Abstract
Objectives
This work aimed at studying in vitro interactions between human tendon‐derived cells (hTDCs) and pre‐osteoblasts (pre‐OBs) that may trigger a cascade of events involved in enthesis regeneration.
Materials and methods
The effect of 5 osteogenic medium (OM) conditions over the modulation of hTDCs and pre‐OBs towards the tenogenic and osteogenic phenotypes, respectively, was studied. Three different medium conditions were chosen for subsequently establishing a direct co‐culture system in order to study the expression of bone, tendon and interface‐related markers.
Results
A higher matrix mineralization and ALP activity was observed in co‐cultures in the presence of OM. Higher transcription levels of bone‐ (ALPL, RUNX2, SPP1) and interface‐related genes (ACAN, COMP) were found in co‐cultures. The expression of aggrecan was influenced by the presence of OM and cell‐cell interactions occurring in co‐culture.
Conclusions
The present work assessed both the influence of OM on cell phenotype modulation and the importance of co‐culture models while promoting cell‐cell interactions and the exchange of soluble factors in triggering an interface‐like phenotype to potentially modulate enthesis regeneration.
1. INTRODUCTION
The enthesis allows a smooth transition between tendon and bone. Typically, 3 continuous regions are distinguished: tendon, non‐mineralized and mineralized fibrocartilage and bone, resulting in a gradient of composition, organization and, therefore, mechanical properties.1, 2 This soft‐hard tissue interface is vulnerable to acute or overuse sports injuries (eg, golfer's elbow, jumper's knee and Achilles insertional tendinopathies).3, 4 Frequently, orthopaedic surgeons face the challenge of partial or complete rupture of Achilles and rotator cuff tendons. Currently, grafts are most often applied but several problems are associated, including donor tissue availability and morbidity, host tissue reaction and time of biological incorporation,5, 6, 7 and high rupture recurrence rates due to neo‐fibrovascular tissue formation that compromises stability and mechanical properties of repaired enthesis.6, 7, 8
Thus, interface tissue engineering (ITE) aims at the recreating different tissues in vitro in order to repair or to regenerate diseased/damaged musculoskeletal interfaces.9, 10 Several attempts have been made to improve enthesis repair and regeneration, considering a need to mimic the nano‐ and micro‐structure of native tissue and replicate spatial distribution of cells and signalling factors.2, 3, 11 Cell‐based therapies provide new possibilities within tissue engineering to generate a functional enthesis, due to the central role of cell‐cell interactions in developing a functional tissue. Particularly, the use of multiple cell types in co‐culture systems is being increasingly explored aiming at mimicking the characteristic cellular environment found in the native tissue. Recent studies have shown that these strategies can affect cell fate and function.12, 13, 14 Indeed, different strategies have been used to mimic the complex cellular composition of enthesis. Normally, in vitro co‐culture approaches rely on the culture of differentiated cells (fibroblasts, osteoblasts, chondrocytes) and mesenchymal stem cells (MSCs).12, 13, 15, 16, 17 Here, compared to stem cells, the use of differentiated cells poses practical challenges for clinical applications given their lower regenerative power and longer in vitro expansion periods to obtain a sufficient cell number before transplantation. Therefore, MSCs, particularly bone marrow MSCs, have been explored in enthesis regeneration.17, 18, 19 However, adipose‐derived stem cells (ASCs) offer some advantages: (i) isolation from lipoaspirates provides an abundant autologous cell source, with relatively lower donor site morbidity and pain20; (ii) ASCs exhibit higher in vitro proliferation capacity, lower senescence and ability to generate tendon,21, 22, 23, 24 bone25, 26 and cartilage‐like27, 28, 29 tissues.
Co‐culture systems constitute interesting platforms to study cellular interactions occurring in enthesis, allowing a close replication of the native cellular environment.30, 31 Several in vitro studies have reported the important role of cell‐cell communication and paracrine factors on the expression of interface‐relevant markers, as well as the formation of fibrocartilage‐like interface.12, 15, 17 Nonetheless, the use of sources such as ASCs in co‐culture is still unexplored, as option for ITE.
Herein, we propose optimizing a co‐culture model to study cellular interactions between human ASCs (hASCs), pre‐differentiated towards the osteogenic lineage (herein called pre‐osteoblasts, pre‐OBs) and human tendon‐derived cells (hTDCs), used to replicate cellular environments found in bone and tendon, respectively. Firstly, the effect of different concentrations of osteogenic culture medium on tendon and bone niches maintenance was explored. Afterwards, a co‐culture model was established to evaluate the effect of different medium conditions and the influence of direct cell contact on the expression of relevant tendon‐, bone‐ and interface‐related markers.
2. MATERIALS AND METHODS
2.1. Cell isolation and culture
Samples were obtained under protocols previously established with Hospital da Prelada (Porto, Portugal) and under informed consent of the patients, according to Helsinki's Declaration and as reviewed and approved by the Ethical Committee of Hospital da Prelada.
Human ASCs were isolated from subcutaneous fat tissue obtained from lipoaspirate samples of healthy female patients32, 33 with ages in the range of 27‐42 years; hTDCs were isolated from tendon surplus samples of healthy male patients undergoing elective orthopaedic surgeries34, 35 with ages in the range of 25‐30 years, as described in Supporting Information. Cells were cultured in basal medium composed of minimum essential medium eagle (α‐MEM; Alfagene, Carcavelos, Portugal) supplemented with 10% (v/v) fetal bovine serum (Alfagene) and 1% (v/v) antibiotic/antimitotic solution (A/A; Alfagene).
2.2. Osteogenic differentiation
Osteogenic precommitment was performed by culturing hASCs in osteogenic medium (OM) (basal medium with 10 mmol/L β‐glycerophosphate (βGP, G9422; Sigma‐Aldrich, St. Louis, MO, USA), 10−8 mol/L dexamethasone (DEX, D2915; Sigma‐Aldrich), and 50 μg/mL l‐ascorbic acid 2‐phosphate sesquimagnesium salt hydrate (AA, 013‐12061; Wako, Neuss, Germany). After selecting a 14‐day precommitment period, hASCs were herein called pre‐OBs and used in both single and co‐culture studies.
2.3. Culture setups
Figure 1 depicts the experimental setup used for the experiments. In single cultures, pre‐OBs and hTDCs were seeded separately at a density of 2 × 103 cells/cm2. Five medium conditions containing different ratios of basal (BM) and osteogenic media (OM) were tested (Table 1). Three conditions were selected for establishing direct contact co‐cultures by seeding pre‐OBs and hTDCs together in a ratio of 1:1 using a final density of 2 × 103 cells/cm2. Single cultures of pre‐OBs and hTDCs were used as controls. Proliferation, alkaline phosphatase activity (ALP), alizarin red (AZ) staining, protein and gene expression were determined after 1, 7 and 14 days of culture.
Figure 1.
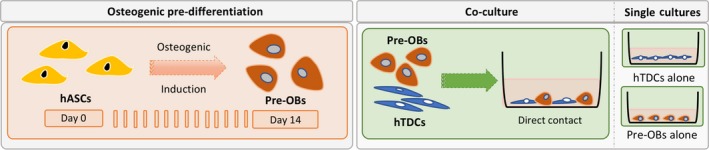
Experimental setup illustrating the temporal approach used to induce the osteogenic differentiation of hASCs and consequent establishment of a direct co‐culture model and correspondent single cultures. To assess the influence of different concentrations of osteogenic supplementation on culture medium, both pre‐OBs and hTDCs were initially seeded separately in single culture systems. For this purpose, cellular responses were studied to simultaneously evaluate the effects on the osteogenic commitment of pre‐OBs and the tenogenic phenotype of hTDCs. Subsequently, co‐culture systems were established using pre‐OBs and hTDCs under direct contact conditions to further investigate the crosstalk occurring between these 2 cell types under selected culture medium conditions. In this case, single culture systems were also performed in parallel as controls
Table 1.
Composition of cell culture media used in single and co‐cultures
| Ratios (%) | ||
|---|---|---|
| Basal medium (BM) | Osteogenic medium (OM) | |
| Condition 1—100 BM: 0 OM | 100 | 0 |
| Condition 2—75 BM: 25 OM | 75 | 25 |
| Condition 3—50 BM: 50 OM | 50 | 50 |
| Condition 4—25 BM: 75 OM | 25 | 75 |
| Condition 5—0 BM: 100 OM | 0 | 100 |
Basal medium (BM): alpha‐MEM supplemented with 10% FBS and 1% antibiotic/antimitotic solution; osteogenic medium (OM): basal medium supplemented with 10 mmol/L βGP, 10−8 mol/L DEX and 50 μg/mL AA.
