Abstract
Objective
We inspected the relevance of CD44, ABCB1 and ADAM17 in OSCC stemness and deciphered the role of autophagy/mitophagy in regulating stemness and chemoresistance.
Material and methods
A retrospective analysis of CD44, ABCB1 and ADAM17 with respect to the various clinico‐pathological factors and their correlation was analysed in sixty OSCC samples. Furthermore, the stemness and chemoresistance were studied in resistant oral cancer cells using sphere formation assay, flow cytometry and florescence microscopy. The role of autophagy/mitophagy was investigated by transient transfection of siATG14, GFP‐LC3, tF‐LC3, mKeima‐Red‐Mito7 and Western blot analysis of autophagic and mitochondrial proteins.
Results
In OSCC, high CD44, ABCB1 and ADAM17 expressions were correlated with higher tumour grades and poor differentiation and show significant correlation in their co‐expression. In vitro and OSCC tissue double labelling confirmed that CD44+ cells co‐expresses ABCB1 and ADAM17. Further, cisplatin (CDDP)‐resistant FaDu cells displayed stem‐like features and higher CD44, ABCB1 and ADAM17 expression. Higher autophagic flux and mitophagy were observed in resistant FaDu cells as compared to parental cells, and inhibition of autophagy led to the decrease in stemness, restoration of mitochondrial proteins and reduced expression of CD44, ABCB1 and ADAM17.
Conclusion
The CD44+/ABCB1+/ADAM17+ expression in OSCC is associated with stemness and chemoresistance. Further, this study highlights the involvement of mitophagy in chemoresistance and autophagic regulation of stemness in OSCC.
Keywords: ABCB1, ADAM17, autophagy, CD44, chemoresistance, stemness
1. INTRODUCTION
Regardless of the development in early detection, diagnosis and multimodal treatment strategies, oral squamous cell carcinomas (OSCC) is often associated with a high rate of locoregional recurrence, ipsilateral and bilateral lymph node metastasis and chemo‐ and radioresistance.1 The marginal improvement in the overall 5‐year survival rate of the OSCC patients and the inefficacy of the cotemporary adjuvant chemo‐radio‐therapeutics is now been ostensible to the arbitrary cytoreductive approaches that are designed to target bulk tumour cells setting aside a small subset of resistant tumour cells which could propagate the tumour post‐treatment. This resistant subpopulation of cells with exclusive self‐renewal capacity, hierarchical differentiation and tumorigenicity is referred as Cancer Stem Cells (CSCs).2
Autophagy is an evolutionary conserved lysosome‐dependent self‐digestive recycling mechanism for providing metabolite for biosynthesis and energy synthesis as a response to stress conditions. Although autophagy functions as a tumour suppressor mechanism, it also shows protumour activity to facilitate cell survival during cancer progression.2 Substantial studies suggest the significant contribution of autophagy in the resistance of tumour cells to chemotherapy.3 Moreover, increased autophagic flux is also reported to induce resistance against therapy‐induced cytotoxicity via maintaining the survival of CSCs through conferring stress tolerance and limiting damages.2 Recently, the concept selective autophagy came forward which emphasizes role of organellar autophagy like selective mitochondrial autophagy or “Mitophagy” in the context of cancer. The failure to properly modulate mitochondrial quality control via mitophagy in response to oncogenic stresses might regulate the tumorigenesis both positively and negatively.4, 5 Furthermore, chemoresistant cancer cells are reported to exhibit stem cell‐like features.6 This study elucidates the role of autophagy in inducing stemness and chemoresistance in oral cancer.
