacent normal kidney tissues and normal kidney cell HK‐2. According to the function assays, ST8SIA4 promoted tumorigenesis of RCC cells in vivo and in vitro. The expression of ST8SIA4 was increased when 786‐O cells transfected with miR‐124 and the inverse results appeared in cells transfected with HOTAIR. HOTAIR/miR‐124/ST8SIA4 axis further modulated the proliferation and invasion of RCC cell lines. These data verified the hypothesis that HOTAIR functioned as a ceRNA for miR‐124 to facilitate the expression of ST8SIA4.
In summary, the research demonstrated that altered level of HOTAIR was associated with RCC progression. Our results provided new insights into the dysregulated in the development of RCC and suggested that HOTAIR/miR‐124/ST8SIA4 axis represented a potential therapeutic target for RCC.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by grants from National Natural Science Foundation of China (81772277).
Pan Y, Wu Y, Hu J, et al. Long noncoding RNA HOTAIR promotes renal cell carcinoma malignancy through alpha‐2, 8‐sialyltransferase 4 by sponging microRNA‐124. Cell Prolif. 2018;51:e12507 10.1111/cpr.12507
Yue Pan, and Yongjin Wu are the two authors contributed equally to this work.
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277‐300. [DOI] [PubMed] [Google Scholar]
- 2. Comperat E, Camparo P. Histological classification of malignant renal tumours at a time of major diagnostic and therapeutic changes. Diagn Interv Imaging. 2012;93:221‐231. [DOI] [PubMed] [Google Scholar]
- 3. Lam JS, Leppert JT, Belldegrun AS, Figlin RA. Novel approaches in the therapy of metastatic renal cell carcinoma. World J Urol. 2005;23:202‐212. [DOI] [PubMed] [Google Scholar]
- 4. Miyazaki J, Ito K, Fujita T, et al. Progression of human renal cell carcinoma via inhibition of RhoA‐ROCK Axis by PARG1. Transl Oncol. 2017;10:142‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heng DY, Xie W, Regan MM, et al. External validation and comparison with other models of the International Metastatic Renal‐Cell Carcinoma Database Consortium prognostic model: a population‐based study. Lancet Oncol. 2013;14:141‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitsui Y, Hirata H, Arichi N, et al. Inactivation of bone morphogenetic protein 2 may predict clinical outcome and poor overall survival for renal cell carcinoma through epigenetic pathways. Oncotarget. 2015;6:9577‐9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell‐cycle gene expression and proliferation in breast cancer cells. Mol Cell. 2015;59:698‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu XD. Non‐coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1:190‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amankwatia EB, Chakravarty P, Carey FA, et al. MicroRNA‐224 is associated with colorectal cancer progression and response to 5‐fluorouracil‐based chemotherapy by KRAS‐dependent and ‐independent mechanisms. Br J Cancer. 2015;112:1480‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang C, Qiu S, Sun F, et al. Long non‐coding RNA HNF1A‐AS1 mediated repression of miR‐34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50‐62. [DOI] [PubMed] [Google Scholar]
- 11. Zhang A, Zhao JC, Kim J, et al. LncRNA HOTAIR enhances the androgen‐receptor‐mediated transcriptional program and drives castration‐resistant prostate cancer. Cell Rep. 2015;13:209‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai WL, Huang WD, Li B, et al. microRNA‐124 inhibits bone metastasis of breast cancer by repressing Interleukin‐11. Mol Cancer. 2018;17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Butz H, Szabo PM, Khella HW, Nofech‐Mozes R, Patocs A, Yousef GM. miRNA‐target network reveals miR‐124 as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget. 2015;6:12543‐12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clerc F, Reiding KR, Jansen BC, Kammeijer GS, Bondt A, Wuhrer M. Human plasma protein N‐glycosylation. Glycoconj J. 2016;33:309‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Somerville RA. Host and transmissible spongiform encephalopathy agent strain control glycosylation of PrP. J Gen Virol. 1999;80:1865‐1872. [DOI] [PubMed] [Google Scholar]
- 17. Audry M, Jeanneau C, Imberty A, Harduin‐Lepers A, Delannoy P, Breton C. Current trends in the structure‐activity relationships of sialyltransferases. Glycobiology. 2011;21:716‐726. [DOI] [PubMed] [Google Scholar]
- 18. Bai Q, Liu L, Xia Y, et al. Prognostic significance of ST3GAL‐1 expression in patients with clear cell renal cell carcinoma. BMC Cancer. 2015;15:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Senda M, Ito A, Tsuchida A, et al. Identification and expression of a sialyltransferase responsible for the synthesis of disialylgalactosylgloboside in normal and malignant kidney cells: downregulation of ST6GalNAc VI in renal cancers. Biochem J. 2007;402:459‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF‐1alpha/AXL signaling through inhibition of miR‐217 in renal cell carcinoma. Cell Death Dis. 2017;8:e2772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Xia M, Yao L, Zhang Q, et al. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up‐regulating histone H3K27 demethylase JMJD3. Oncotarget. 2017;8:19795‐19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dasgupta P, Kulkarni P, Majid S, et al. MicroRNA‐203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial‐to‐mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018;17:1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katayama H, Tamai K, Shibuya R, et al. Long non‐coding RNA HOTAIR promotes cell migration by upregulating insulin growth factor‐binding protein 2 in renal cell carcinoma. Sci Rep. 2017;7:12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710‐718. [DOI] [PubMed] [Google Scholar]
- 28. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren K, Li Y, Lu H, et al. Long noncoding RNA HOTAIR controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. Transl Oncol. 2016;9:489‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park SY, Kim H, Yoon S, et al. KITENIN‐targeting microRNA‐124 suppresses colorectal cancer cell motility and tumorigenesis. Mol Ther. 2014;22:1653‐1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94:887‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baskakov IV, Katorcha E. Multifaceted role of sialylation in Prion diseases. Front Neurosci. 2016;10:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortiz‐Soto ME, Seibel J. Expression of functional human sialyltransferases ST3Gal1 and ST6Gal1 in Escherichia coli . PLoS ONE. 2016;11:e0155410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saito S, Yamashita S, Endoh M, et al. Clinical significance of ST3Gal IV expression in human renal cell carcinoma. Oncol Rep. 2002;9:1251‐1255. [PubMed] [Google Scholar]
- 35. Ma X, Dong W, Su Z, et al. Functional roles of sialylation in breast cancer progression through miR‐26a/26b targeting ST8SIA4. Cell Death Dis. 2016;7:e2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