2.4. dsDNA quantification, alizarin red staining and ALP activity
Cellular proliferation was evaluated using a fluorometric dsDNA quantification kit (PicoGreen; Molecular Probes, Invitrogen, Carlsbad, CA, USA) to determine DNA content. Calcium deposition was assessed by AZ staining and quantitative analysis after 7 and 14 days of culture. ALP activity was determined using a colorimetric p‐nitrophenol (pNp) assay. Details are provided in Supporting Information.
2.5. RT‐PCR analysis
Expression of tendon‐, bone‐ and interface‐related markers was analysed. Total mRNA was extracted from single cultures (pre‐OBs and hTDCs) and co‐culture at days 0, 1, 7 and 14 days using RIBOZOL™ RNA extraction reagent (VWRCN580, VWR). Primer sequences (Table 2) were designed using Primer‐BLAST tool and synthesized by Eurofins Genomics. The evaluation of the relative expression level was performed using the 2−ΔΔCt method. Transcript levels of selected genes were analysed and normalized to the expression of the selected housekeeping gene, glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH). All values were firstly normalized against GAPDH housekeeping gene transcript values, and then to the basal condition (100BM:0OM, calibrator sample) of the respective collection day. Three samples were used for each condition. Results are represented as fold change.
Table 2.
Primers used for quantitative RT‐PCR analysis
| Target gene | Gene abbreviation | Primer sequence | NBCI reference | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Glyceraldehyde 3‐phosphate dehydrogenase | GAPDH | AGCCTCAAGATCATCAGCAA | GTCATGAGTCCTTCCAGGAT | NM_002046.5 |
| Collagen, type I, α1 | COL1A1 | GTCACAGATCACGTCATCGC | CGAAGACCCACCAATCAC | NM_000088.3 |
| Collagen, type III, α1 | COL3A1 | TTGGCATGGTTCTGGCTTCC | GCTGGCTACTTCTCGTG | NM_000090.3 |
| Osteogenic‐related markers | ||||
| Secreted phosphoprotein 1 (Osteopontin) | SPP1 | CAGACCTGACATCCAGTACCC | GGTCATCCAGCTGACTCGTT | NM_001040058.1 |
| Run‐ related transcription factor 2 | RUNX2 | TGTCTGTGCCTTCTGGGTTC | CCGGCCTGCCTATGCTGTTA | NM_001024630.3 |
| Alkaline Phosphatase | ALPL | GAAGGAAAAGCCAAGCAGGC | GGGGGCCAGACCAAAGATAG | NM_000478.5 |
| Tenogenic‐related markers | ||||
| Scleraxis | SCX | CGAGAACACCCAGCCCAAAC | CTCCGAATCGCAGTCTTTCTGTC | NM_001717912 |
| Mohawk | MKX | TGTTAAGGCCATAGCTGCGT | TCGCACAGACACCTGGAAAA | NM_173576.5 |
| Decorin | DCN | CAGCATTCCTCAAGGTCTTCCT | GAGAGCCATTGTCAACAGCA | NM_001920.3 |
| Tenascin‐c | TNC | ACTGCCAAGTTCACAACAGACC | CCCACAATGACTTCCTTGACTG | NM_002160.3 |
| Chondrogenic‐related markers | ||||
| Cartilage oligomeric matrix protein | COMP | AGGATGGAGACGGACATCAG | TCTGCATCAAAGTCGTCCTG | NM_000095.2 |
| Aggrecan | ACAN | TGGTCTTGCAGCAGTTGATTC | TAGAGTCCTCAAGCCTCCTGT | NM_013227.3 |
2.6. Immunocytochemistry
Protein expression of tendon‐, bone‐ and interface‐related markers was evaluated using mouse anti‐human Tenascin‐C (TNC, 1:3000; Abcam), mouse anti‐human collagen I (COL1, 1:500; Abcam), rabbit anti‐human collagen III (COL3, 1:100; Abcam), rabbit anti‐human osteopontin (OPN, 1:100; Abcam), mouse anti‐human osteocalcin (OCN, 1:50; Abcam), mouse anti‐human Decorin (DCN, 1:100; Abcam), mouse anti‐human Aggrecan (ACAN, 1:200; Alfagene) and rabbit anti‐human Collagen II (COL2, 1:200; Abcam). Donkey anti‐mouse AlexaFluor 488, mouse anti‐rabbit AlexaFluor 488 or rabbit anti‐goat AlexaFluor 488 were used as secondary antibodies. Nuclei were counterstained with 4,6‐diamidino‐2‐phenyindole dilactate (DAPI; Invitrogen). All samples were visualized and images acquired by fluorescence microscopy (Axio Imager Z1 m; Zeiss, Deutschland, Germany). Detailed protocol is provided in Supporting Information.
2.7. Quantitative analysis of fluorescence images
All images were acquired by fluorescence microscope and the signal emitted from the expression of the intended proteins was, afterwards, quantified using ImageJ software. Briefly, proteins expression was quantitatively analysed in stained captions of Phalloidin/DAPI and the protein of interest at 7 and 14 days of culture. The images were split in 3 channels, being only the green analysed and the fluorescence intensity measured. A total of 4 images were analysed and results are expressed as mean ± SEM.
2.8. Statistical analysis
Results were obtained from 3 independent experiments with a minimum of 3 replicates for each condition. Results are presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism 7.0 software. Two‐way analysis of variance with Sidak and Tukey tests was performed. Differences between experimental groups were considered significant with a confidence interval of 95%, whenever P < .05.
3. RESULTS
3.1. Influence of increasing osteogenic medium concentrations at single culture level
A 14‐day osteogenesis precommitment period was first defined for obtaining pre‐OBs (Supporting Information Figure S1).
3.1.1. Effect on osteogenic and tenogenic phenotype of pre‐OBs and hTDCs: proliferation, ALP activity and mineralization
The influence of different media on proliferation, ALP activity and mineralization of single cultures was studied (Figure 2). At 14 days, the presence of OM in conditions 25BM:75OM and 0BM:100OM increased hTDCs proliferation, in comparison with the intermediate medium condition 50BM:50OM (0BM:100OM, P < .007) and lower concentrations in conditions 100BM:0OM (25BM:75OM, P < .05; 0BM:100OM, P < .007) and 75BM:25OM (0BM:100OM, P < .007) (Figure 2A); whereas no significant differences were found for pre‐OBs between medium conditions.
Figure 2.
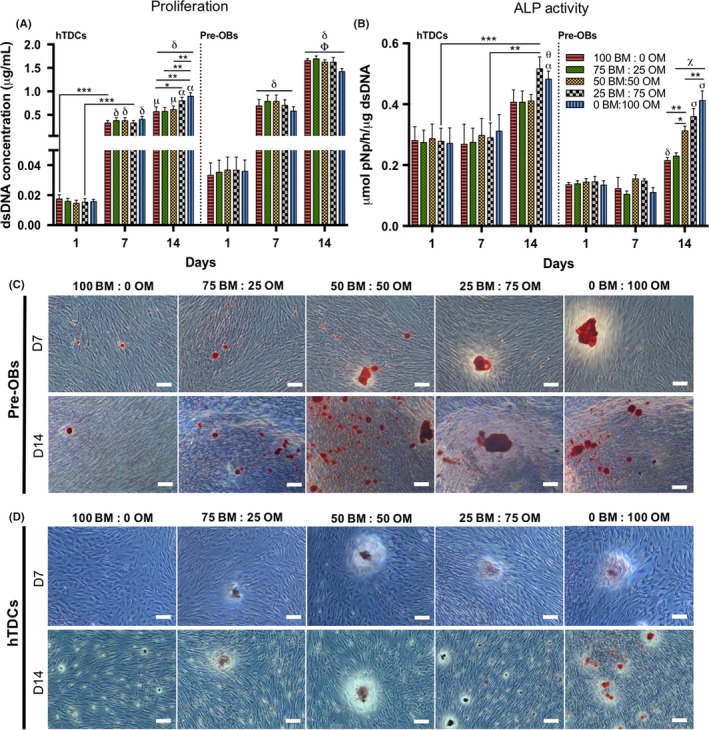
Evaluation of cell proliferation, alkaline phosphatase activity and matrix mineralization in single cultures of pre‐OBs and hTDCs in the presence of increasing concentrations of osteogenic medium. (A) Cell proliferation, evaluated by dsDNA concentration. Results are presented as mean ± SEM and statistically significant differences are *, P < .05; **, P < .007; ***, P < .0008; μ, P < .02 where μ is statistically significant in comparison with the same conditions at day 7; α, P < .0001, α is statistically significant in comparison with the same conditions at day 7; δ, P < .0001, δ is statistically significant in comparison with the same conditions at day 1; Φ, P < .0001, Φ is statistically significant in comparison with the same conditions at day 7. (B) Quantification of alkaline phosphatase (ALP) activity by the colorimetric p‐nitrophenol (pNP) assay. Results were normalized against the amount of dsDNA of the correspondent sample and are presented as mean ± SEM. Statistically significant differences are shown as *, P < .02; **, P < .003; ***, P < .0009; α, P < .02, α is statistically significant in comparison with the same condition at day 7; δ, P < .004, δ is statistically significant in comparison with the same conditions at day 1 and 7; θ, P < .003, θ is statistically significant in comparison with the same condition at day 1; χ, P < .0001, χ is statistically significant in comparison with the same conditions at days 1 and 7; σ, P < .0001 σ is statistically significant in comparison with the conditions 100 BM: 0 OM and 75 BM: 25 OM. (C, D) Alizarin red staining, demonstrating matrix mineralization in both (C) pre‐OBs and (D) hTDCs (scale bars, 100 μm). Legend: BM: basal medium; OM: osteogenic medium
Regarding ALP activity (Figure 2B), at 14 days, pre‐OBs exhibited the highest activity in conditions 25BM:75OM and 0BM:100OM (P < .0001), followed by the intermediate condition 50BM:50OM (P < .02). Although hTDCs exhibited a significant increased ALP activity with higher ratios of OM (25BM:75OM; 0BM:100OM) between days 7 and 14, no significant changes were observed between conditions.