CD44, most commonly studied CSC surface marker in oral cancer, is a transmembrane receptor which binds with hyaluronic acid (HA) and signals many cancer‐related events such as proliferation, invasion and metastasis. CD44 is also associated with apoptosis resistance, drug resistance and stemness.7 The ABCB1 subfamily of ABC transporters or the P‐glycoproteins responsible for reduced intracellular accumulation of chemotherapeutics is highly expressed in CSCs. But the expressions of ABCB1 in oral cancer are not well studied.8 ADAM17 (A disintegrin and metalloproteinase) or TACE (TNF‐alpha converting enzyme), a membrane protein responsible for the cleavage CD44 ectodomain, extracellular matrix remodelling, invasion and metastasis of cancer, is also associated with inducing stemness and driving tumorigenesis in head and neck cancer.9 The purpose of this study is to investigate the clinical significance of CD44+/ABCB1+/ADAM17+ cells in resected tumour specimens of OSCC using the immunohistochemical assay and highlight the relevance of the expression profile of CD44+/ABCB1+/ADAM17+ in context of stem cell hypothesis, autophagic regulation of stemness and chemoresistance in OSCC. More importantly, this study emphasizes that selective autophagy like mitophagy might be responsible for chemoresistance in cancer.
2. MATERIALS AND METHODS
2.1. Patients and tumour specimen
Archival formalin‐fixed and paraffin‐embedded (FFPE) tumour specimens from sixty cases of OSCC were collected from Drs. Tribedi and Roy Diagnostic Laboratory, Kolkata, local hospitals, nursing homes and clinics near the Ranchi area and Bangalore, India. The material had been stored with permission of the local ethics committee after informed consent obtained from the patients prior to surgical resection. Tumour and patient characteristics are summarized in Table S1. The tumour blocks of paraffin‐embedded tissue were selected by experienced pathologists, evaluating the routine H.E.‐stained sections. Sections from all available tumours underwent intensive histopathologic assessment, blinded to the prior histopathology report. Serial tissue sections (5 μm thickness) were cut from FFPE blocks on a microtome and mounted from warm water onto adhesive microscope slides. Tumour Grading was performed according to WHO criteria. Tumour characteristics and patient characteristics were collected in a database (Excel, Microsoft) for further analysis.
2.2. Immunohistochemistry
2.2.1. Immunostaining
For immunohistochemical analysis, unconjugated specific ABCB1 (1:100, abcam # ab155421), ADAM17 (1:100, abcam # ab574821) (Abcam, Inc., Cambridge, UK) and CD44 antibody (1:100, Immunotools # 21810441) were used. Pretreatment and immunohistochemical single staining procedure were performed as described in the Super Sensitive™ Polymer HRP IHC Detection System procured from Biogenex, Fremont, CA, USA.
2.2.2. Assessment standard
Each stained tumour specimen was photographed for 6 randomly selected fields with Olympus IX71 microscope, and all assessment were made at a magnification of ×200. The average of their evaluations was calculated for statistical analysis. Each slide was evaluated independently by 2 independent observers who did not know the clinical outcomes. The expression of each antibody was evaluated for each tissue sample by calculating the total Immuno‐reactive Score (IRS) as the product of the proportion and intensity scores. The proportion score reflected the estimated fraction of positive‐stained tumour cells (0, none; 1, 1%‐25%; 2, 26%‐50%; 3, 51%‐75%; 4, 76%‐100%). The intensity score represented the estimated staining intensity (0, no staining; 1, weak/light yellow; 2, moderate/buffy; 3, strong/brown).10 The total IRS ranged from 0 to 12, and the scores were averaged. Scores of 0‐5 were designated as low expression, while scores of 6‐12 were designated as high expression. Moreover, IRS scores: 0 is considered (−); 1 to 4 (+); 5 to 8 (++); and 9 to 12 (+++).
2.3. Cell culture
The human hypopharyngeal cancer cell line FaDu was procured from American Type Culture Collection ([ATCC], Manassas, USA). FaDu cells were cultured in MEM complemented with 10% fetal bovine serum and antibiotic‐antimycotic solution followed by incubation at 37°C in a humidified (95% air: 5% CO2) incubator.
2.4. Establishment of CDDP resistance in OSCC cell line
FaDu cells were repeatedly subcultured in MEM containing increasing concentrations cis‐Diammineplatinum (II) dichloride (CDDP/cisplatin) (Sigma‐Aldrich, St. Louis, MO, USA) over a 3‐month period. FaDu cells that grew in 0.5 μM CDDP were designated as FaDu CDDP resistant (FaDu CDDP‐R) and maintained in MEM containing 0.5 μM cisplatin, whereas the parental FaDu cells were designated as FaDu‐P.