Additionally, AZ staining confirmed that mineralized calcium deposits increased with increasing OM ratios, which was more evident in pre‐OBs, compared to hTDCs, as shown by staining intensity (Figure 2C,D).
3.1.2. Effect on osteogenic and tenogenic phenotype of pre‐OBs and hTDCs: expression of bone‐ and tendon‐related markers
Maintenance of pre‐OBs phenotype was confirmed by analysing the expression of COL1A1, COL3A1 and bone‐related genes (SPP1, ALPL, RUNX2) by real‐time reverse transcription polymerase chain reaction (RT‐PCR) (Figure 3A). No significant changes in the expression of the studied markers were detected after 7 days. Nonetheless, SPP1 transcript levels (Figure 3A‐i) increased significantly from 7 to 14 days (P < .0002) in the highest concentration of OM and were significantly higher in cells cultured for 14 days in this condition (0BM:100OM), comparing to lower ratios of osteogenic supplementation (75BM:25OM, P < .004; 50BM:50OM, P < .004) and basal medium (100BM:0OM, P < .004). Similarly, a significant increase of ALPL transcript levels (Figure 3A‐ii) was observed from day 7 to 14 in 0BM:100OM, which were significantly higher in cells cultured for 14 days in this condition, compared to cells in basal medium (100BM:0OM, P < .004) and in the intermediate condition (50BM:50OM, P < .04). Also, RUNX2 transcript levels (Figure 3A‐iii) were significantly increased between 7 and 14 days in cells cultured with higher amounts of osteogenic supplementation (50BM:50OM, P < .004; 25BM:75OM, P < .004; 0BM:100OM, P < .0002). Cells cultured for 14 days in 0BM:100OM showed an up‐regulation of RUNX2 in comparison with cells in BM (100BM:0OM, P < .004) and with lower osteogenic supplementation (75BM:25OM, P < .004; 50BM:50OM, P < .04) (Figure 3A‐iii). Concerning COL1A1 expression, no significant changes were observed in the transcription levels between conditions (Figure 3A‐iv). Similarly to SPP1, ALPL and RUNX2, from day 7 to day 14, cells cultured in conditions with the highest concentration of osteogenic supplementation presented significantly higher COL3A1 expression (75BM:25OM, P < .04; 0BM:100OM, P < .004, Figure 3A‐v). After 14 days, COL3A1 transcript levels in cells cultured in 0BM:100OM were significantly higher, compared to cells cultured in medium containing lower osteogenic supplementation (25BM:75OM, P < .04; 50BM:50OM, P < .04) and in BM (100BM:0OM, P < .004) (Figure 3A‐v). Overall, higher concentrations of OM resulted in increased transcript levels of the studied bone‐related markers, supporting osteogenic differentiation of pre‐OBs. Furthermore, no significant differences were observed in OPN expression, an early osteogenic differentiation marker, in cells cultured in the presence of the different ratios of OM:BM at 7 and 14 days of culture (Figure 3B,C).
Figure 3.
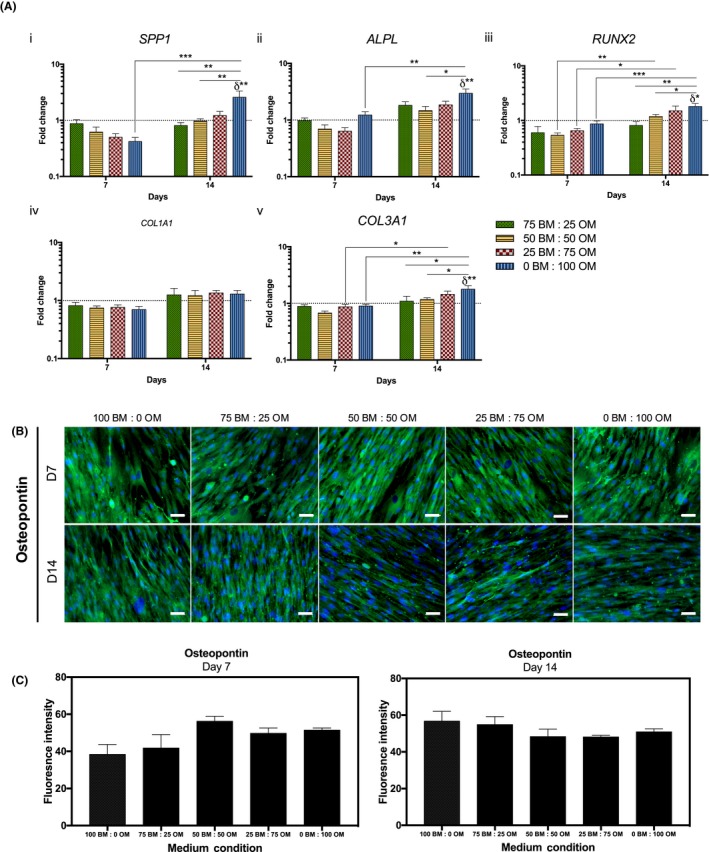
Maintenance of osteogenic phenotype of pre‐osteoblasts through culture time in the presence of different osteogenic medium conditions. (A) Gene expression of osteogenic‐related markers, (i) bone sialoprotein 1/osteopontin (SPP1), (ii) alkaline phosphatase (ALPL), (iii) runt‐related transcription factor 2 (RUNX2), (iv) collagen type I (COL1A1) and (v) collagen type III (COL3A1) evaluated by RT‐PCR. Expression of target genes was normalized against GAPDH housekeeping gene and gene expression in all conditions was normalized to the basal condition (100 BM: 0 OM) of the correspondent day. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .04; **, P < .004; ***, P < .0002; δ statistically significant differences in comparison to the basal condition of the correspondent day. (B) Fluorescence microscopy images of osteopontin (scale bars, 50 μm). (C) Fluorescence intensity quantification of osteopontin at days 7 and 14
Maintenance of the tenogenic phenotype of hTDCs was also evaluated. The expression of tenogenic genes (MKX, SCX, DCN, TNC), as well as COL1A1 and COL3A1 and the bone‐related marker RUNX2 was assessed by RT‐PCR (Figure 4). Strikingly, MKX was down‐regulated in cells cultured with the highest concentration of OM after 7 days (0BM:100OM, P < .05, compared to control) and at 14 days, conditions 25BM:75OM and 0BM:100OM resulted in a significant decrease of MKX transcript levels, compared to control (P < .01; P < .05, respectively, Figure 4A‐i). The highest MKX expression was found for cells cultured with the lowest concentration of OM tested (75BM:25OM, P < .01 comparing to 25BM:75OM; P < .05 comparing to 0BM:100OM), with no differences compared to BM condition. Nonetheless, no differences were observed for SCX transcript levels between conditions (Figure 4A‐ii). Cells cultured in the presence of only OM (0BM:100OM) presented significantly higher DCN and TNC transcript levels from days 7 to 14 (Figure 4Aiii‐iv). At 14 days, TNC was up‐regulated in cells cultured in 0BM:100OM, compared to basal condition (P < .01) and TNC transcript levels in cells cultured in condition 0BM:100OM were significantly higher in comparison to those in lower ratios of osteogenic supplementation (75BM:25OM, P < .01; 50BM:50OM, P < .01; 25BM:75OM, P < .05, Figure 4A‐iv). Additionally, COL1A1 was up‐regulated in cells cultured in the highest concentration of osteogenic supplementation, as soon as 7 days of culture (0BM:100OM, P < .0005, compared to basal condition) and in comparison with cells cultured in the presence of lower OM ratios (50BM:50OM, P < .05; 25BM:75OM, P < .01, Figure 4A‐v). COL3A1 was up‐regulated in cells cultured with 50BM:50OM (P < .05) and 25BM:75OM (P < .0001) in comparison with cells cultured in basal condition (100 BM:0OM, Figure 4A‐vi). For both COL1A1 and COL3A1, a significant decrease in the transcription levels of cells cultured in different conditions was noticed from day 7 to 14 (Figure 4Av‐vi). RUNX2 was up‐regulated in cells cultured in 50BM:50OM (P < .05), 25BM:75OM (P < .05) and 0BM:100OM (P < .01) after 14 days, in comparison to basal condition (Figure 4A‐vii).