2.5. Drug sensitivity assay
FaDu‐P and FaDu‐CDDP‐R cell concentration was adjusted to 5 × 104 cells/mL, plated in 96‐well flat‐bottom culture plates and incubated with various concentrations of CDDP for 72 h. The effect of CDDP on cancer cell viability was studied using MTT dye reduction assay by measuring the optical density at 595 nm using a micro‐plate reader spectrophotometer (Perkin‐ Elmer, Walthman, MA, USA).
2.6. Sphere formation assay
FaDu‐P and FaDu‐CDDP‐R cells were cultured in serum‐free medium supplemented with 2% B27 supplement (Gibco, Gaithersburg, MD 20877, USA), 1% N2 supplement (Gibco, Gaithersburg, MD 20877, USA), 20 ng/mL epidermal growth factor (EGF; BD Biosciences, Franklin Lakes, NJ, USA) and 20 ng/mL basic fibroblast growth factor‐2 (bFGF‐2; BD Biosciences Franklin Lakes, NJ, USA) and maintained in ultralow attachment plates (Corning, NY 14831, USA) at 37°C in a humidified 5% CO2 incubator. After 10 days of culture, the number of non‐adherent spherical clusters of cells or orospheres was counted under a microscope.
2.7. Immunocytofluorescence staining and analysis
FaDu‐P and FaDu‐CDDP‐R cells were fixed with 10% formaldehyde, permeabilized by 0.1% Triton X‐100, and then incubated with the primary antibodies for CD44 (1:500, Immunotools # 21810441), ABCB1 (1:500, abcam # ab155421), ADAM17 (1:500, abcam # ab574821), β‐catenin (1:500, BD Biosciences # 610153), LC3B (1:500; Novus Biologicals # NB‐100‐2220), TOM20 (1:500, BD Biosciences # 612278). The secondary anti‐rabbit and/or anti‐mouse antibodies conjugated with Alexa Fluor (1: 500; A11001, A11004, A11011 and A11008) from Invitrogen were used to study the fluorescence of our desired proteins, which were detected using an Olympus IX71 fluorescent inverted microscope and the cellSens Standard software (version 1.6, Olympus Soft Imaging Solutions GmbH, Johann‐ Krane‐Weg, Münster, Germany). Immunofluorescence double staining tissue for CD44, ABCB1 and ADAM17 were done as described in Grimm et al11. The immunocytofluorescent intensities were quantified by measuring the integrated optical density and area fraction with the ImageJ software (National Institute of Mental Health, Bethesda, MA, USA).12
2.8. Flow cytometry
The FaDu‐P cells and FaDu‐CDDP‐R cells were harvested and incubated with PE‐labelled CD44 antibodies (BD Biosciences # 550989) for 30 min at room temperature in dark. Then, the cells were washed with ice‐cold PBS (10 mM, pH 7.4) and re‐suspended in 500 μL of PBS. The expression of CD44 and percentage of CD44+ population were analysed with the help of BD ACCURI C6 flow cytometer and FCS EXPRESS software.
2.9. Western blot
Samples containing equal amounts of protein were subjected to SDS‐PAGE and transferred onto PVDF membrane. The blots were incubated with primary antibody against LC3B (1:1000; Novus Biologicals # nb‐100‐2220), SQSTM1/p62 (1:1000, BD Biosciences # 610832), ULK1 (1:1000, Cell Signaling and Technology # 8054S), Beclin 1 (1:1000, Cell Signaling and Technology # 3738), Atg5 (1:1000, Cell Signaling and Technology # 2630S), Atg7 (1:1000 Biogenesis‐R‐161‐100), ATG14 (1:1000, Cell Signaling and Technology # 5504BC), COX‐IV (1:1000, Cell Signaling and Technology # 4844S), actin (1:1000, Sigma‐Aldrich # A2066) followed to peroxidase labelled anti‐mouse (1:1000, Cell Signaling and Technology # 7076)/anti‐rabbit (1:1000, Cell Signaling and Technology # 7074) secondary antibodies. The Western blot band intensities were quantified by measuring the integrated optical density and area fraction with the ImageJ software (National Institute of Mental Health).12
2.10. Small‐interfering RNA and plasmid transfection
The indicated FaDu cells were transfected with siRNA for ATG14 by using Lipofectamine 2000 reagent following the manufacturer's instructions. Control‐siRNA was used as a negative control. The tandem GFP‐RFP‐LC3 transfection was performed to study the autophagic flux in parental and resistance FaDu cells, and mKeima‐Red‐Mito7plasmid transfection was carried out to investigate the lysosomal delivery of mitochondria to be degraded by autophagy. A corresponding backbone plasmid was used as negative control for each plasmid transfection.