Figure 4.
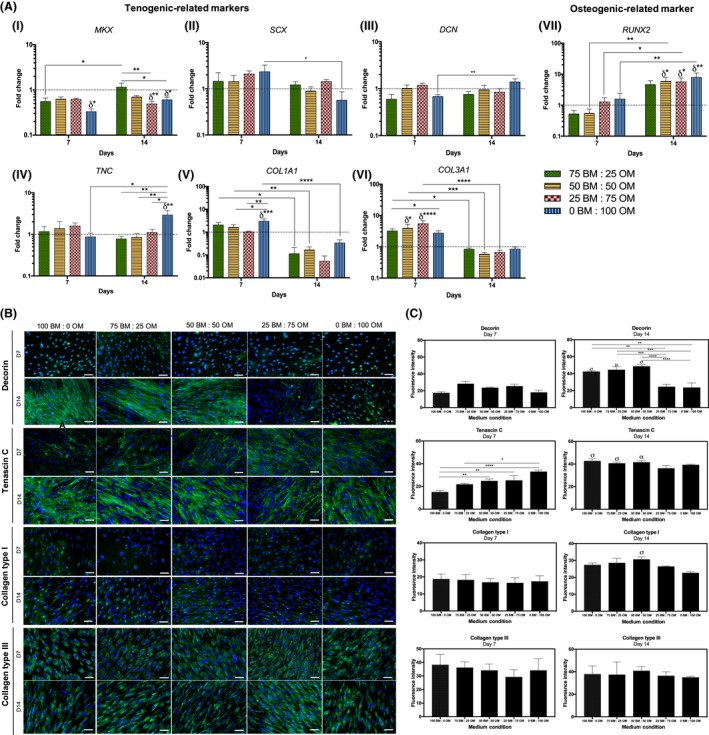
Assessment of the expression of tenogenic and osteogenic markers by hTDCs through culture time in different osteogenic medium conditions after 7 and 14 days. (A) Gene expression analysis of (i) mohawk (MKX), (ii) scleraxis (SCX), (iii) decorin (DCN), (iv) tenascin C (TNC), (v) collagen type I (COL1A1), (vi) collagen type III (COL3A1) and (vii) runt‐related transcription factor 2 (RUNX2) by RT‐PCR. Expression of target genes was normalized against GAPDH housekeeping gene and gene expression in all conditions was normalized to basal condition (100 BM: 0 OM) of the correspondent day. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .05; **, P < .01; ***, P < .0005; ****, P < .0001; δ statistically significant differences in comparison to the basal condition of the correspondent day. (B) Fluorescence microscopy of tenogenic‐related markers, decorin, tenascin‐C, collagen type I and collagen type III (scale bars, 50 μm). Nuclei were counterstained with DAPI. (C) Fluorescence intensity quantification of decorin, tenascin‐C, collagen type I and collagen type III at days 7 and 14. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .04; **, P < .006; ***, P < .0006; ****, P < .0001; σ, P < .0001, is statistically significant in correspondence to the same condition at day 7; α, P < .0005, is statistically significant in correspondence to the same condition at day 7; μ, P < .009, is statistically significant correspondence to the same cor>739572ndition at day 7
Tendon‐related ECM markers DCN, TNC, COL1 and COL3 were further studied at the protein level (Figure 4B,C). Interestingly, DCN was highly expressed at 14 days in cells cultured in the presence of lower OM concentrations, namely 100BM:0OM, 75BM:25OM and 50BM:50OM, in comparison with cells cultured for 7 days. TNC was detected at 7 days, but strongly expressed after 14 days by cells cultured in the presence of 100BM:0OM (P < .0001), 75BM:25OM (P < .0001) and 50BM:50OM (P < .0005) (Figure 4B,C). Furthermore, at both 7 and 14 days of culture, cells expressed COL1 intracellularly and were able to synthesize and produce COL3, being only observed a significant increase in the expression of COL1 in cells cultured in the presence of 50BM:50OM (P < .0001) for a period of 14 days. Overall, increasing concentrations of osteogenic supplementation affected the expression of tendon‐related markers.
3.2. Influence of direct pre‐Obs‐hTDCs crosstalk in co‐culture systems
Based on obtained results, 3 medium conditions were chosen, maximizing cell growth and balancing tendon‐ and bone‐related markers expression and matrix mineralization. Condition 50BM:50OM seemed to better maintain the osteogenic phenotype of pre‐OBs without inducing rapid cell maturation and the tenogenic phenotype of hTDCs, without triggering phenotypic drift towards osteogenesis. Conditions 100BM:0OM and 0BM:100OM were used for comparison.
3.2.1. Effects of direct contact co‐culture: cell proliferation, ALP activity and matrix mineralization
DNA content, ALP activity and mineralization were analysed in co‐cultures to evaluate the combined influence of direct contact and medium (Figure 5). Condition 50BM:50OM resulted in a significant increase in DNA content, in comparison with the basal condition (P < .03) at 14 days, while no differences were observed for condition 0BM:100OM (Figure 5A). A significantly higher proliferation was observed in co‐cultures compared to hTDCs in BM (P < .0002) and condition 50BM:50OM (P < .0001, Figure S2). At day 14, cells cultured with 0BM:100OM exhibited significantly higher ALP activity, compared to 50BM:50OM (P < .0001) and 100BM:0OM (P < .0006, Figure 5B). Strikingly, after 14 days, ALP activity was significantly higher in co‐cultures, when compared to single cultures (pre‐OBs and hTDCs) in all conditions (Figure S3). Moreover, increasing OM ratios increased mineral deposition (Figures 5C, S4). After 7 days, higher concentrations were observed for pre‐OBs and co‐cultures in comparison with hTDCs in all conditions (Figure 5Di). Interestingly, cells co‐cultured for 14 days in the presence of 100% OM exhibited significantly increased mineralization, in comparison to pre‐OBs and hTDCs alone (P < .0001, Figure 5Dii), suggesting a synergistic effect between osteogenic supplementation and direct cell‐cell interactions.
Figure 5.