2.11. Statistical analysis
The data are presented as the mean ± SD. Statistical comparisons between subgroups were made using the appropriate statistical test (chi‐square, chi‐square for linear trend tests and Fisher exact test). The correlation between the marker proteins was analysed by Spearman rank test. Student's t test was used for evaluating statistical differences between experimental groups. The analysis was performed by GraphPad Prism 4.0 software. A 2‐tailed P‐value test was used in all analyses, and the P value was defined as follows: not significant (n.s.): P > 0.05;*: P ≤ 0.05; **: P ≤ 0.01; ***: P ≤ 0.001; ****: P ≤ 0.0001 were considered statistically significant.
3. RESULTS
3.1. Clinical presentation of disease in this cohort
Numerous clinical characteristics of our cohort are described in Table S1. The median age of the patients was 51 years. Out of 60 cases, 27 (45%) patients were ≤51 years, 22 (36.6%) patients were above 51 years, whereas the age details of 11 (18.33%) patients were not available. Our cohort included 30 (50%) male patients and 25 (41.66%) female patients and the gender details of 5 (8.33%) cases were not available. With respect to the histological characteristics and grading of the cancers showed that within our cohort, 32 (53.33%) patients had Grade I (well differentiated) OSCC, 19 (31.66%) had Grade II (moderately differentiated) and 9 (15%) patients had Grade III (poorly differentiated) tumours. We concluded that this cohort was suitable for determining the relative usefulness of CD44, ABCB1 and ADAM17 expression as an indicator in CSC hypothesis and chemoresistance.
3.2. CD44, ABCB1 and ADAM17 expressions and their association with clinico‐pathological features of OSCC
Expression of CD44 was mostly membranous, and the positive expression rate of CD44 in OSCC was 90% (54/60). The IRS of cancer cells with CD44 expression ranged from 0 to 12 (mean 7.73, median 8). The 85% (51/60) cases were considered positive for the expression of ABCB1. The IRS of the cancer cells with ABCB1 expression ranged from 0 to 12 (mean 4.5, median 4). Likewise, 91.7% (55/60) cases were ADAM17 positive. The IRS of the cancer cells with ADAM17 expression ranged from 0 to 12 (mean 6.11, median 8) (Table S2). The expression of CD44 (Figure 1A), ABCB1 (Figure 1B) and ADAM17 (Figure 1C) apparently increased in a grade‐wise manner in OSCC tissue samples. The expression profiles of CD44, ABCB1 and ADAM17 were similar in OSCC tissue. The high expression percentages of CD44, ABCB1 and ADAM17 in Grade I (well differentiated) OSCC tumours were 31.25% (19/32), 15.62% (5/32) and 40.62% (13/32), respectively, which were significantly lower than the Grade II (moderately differentiated) OSCC tumours: 78.94% (15/19), 52.63% (10/19) and 73.68% (14/19), respectively. The CD44, ABCB1 and ADAM17 high expression in Grade III (poorly differentiated) OSCC tumours were 100% (9/9), 88.88% (8/9) and 88.88% (8/9), respectively, which were considerably high than Grade I and Grade II OSCC tumours with all P < 0.05 (P = 0.0129, P = 0.0001, P = 0.0026). However, expression profiles of CD44, ABCB1 and ADAM17 did not associate with age and sex of the patients (P all > 0.05) (Table 1).
Figure 1.