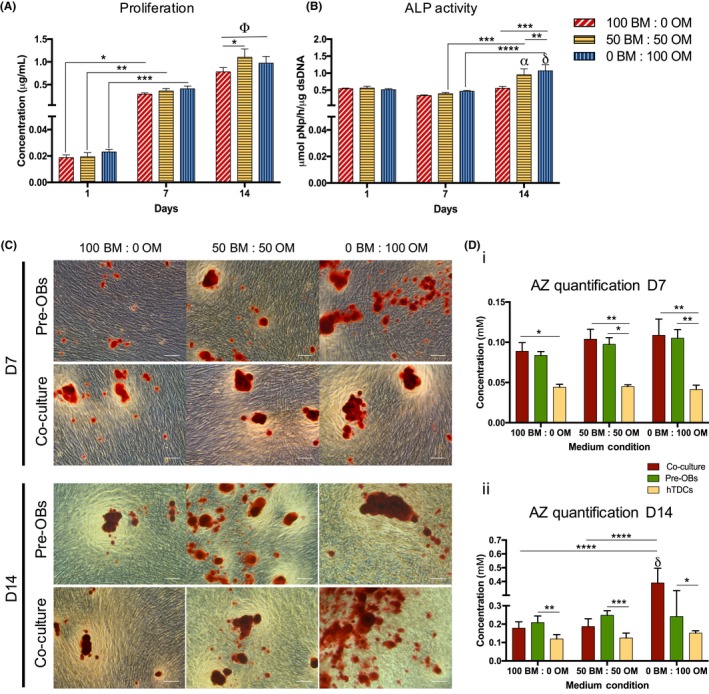
Evaluation of cell proliferation, alkaline phosphatase activity and matrix mineralization in direct co‐culture. (A) Cell proliferation was quantified by a fluorimetric dsDNA quantification kit. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .03; **, P < .004; ***, P < .0009; Φ, P < .0001, Φ is statistically significant in correspondence to all the conditions at day 1 and 7. (B) Alkaline phosphatase (ALP) activity was quantified by the colorimetric p‐nitrophenol (pNP). Results are presented as mean ± SEM and statistically significant differences are shown as **, P < .0001; ***, P < .0006; ****, P < .0001; α, P < .0002, α is statistically significant in correspondence to the same condition at day 1; δ, P < .0001, δ is statistically significant in correspondence to the same condition at day 1. (C) Alizarin red (AZ) staining was performed for both pre‐OBs and direct co‐cultures after 7 and 14 days of culture (scale bars, 100 μm). (D) Quantification of AZ staining was performed using cetylpyridinium chloride solution at 7 and 14 days of culture. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .05; **, P < .009; ***, P < .0003; ****, P < .0001; δ, P < .0001, δ is statistically significant in correspondence to pre‐OBs and hTDCs in the same condition
3.2.2. Effects of direct contact co‐culture: expression of bone‐, tendon‐ and interface‐related markers
The expression of bone‐ (ALPL, RUNX2 and SPP1), tendon‐ (SCX, MKX and TNC) and interface‐related markers (ACAN, COMP), as well as COL1A1 and COL3A1 was analysed by RT‐PCR after 7 and 14 days of culture (Figure 6). Although no differences were found for ALPL transcript levels (Figure 6A), after 7 days, co‐cultured cells in 50BM:50OM presented significantly higher RUNX2 transcript levels (P < .0007, vs pre‐OBs; P < .009 vs hTDCs, Figure 6B), but without differences comparing to basal condition. Interestingly, co‐cultured cells expressed significantly higher SPP1 transcript levels as soon as 7 days in 0BM:100OM comparing with pre‐OBs in the same condition (P < .0007, Figure 6C). From day 7 to 14, SPP1 expression significantly decreased in co‐cultures in condition 0BM:100OM (P < .0007), but no differences were found comparing to basal condition.
Figure 6.
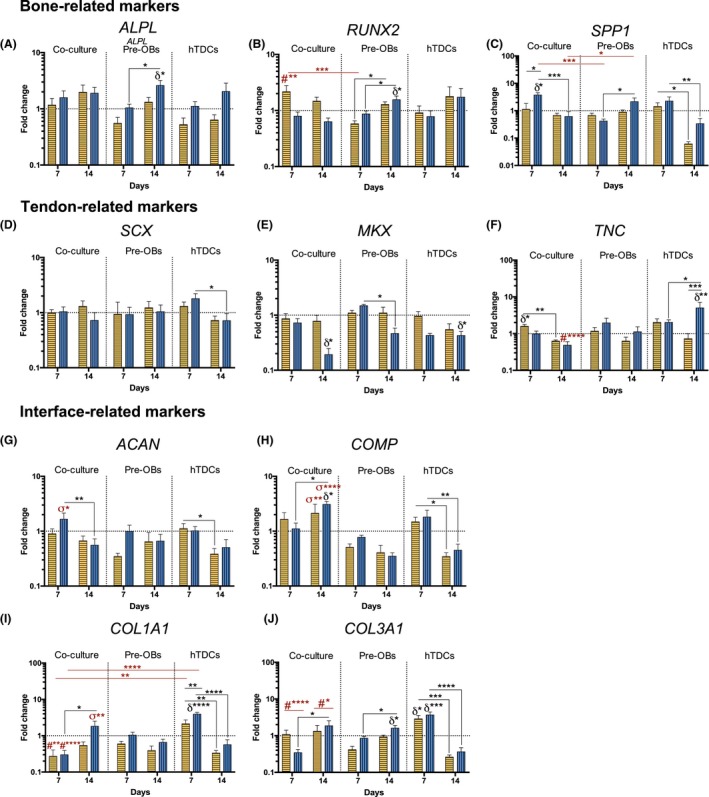
Evaluation of the gene expression of bone‐, tendon‐ and interface‐related markers in single cultures and direct co‐cultures up to 14 days in culture. Gene expression of (A) alkaline phosphatase (ALPL), (B) runt‐related transcription factor 2 (RUNX2), (C) bone sialoprotein 1/ osteopontin (SPP1), (D) scleraxis (SCX), (E) homeobox mohawk (MKX), (F) tenascin C (TNC), (G) aggrecan (ACAN), (H) cartilage oligomeric matrix protein (COMP), (I) collagen type I (COL1A1) and (J) collagen type III (COL3A1) was analysed by RT‐PCR. Target genes were normalized against GAPDH housekeeping gene. The gene expression in all conditions was normalized to basal condition (100 BM: 0 OM) of the correspondent day. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .05; **, P < .009; ***, P < .0007; ****, P < .0001; δ statistically significant differences in comparison to the basal condition of the correspondent day; # statistically significant differences in comparison with hTDCs in the same condition; σ statistically significant differences in comparison pre‐OBs and hTDCs in the same conditions
Additionally, no significant differences were observed for SCX expression in co‐cultures when comparing to pre‐OBs and hTDCs single cultures, nor between medium conditions (Figure 6D). Strikingly, similarly to hTDCs alone, MKX transcript levels were significantly lower in co‐cultures in 0BM:100OM after 14 days comparing to BM (P < .05, Figure 6E). TNC was up‐regulated in co‐cultures after 7 days in 50BM:50OM (P < .05, vs BM), decreasing significantly from day 7 to 14 (P < .009, Figure 6F). Interestingly, at day 14, the presence of only OM significantly decreased TNC transcript levels in co‐cultures, comparing with hTDCs in the same medium condition (P < .0007).
Furthermore, ACAN was up‐regulated in co‐cultures as soon as 7 days in 0BM:100OM (P < .05, compared to pre‐OBs and hTDCs, Figure 6G) and COMP was up‐regulated in co‐cultures in 0BM:100OM after 14 days (P < .05, compared to 100BM:0OM Figure 6H). Strikingly, co‐cultured cells expressed significantly higher COMP transcript levels after 14 days compared with pre‐OBs and hTDCs cultured in the same conditions (50BM:50OM, P < .009; 0BM:100OM, P < .0001, Figure 6H). In pre‐OBs, no differences were observed for ACAN and COMP expression; whereas OM led to a significant decrease of COMP expression in conditions 50BM:50OM (P < .05) and 0BM:100OM (P < .009) in hTDCs from day 7 to 14.
Higher COL1A1 (Figure 6I) and COL3A1 (Figure 6J) transcript levels were noticed at 7 days in hTDCs in comparison with co‐cultures in both medium conditions (COL1A1: 50BM:50OM, P < .009; 0BM:100OM, P < .0001; COL3A1: 50BM:50OM, P < .0001; 0BM:100OM, P < .0001). Nevertheless, after 14 days, co‐cultured cells in 0BM:100OM expressed significantly higher COL1A1 transcript levels (P < .009, comparing to pre‐OBs and hTDCs Figure 6I) and both medium conditions led to significantly higher COL3A1 expression levels in co‐cultures (P < .05, comparing to pre‐OBs and hTDCs Figure 6J). No differences were observed in comparison to basal condition.
Immunocytochemistry analysis was also performed against OCN, TNC, ACAN and COL2 (Figure 7). A higher OCN deposition was observed in co‐cultures after both 7 and 14 days, whereas in pre‐OBs, OCN was highly detected after 7 days in comparison with 14 days of culture (100BM:0OM, P < .006; 50BM:50OM, P < .0001; and 0BM:100OM, P < .006) (Figure 7A). After 14 days, OM present in conditions 50BM:50OM and 0BM:100OM affected TNC expression, which was observed in hTDCs in all medium conditions, but only observed in basal condition (100BM:0OM) in co‐cultures (P < .0001) (Figure 7B). No ACAN expression was found in pre‐OBs and hTDCs, while, in co‐cultures, an abundant deposition was observed in conditions containing OM as soon as 7 days and maintained up to 14 days (P < .0001) (Figure 7C). COL2 was deposited at a higher extent in co‐cultures as soon as after 7 days in all conditions, whereas hTDCs expressed COL2 at 7 days mainly in the presence of OM and at 14 days in all medium conditions, being highly expressed in condition 50BM:50OM in comparison with conditions 100BM:0OM (P < .006) and 0BM:100OM (P < .04). COL2 expression was observed in pre‐OBs at 14 days in condition 50BM:50OM and 0BM:100OM (Figure 7D), even though COL2 expression was significantly lower in comparison with co‐cultures (P < .0001). Altogether, these results support the influence of direct contact occurring between 2 types of cells in the modulation of cellular phenotype.
Figure 7.
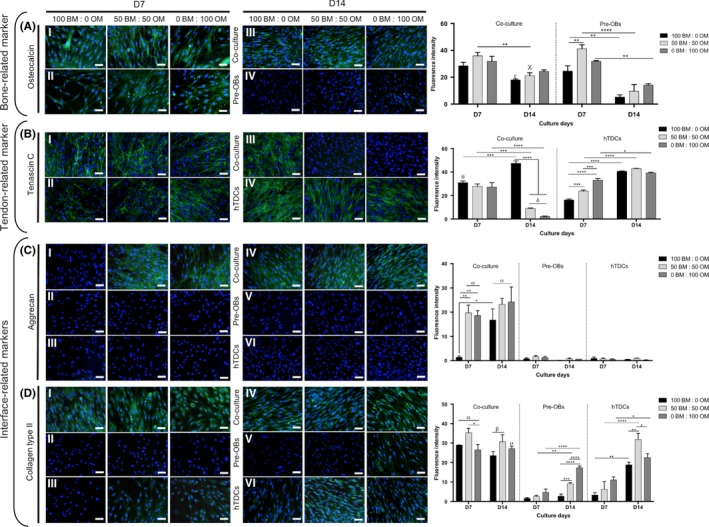
Expression of bone, tendon‐ and interface‐related proteins in single cultures of pre‐OBs and hTDCs and in direct contact co‐cultures. Fluorescence microscopy images of (A) osteocalcin, (B) tenascin C, (C) aggrecan and (D) collagen type II after 7 and 14 days of culture (scale bars, 50 μm). Nuclei were counterstained with DAPI (blue). Fluorescence intensity quantification was performed. Results are presented as mean ± SEM and statistically significant differences are shown as *, P < .05; **, P < .006; ***, P < .0007; ****, P < .0001; δ, P < .0001, δ is statistically significant in comparison with hTDCs in the same day; α, P < .0001, is statistically significant in comparison with pre‐OBs and hTDCs in the same day; β, P < .0001, is statistically significant in comparison with pre‐OBs in the same day; σ, P < .0001, is statistically significant in comparison with hTDCs in the same day; χ, P < .0003, is statistically significant in comparison with pre‐OBs in the same day; θ, P < .0006, is statistically significant in comparison with hTDCs in the same day; μ, P < .04, is statistically significant in comparison with pre‐OBs in the same day; ε, P < .05, is statistically significant in comparison with pre‐OBs in the same day
4. DISCUSSION
The success of ITE depends on optimal cell source selection and on the understanding of cellular interactions within native tissues. Strategies for enthesis tissue engineering and regeneration frequently disregard the existence of different cellular phenotypes and, thus, the need to establish adequate culture conditions to balance cellular behaviour. Co‐culture systems constitute platforms to study cellular communication through direct or indirect interactions between different cell types.30, 31, 36, 37 Herein, we explored an in vitro strategy to modulate cellular phenotype for tissue engineering approaches, envisioning the combination of different cell types with medium supplementation. We first studied the effect of different OM concentrations on the modulation of hASCs (pre‐OBs) and hTDCs cellular responses and, afterwards, the influence of establishing a direct contact co‐culture model on the expression of interface‐relevant markers. Human ASCs constitute a promising cell source for tissue regeneration38 and their osteogenic commitment has been well described.39 Hence, this cell population was herein precommitted towards the osteogenic lineage to obtain pre‐OBs, aiming at mimicking, as close as possible, the cellular environment found in bone. Additionally, to mimic the cellular environment found in tendon counterpart of enthesis and given the lack of well‐established protocols for tenogenic differentiation, hTDCs were used, which are composed by tenocytes/tenoblasts and stem/progenitor cells40, 41 and hold potential for musculoskeletal tissue engineering.41, 42 Both cell populations were chosen based on evidences that in vitro interactions between different cell types may modulate cellular trans‐differentiation and lead to an eventual fibrocartilage formation,15 suggesting that heterotypic cellular interactions occurring between tendon and bone cells may initiate important events leading to interfacial regeneration.
Previous studies have addressed the formulation of optimal medium conditions in co‐culture models, reporting that 1 mmol/L β‐GP maximized cell growth and osteoblast mineralization, while minimizing ectopic fibroblast mineralization,15 whereas 3 mmol/L β‐GP concentration promoted low fibroblast‐osteoblast mineralization.16 Therefore, co‐culture medium should be tuned to each unique culture system towards improving tissue engineering strategies currently explored.
Our results clearly showed an osteogenic commitment of hASCs and the phenotype maintenance of pre‐OBs during culture time, by an up‐regulation of SPP1, RUNX2 and ALPL. During osteoblast‐lineage differentiation and maturation, RUNX2 expression is essential, as an important osteogenic master switch and regulator of ECM‐related genes expression, like SPP1, and even ALPL.43, 44 Increasing concentrations of OM promoted the expression of bone‐related markers at the gene and protein level, simultaneously inducing matrix mineralization.
Additionally, maintenance of hTDCs tenogenic phenotype was also assessed. The intermediate condition (50BM:50OM) demonstrated a possible phenotypic modulation of hTDCs towards tenogenesis. Remarkably, increasing concentrations of OM clearly down‐regulated MKX and SCX, transcription factors essential during tendon differentiation,45, 46 as well as COL1A1 and COL3A1, while up‐regulating DCN and TNC expression after 14 days of culture. Collagen types I and III are the most abundant collagens found in tendon tissue; decorin and tenascin‐C are known to be the major non‐collageneous components of tendon ECM, and several authors reported their expression in bone cells.47, 48, 49, 50 At protein level, DCN and TNC were highly expressed in the presence of medium conditions containing lower ratios of OM. Furthermore, RUNX2 up‐regulation in hTDCs may be strongly associated with the isolated mixed population of cells that could undergo osteogenic differentiation.41 Thus, increasing concentrations of OM may lead to a phenotypic drift of this population towards the osteogenic lineage. Nonetheless, condition 50BM:50OM seemed to modulate pre‐OBs osteogenic commitment, while maintaining hTDCs tenogenic phenotype, without significant osteogenic drift. Thus, this intermediate medium condition was selected for further studies.
Subsequently, a direct co‐culture system was established using 3 predetermined medium conditions. The co‐culture system with different medium conditions offered a specific environment for the 2 cell types, not affecting cell proliferation, increasing matrix mineralization and ALP activity. The expression of bone‐related markers was higher in co‐cultures in the presence of OM, as early as 7 days of culture. Particularly, the highest concentration of OM (0BM:100OM) induced an up‐regulation of SPP1 at 7 days. Strikingly, the higher expression of RUNX2 in condition 50BM:50OM may indicate the dynamic and evolving osteogenic process that is still occurring and explain higher ECM matrix mineralization observed in co‐cultures after 14 days in the presence of OM.51 Higher SPP1 and RUNX2 transcript levels found in co‐cultures after 7 days in comparison to pre‐OBs alone suggest the existence of bi‐directional cellular communication events orchestrating osteogenesis.
In turn, SCX and MKX expression in co‐culture conditions exhibited no significant differences over time in the intermediate medium condition (50BM:50OM), when compared to BM, suggesting that a tenogenic phenotype may still be rescued in the presence of OM, possibly as a result of heterotypic cellular interactions.