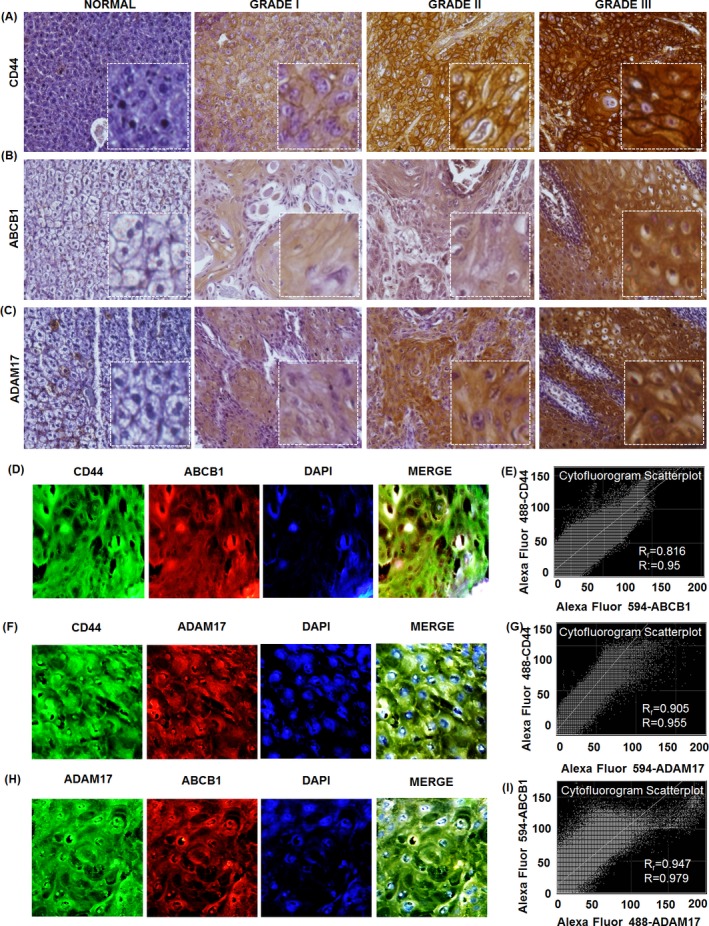
Expression of CD44, ABCB1 and ADAM17 in normal oral tissue and oral squamous cell carcinoma tissue and their co‐expression. Slide shows representative images of CD44 (A), ABCB1 (B) and ADAM17 (C) staining in normal oral tissue and different grades of OSCC tissue samples. Brown chromogen colour (3,3‐Diaminobenzidine) indicates positive CD44, ABCB1 and ADAM17 staining and the purple colour indicates the nuclear counterstaining by haematoxylin. The square box demonstrates the area of interest shown in larger magnification. Images demonstrate a representative immunofluorescent double labelling of indicated proteins and their cytofluorogram scatter plot depicting the co‐expression (D‐I)
Table 1.
Relationship between CD44, ABCB1 and ADAM17 and the clinico‐pathological features OSCC
| Groups | Cases | CD44 | ABCB1 | ADAM17 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | Low | High | P value | ||
| Gender | 55a | — | — | 0.5616 | — | — | 0.5946 | — | — | 0.7903 |
| Male | 30 | 8 | 22 | 19 | 11 | 12 | 18 | |||
| Female | 25 | 9 | 16 | 14 | 11 | 11 | 14 | |||
| Age | 49b | — | — | 0.1317 | — | — | 1.000 | — | — | 0.5675 |
| ≤51 | 26 | 12 | 14 | 17 | 9 | 13 | 13 | |||
| >51 | 23 | 5 | 18 | 15 | 8 | 9 | 14 | |||
| Tumour Grade | 60 | — | — | 0.0129* | 0.0001**** | — | — | 0.0026** | ||
| Grade I | 32 | 13 | 19 | 27 | 5 | 19 | 13 | |||
| Grade II | 19 | 4 | 15 | 9 | 10 | 5 | 14 | |||
| Grade III | 9 | 0 | 9 | 1 | 8 | 1 | 8 | |||
Five cases of OSCC patients without gender details.
Eleven cases of OSCC patients without age details.
*P ≤ .05; **P ≤ .01; ****P ≤ .0001.