Interestingly, the presence of OM significantly increased the expression of interface‐relevant markers, ACAN and COMP, in co‐cultures, in comparison with pre‐OBs and hTDCs single cultures. The production of extracellular molecules, such as tenascin and cartilage oligomeric matrix protein (COMP), activates intracellular signalling pathways of kinase and paxilin (focal adhesion enzymes) leading to the initiation of a transition step from chondroprogenitor cells to fully committed chondrocytes.52 Moreover, the presence of OM also influenced the deposition of ACAN and COL2. ACAN production was influenced by pre‐OBs and hTDCs direct cell contact, given that it was only observed in co‐culture conditions. Increasing ACAN expression in the matrix is normally observed in early differentiation periods where chondrocytes start to become hypertrophic.53 Moreover, high levels of COL2 and ACAN are expressed by chondroprogenitor cells and chondrocytes in the extracellular matrix,53 being their deposition governed by the nuclear transcription factor Sox9,53, 54 being an important transcription factor to be considered in future studies. Interestingly, in co‐cultures, Runx2 is significantly increased as soon as 7 days of culture in comparison with hTDCs and Pre‐OBs. Runx2 is expressed during the differentiation of chondrocytes to hypertrophic chondrocytes, being the BMP‐induced Smad1 and Smad1 and Runx2/Cbfa1 interactions important for the hypertrophy.52, 55, 56 Nevertheless, more studies at the molecular level will be useful to help clarifying the role of each cell type within the proposed system of heterotypic cell interactions. Altogether, these results suggest a possible targeting towards an osteochondral lineage, where the expression of both bone and chondrogenic‐related markers is increased. Furthermore, since the expression of COL2 was observed in all medium conditions and the expression of ACAN was only observed after 14 days in basal condition, OM ratios and cell‐cell interactions occurring in co‐culture seem to play an important role in the maintenance of a chondrogenic‐like ECM in longer culture periods.57
Overall, our work suggests that a balanced process of osteogenic and tenogenic commitment of distinct cell populations may be achieved through the use of intermediate concentrations of osteogenic supplementation. Furthermore, interactions occurring in co‐cultures play an important role in the expression of tendon‐, bone‐ and interface‐related markers. These findings serve as basis for understanding the effect of medium conditions on the modulation of cell phenotype along with the importance of cell‐cell contact on the regulation of the biological environment with future application in studies involving more complex 3D systems in combination with dynamic platforms, such as bioreactors, to promote the formation of a new engineered enthesis‐like tissue.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Hospital da Prelada (Porto, Portugal) for providing tendon tissue (Orthopaedics Department) and adipose tissue (Plastic Surgery Department) samples; the financial support from the European Union Framework Programme for Research and Innovation HORIZON2020, under the TEAMING Grant agreement No 739572—The Discoveries CTR, FCT—Fundação para a Ciência e Tecnologia for the PhD grant of IC (PD/BD/128088/2016); and the Project NORTE‐01‐0145‐FEDER‐000021:”Accelerating tissue engineering and personalized medicine discoveries by the integration of key enabling nanotechnologies, marine‐derived biomaterials and stem cells”, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
Calejo I, Costa‐Almeida R, Gonçalves AI, Berdecka D, Reis RL, Gomes ME. Bi‐directional modulation of cellular interactions in an in vitro co‐culture model of tendon‐to‐bone interface. Cell Prolif. 2018;51:e12493 10.1111/cpr.12493
REFERENCES
- 1. Galatz L, Rothermich S, VanderPloeg K, Petersen B, Sandell L, Thomopoulos S. The development and morphogenesis of the tendon‐to‐bone insertion What development can teach us about healing. J Musculoskelet Neuronal Interact. 2010;10:35‐45. [PMC free article] [PubMed] [Google Scholar]
- 2. Zelzer E, Blitz E, Killian ML, Thomopoulos S. Tendon‐to‐bone attachment: from development to maturity. Birth Defects Res C Embryo Today. 2014;102:101‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benjamin M, Kumai T, Milz S, Boszczyk BM, Boszczyk AA, Ralphs JR. The skeletal attachment of tendons—tendon ‘entheses’. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:931‐945. [DOI] [PubMed] [Google Scholar]
- 4. Benjamin M, Toumi H, Ralphs JR, Bydder G, Best TM, Milz S. Where tendons and ligaments meet bone‐ attachment sites (‘entheses’) in relation to exercise and:or mechanical load. J Anat. 2006;208:471‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson A, Nutton RW, Keating JF. Current trends in the use of tendon allografts in orthopaedic surgery. J Bone Joint Surg Br. 2006;88:988‐992. [DOI] [PubMed] [Google Scholar]
- 6. Kaeding CC, Aros B, Pedroza A, et al. Allograft versus autograft anterior cruciate ligament reconstruction: predictors of failure from a MOON prospective longitudinal cohort. Sports Health. 2011;3:73‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thomopoulos S. Tendon to bone healing: differences in biomechanical, structural, and compositional properties due to a range of activity levels. J Biomech Eng. 2003;125:106‐113. [DOI] [PubMed] [Google Scholar]
- 8. Lui PP, Zhang P, Chan K, Qin L. Biology and augmentation of tendon‐bone insertion repair. J Orthop Surg Res. 2010;5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seidi A, Ramalingam M, Elloumi‐Hannachi I, Ostrovidov S, Khademhosseini A. Gradient biomaterials for soft‐to‐hard interface tissue engineering. Acta Biomater. 2011;7:1441‐1451. [DOI] [PubMed] [Google Scholar]
- 10. Paxton JZ, Baar K, Grover LM. Current progress in enthesis repair: strategies for interfacial tissue engineering. Orthop Mus Sys. 2013;01:003. [Google Scholar]
- 11. Lu HH, Thomopoulos S. Functional attachment of soft tissues to bone: development, healing, and tissue engineering. Annu Rev Biomed Eng. 2013;15:201‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang IE, Bogdanowicz DR, Mitroo S, Shan J, Kala S, Lu HH. Cellular interactions regulate stem cell differentiation in tri‐culture. Connect Tissue Res. 2016;57:476‐487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang J, Nicoll SB, Lu HH. Co‐culture of osteoblasts and chondrocytes modulates cellular differentiation in vitro. Biochem Biophys Res Commun. 2005;338:762‐770. [DOI] [PubMed] [Google Scholar]
- 14. Veronesi F, Torricelli P, Della Bella E, Pagani S, Fini M. In vitro mutual interaction between tenocytes and adipose‐derived mesenchymal stromal cells. Cytotherapy. 2015;17:215‐223. [DOI] [PubMed] [Google Scholar]
- 15. Wang IE, Shan J, Choi R, et al. Role of osteoblast‐fibroblast interactions in the formation of the ligament‐to‐bone interface. J Orthop Res. 2007;25:1609‐1620. [DOI] [PubMed] [Google Scholar]
- 16. Cooper JO, Bumgardner JD, Cole JA, Smith RA, Haggard WO. Co‐cultured tissue‐specific scaffolds for tendon/bone interface engineering. J Tissue Eng. 2014;5:2041731414542294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He P, Ng KS, Toh SL, Goh JC. In vitro ligament‐bone interface regeneration using a trilineage coculture system on a hybrid silk scaffold. Biomacromol. 2012;13:2692‐2703. [DOI] [PubMed] [Google Scholar]
- 18. Chen B, Li B, Qi YJ, et al. Enhancement of tendon‐to‐bone healing after anterior cruciate ligament reconstruction using bone marrow‐derived mesenchymal stem cells genetically modified with bFGF/BMP2. Sci Rep. 2016;6:25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gulotta LV, Kovacevic D, Ehteshami JR, Dagher E, Packer JD, Rodeo SA. Application of bone marrow‐derived mesenchymal stem cells in a rotator cuff repair model. Am J Sports Med. 2009;37:2126‐2133. [DOI] [PubMed] [Google Scholar]