Next, we investigated whether there is any link between CD44, ABCB1 and ADAM17 with STRING 10.5 (https://string-db.org/) protein‐protein interaction online software. Protein‐protein interaction (PPI) enrichment P‐value for CD44, ABCB1 and ADAM17 interaction is 0.00523, indicating that these proteins have more interactions among themselves than what would be expected for a random set of proteins of similar size, drawn from the genome (Figure S1). From the statistical analysis, we found a positive correlation between CD44 (or ADAM17) and ABCB1, as well as between CD44 and ADAM17 in OSCC. The CD44 and ABCB1 share a moderate positive correlation (Spearman correlation coefficient or r s: 0.574), whereas CD44 and ADAM17 and CD44 and ABCB1 share a weak positive relation with r s 0.428 and 0.346, respectively, all P < 0.01) (Table S3). Furthermore, pairwise correlation analysis of these 3 proteins shows that the high expression of CD44/ABCB1, CD44/ADAM17 and ABCB1/ADAM17 was correlated with tumour grades and tumour differentiation (all P < 0.05). Also, the pairwise correlation analysis of these 3 proteins did not show a relationship with age and sex of the patients (all P > 0.05) (Table 2). Further, we analysed the correlation between the triple high expression (tissue samples having high expression status for all 3 proteins) and triple non‐high expression (tissue sample having at least one low expression status among the 3 proteins). It is observed that triple high expression of CD44/ABCB1/ADAM17 was remarkably correlated with tumour grades and tumour differentiation (all P < 0.05). However, it did not show any relation with age and sex of the patient (all P > 0.05) (Table 3).
Table 2.
Relationship between CD44/ABCB1, ABCB1/ADAM17 and CD44/ADAM17 expression and the clinico‐pathological features of OSCC
| Groups | Age in yearsa | Genderb | Tumour grades | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤51 | >51 | P value | Male | Female | P value | Grade I | Grade II | Grade III | P value | |
| CD44/ABCB1 | ||||||||||
| CD44HighABCB1High | 7 | 6 | 0.1769 | 11 | 8 | 0.7207 | 4 | 8 | 8 | 0.0007*** |
| CD44HighABCB1Low | 7 | 12 | 11 | 8 | 15 | 7 | 1 | |||
| CD44LowABCB1High | 2 | 1 | 0 | 3 | 1 | 2 | 0 | |||
| CD44LowABCB1Low | 10 | 4 | 8 | 6 | 12 | 2 | 0 | |||
| ABCB1/ADAM17 | ||||||||||
| ABCB1HighADAM17High | 6 | 4 | 0.6950 | 9 | 6 | 0.7359 | 1 | 7 | 7 | 0.0001*** |
| ABCB1HighADAM17Low | 3 | 3 | 2 | 5 | 4 | 3 | 1 | |||
| ABCB1LowADAM17High | 7 | 10 | 9 | 8 | 12 | 7 | 1 | |||
| ABCB1LowADAM17Low | 10 | 6 | 10 | 6 | 15 | 2 | 0 | |||
| CD44/ADAM17 | ||||||||||
| CD44HighADAM17High | 8 | 14 | 0.9369 | 16 | 10 | 0.4762 | 9 | 12 | 8 | 0.0346* |
| CD44HighADAM17Low | 6 | 4 | 6 | 6 | 10 | 3 | 1 | |||
| CD44LowADAM17High | 5 | 1 | 2 | 4 | 4 | 2 | 0 | |||
| CD44LowADAM17Low | 7 | 4 | 6 | 5 | 9 | 2 | 0 | |||
| Cases | 26 | 23 | 30 | 25 | 32 | 19 | 9 | |||
Eleven cases of OSCC patients without age details.
Five cases of OSCC patients without gender details.
*P ≤ .05; ***P ≤ .001.
Table 3.
Relationship between triple high expression and triple non‐high expression of CD44/ABCB1/ADAM17 and the clinico‐pathological features of OSCC
| Groups | Age in years | Gender | Tumour grades | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤51 | >51 | P value | Male | Female | P value | Grade I | Grade II | Grade III | P value | |
| CD44/ABCB1/ADAM17 | ||||||||||
| Triple high cases | 22 | 19 | 1.000 | 21 | 21 | 0.3406 | 30 | 14 | 2 | 0.0001**** |
| Triple non‐high cases | 4 | 4 | 9 | 4 | 2 | 5 | 7 | |||
| Cases | 26 | 23 | 30 | 25 | 32 | 19 | 7 | |||
Five cases of OSCC patients without gender details; 11 cases of OSCC patients without age details.