- 20. Burrow KL, Hoyland JA, Richardson SM. Human adipose‐derived stem cells exhibit enhanced proliferative capacity and retain multipotency longer than donor‐matched bone marrow mesenchymal stem cells during expansion in vitro. Stem Cells Int. 2017;2017:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng X, Tsao C, Sylvia VL, et al. Platelet‐derived growth‐factor‐releasing aligned collagen–nanoparticle fibers promote the proliferation and tenogenic differentiation of adipose‐derived stem cells. Acta Biomater. 2014;10:1360‐1369. [DOI] [PubMed] [Google Scholar]
- 22. Deng D, Wang W, Wang B, et al. Repair of Achilles tendon defect with autologous ASCs engineered tendon in a rabbit model. Biomaterials. 2014;35:8801‐8809. [DOI] [PubMed] [Google Scholar]
- 23. Uysal CA, Tobita M, Hyakusoku H, Mizuno H. Adipose‐derived stem cells enhance primary tendon repair: biomechanical and immunohistochemical evaluation. J Plast Reconstr Aes Surg. 2012;65:1712‐1719. [DOI] [PubMed] [Google Scholar]
- 24. Yang G, Rothrauff BB, Lin H, Yu S, Tuan RS. Tendon‐derived extracellular matrix enhances transforming growth factor‐beta3‐induced tenogenic differentiation of human adipose‐derived stem cells. Tissue Eng Part A. 2017;23:166‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mihaila SM, Frias AM, Pirraco RP, et al. Human adipose tissue‐derived SSEA‐4 subpopulation multi‐differentiation potential towards the endothelial and osteogenic lineages. Tissue Eng Part A. 2013;19:235‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cowan CM, Shi YY, Aalami OO, et al. Adipose‐derived adult stromal cells heal critical‐size mouse calvarial defects. Nat Biotechnol. 2004;22:560‐567. [DOI] [PubMed] [Google Scholar]
- 27. Lima J, Gonçalves AI, Rodrigues MT, Reis RL, Gomes ME. The effect of magnetic stimulation on the osteogenic and chondrogenic differentiation of human stem cells derived from the adipose tissue (hASCs). J Magn Magn Mater. 2015;393:526‐536. [Google Scholar]
- 28. Moradi L, Vasei M, Dehghan MM, Majidi M, Farzad Mohajeri S, Bonakdar S. Regeneration of meniscus tissue using adipose mesenchymal stem cells‐chondrocytes co‐culture on a hybrid scaffold: in vivo study. Biomaterials. 2017;126:18‐30. [DOI] [PubMed] [Google Scholar]
- 29. Rada T, Reis RL, Gomes ME. Distinct stem cells subpopulations isolated from human adipose tissue exhibit different chondrogenic and osteogenic differentiation potential. Stem Cell Rev. 2011;7:64‐76. [DOI] [PubMed] [Google Scholar]
- 30. Font Tellado S, Balmayor ER, Van Griensven M. Strategies to engineer tendon/ligament‐to‐bone interface: biomaterials, cells and growth factors. Adv Drug Deliv Rev. 2015;94:126‐140. [DOI] [PubMed] [Google Scholar]
- 31. Costa‐Almeida R, Soares R, Granja PL. Fibroblasts as maestros orchestrating tissue regeneration. J Tissue Eng Regen Med. 2017;12:240‐251. [DOI] [PubMed] [Google Scholar]
- 32. Carvalho PP, Wu X, Yu G, et al. The effect of storage time on adipose‐derived stem cell recovery from human lipoaspirates. Cells Tissues Organs. 2011;194:494‐500. [DOI] [PubMed] [Google Scholar]
- 33. Costa‐Almeida R, Calejo I, Reis RL, Gomes ME. Crosstalk between adipose stem cells and tendon cells reveals a temporal regulation of tenogenesis by matrix deposition and remodeling. J Cell Physiol. 2018;233:5383‐5395. [DOI] [PubMed] [Google Scholar]
- 34. Costa‐Almeida R, Gasperini L, Borges J, et al. Microengineered multicomponent hydrogel fibers: combining polyelectrolyte complexation and microfluidics. ACS Biomater Sci Eng. 2016;3:1322‐1331. [DOI] [PubMed] [Google Scholar]
- 35. Pesqueira T, Costa‐Almeida R, Mithieux SM, et al. Engineering magnetically responsive tropoelastin spongy‐like hydrogels for soft tissue regeneration. J Mater Chem B. 2018;6:1066‐1075. [DOI] [PubMed] [Google Scholar]
- 36. Costa‐Almeida R, Berdecka D, Rodrigues MT, Reis RL, Gomes ME. Tendon explant cultures to study the communication between adipose stem cells and native tendon niche. J Cell Biochem. 2018;119:3653‐3662. [DOI] [PubMed] [Google Scholar]
- 37. Costa‐Almeida R, Calejo I, Reis RL, Gomes ME. Crosstalk between adipose stem cells and tendon cells reveals a temporal regulation of tenogenesis by matrix deposition and remodeling. J Cell Physiol. 2017;233:5383‐5395. [DOI] [PubMed] [Google Scholar]
- 38. Li CY, Wu XY, Tong JB, et al. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno‐free conditions for cell therapy. Stem Cell Res Ther. 2015;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grottkau BE, Lin Y. Osteogenesis of adipose‐derived stem cells. Bone Res. 2013;1:133‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costa‐Almeida R, Gonçalves AI, Gershovich P, Rodrigues MT, Reis RL, Gomes ME. Tendon stem cell niche In: Tissue‐Specific Stem Cell Niche. Cham: Springer; 2015:221‐244. [Google Scholar]
- 41. Bi Y, Ehirchiou D, Kilts TM, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219‐1227. [DOI] [PubMed] [Google Scholar]
- 42. Tan Q, Lui PPY, Rui YF, Wong YM. Comparison of potentials of stem cells isolated from tendon and bone marrow for musculoskeletal tissue engineering. Tissue Eng Part A. 2012;18:840‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Komori T. Regulation of osteoblast differentiation by Runx2 In: Choi Y, ed. Osteoimmunology: Interactions of the Immune and Skeletal Systems II. Boston, MA: Springer US; 2010:43‐49. [Google Scholar]
- 44. Stein GS, Lian JB, van Wijnen AJ, et al. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315‐4329. [DOI] [PubMed] [Google Scholar]
- 45. Howell K, Chien C, Bell R, et al. Novel model of tendon regeneration reveals distinct cell mechanisms underlying regenerative and fibrotic tendon healing. Sci Rep. 2017;7:45238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y, Ramcharan M, Zhou Z, et al. The role of scleraxis in fate determination of mesenchymal stem cells for tenocyte differentiation. Sci Rep. 2015;5:13149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Webb CM, Zaman G, Mosley JR, Tucker RP, Lanyon LE, Mackie EJ. Expression of tenascin‐C in bones responding to mechanical load. J Bone Min Res. 1997;12:52‐58. [DOI] [PubMed] [Google Scholar]
- 48. Bi Y, Stuelten CH, Kilts T, et al. Extracellular matrix proteoglycans control the fate of bone marrow stromal cells. J Biol Chem. 2005;280:30481‐30489. [DOI] [PubMed] [Google Scholar]
- 49. Mackie EJ, Abraham LA, Taylor SL, Tucker RP, Murphy LI. Regulation of tenascin‐C expression in bone cells by transforming growth factor‐β. Bone. 1998;22:301‐307. [DOI] [PubMed] [Google Scholar]
- 50. Silva ED, Babo PS, Costa‐Almeida R, et al. Multifunctional magnetic‐responsive hydrogels to engineer tendon‐to‐bone interface. Nanomed Nanotechnol Biol Med. 2017; 10.1016/j.nano.2017.06.00. [DOI] [PubMed] [Google Scholar]
- 51. Byers BA, Garcia AJ. Exogenous Runx2 expression enhances in vitro osteoblastic differentiation and mineralization in primary bone marrow stromal cells. Tissue Eng. 2004;10:1623‐1632. [DOI] [PubMed] [Google Scholar]
- 52. DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthr Cartil. 2000;8:309‐334. [DOI] [PubMed] [Google Scholar]
- 53. Tuan RS. Biology of developmental and regenerative skeletogenesis. Clin Orthop Relat Res. 2004;427(Suppl):S105‐S117. [DOI] [PubMed] [Google Scholar]
- 54. Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33‐44. [DOI] [PubMed] [Google Scholar]
- 55. Leboy P, Grasso‐Knight G, D'Angelo M, et al. Smad‐Runx interactions during chondrocyte maturation. J Bone Jt Surg Am. 2001;83–A Suppl:S15‐S22. [PubMed] [Google Scholar]
- 56. Zheng Q, Zhou G, Morello R, Chen Y, Garcia‐Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte‐specific expression in vivo. J Cell Biol. 2003;162:833‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rodrigues MT, Lee SJ, Gomes ME, Reis RL, Atala A, Yoo JJ. Bilayered constructs aimed at osteochondral strategies: the influence of medium supplements in the osteogenic and chondrogenic differentiation of amniotic fluid‐derived stem cells. Acta Biomater. 2012;8:2795‐2806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