****P ≤ .0001.
To evaluate the hypothesis that putative CD44+ CSC are associated with ABCB1 and ADAM17 expression, immunofluorescent double‐labelling experiments were operated in OSCC tissue sections and cell lines. We found a dominant population of CD44+/ABCB1+ tumour cells (Figure 1D,E) with Pearson's coefficient of 0.816 and overlap coefficient of 0.95 and CD44+/ADAM17+ tumour cells with Pearson's coefficient of 0.905 and overlap coefficient of 0.955 (Figure 1F,G) in OSCC tissues indicating that CD44+ cells highly co‐expresses ABCB1 and ADAM17. Moreover, we observed the co‐expression of ABCB1 and ADAM17 in OSCC tissue samples (Figure 1H,I) with Pearson's coefficient of 0.947 and overlap coefficient of 0.979. Further, immunohistochemical double staining was revaluated in FaDu cells and CD44+/ABCB1+ (Figure S2A,B) and CD 44+/ADAM17+ (Figure S2C,D) co‐expressing population as well as ABCB1+/ADAM17+ co‐expression in FaDu cell (Figure S2E,F) was observed.
3.3. CDDP‐resistant cells are bestowed with cancer stem‐like features and increased expression of CD44, ABCB1 and ADAM17
Therapeutic resistance is a major concern encountered during the treatment of OSCC. To gain further insights into the mechanisms of chemoresistance and its correlation with stemness, we established the cisplatin (CDDP)‐resistant FaDu cell lines (FaDu‐CDDP‐R). The parental FaDu (FaDu‐P) cells were treated with incremental concentration of cisplatin ranging from 0.01 μM to a final concentration 0.5 μM for a period of 3 months to generate FaDu‐CDDP‐R cells. Once the resistant phenotype was established, the cells were maintained by continuous treatment of 0.5 μM of CDDP. To confirm the sensitivity of FaDu‐CDDP‐R to CDDP exposure, we performed MTT assay to assess the drug sensitivity in terms of cell viability of parental and resistant cell line against CDDP treatment (1‐5 μM). As shown in Figure 2A, parental FaDu cells (FaDu‐P) were found to be significantly more sensitive to CDDP than the resistant FaDu cells (FaDu‐CDDP‐R). Moreover, it is reported that mild therapeutic stress can induce stem‐like, drug‐tolerant stress‐response states.13 To further investigate the effect of CDDP exposure on acquisition of stemness in OSCC, the FaDu‐P and FaDu‐CDDP‐R cells were plated onto ultralow attachment plates and sphere forming activity was observed and quantified after for 10 days. Notably, the resistant FaDu cells produced larger spheres than the parental FaDu cells (Figure 2B). We further noticed that FaDu‐CDDP‐R formed more orospheres (117 ± 11) when cultured in ultralow attachment plates, as compared with the parental FaDu P cells (44 ± 5) (Figure 2C). Furthermore, we analysed the expression of stemness marker and self‐renewal marker β‐catenin expression in both resistant and parental cell line through immunofluorescence microscopy and detected that FaDu‐CDDP‐R cells displayed higher β‐catenin expression than the FaDu‐P cell (Figure 2D). Figure 2E represents the corrected total cell fluorescence (CTCF) of β‐catenin expression in parental FaDu cells and FaDu‐CDDP‐R cells showed significantly higher β‐catenin expression in terms of florescence intensity as compared with FaDu‐P cells. The higher CD44 expression as examined by flow cytometric analysis in FaDu‐CDDP‐R cells further confirmed the CDDP induced stemness in oral cancer (Figure 2F). In order to further advocate our hypothesis, we studied the changes in CD44+ population in FaDu‐P and FaDu‐CDDP‐R group. CD44+ cells were reported to possess stem‐like properties in various experimental setups. Excitingly, our flow cytometric analysis indicated an increase in percentage of CD44+ population in FaDu‐CDDP‐R group against FaDu‐P which goes as 90 ± 2.6 and 36 ± 6.2, respectively. Similarly, the percentage of CD44− population decreased from 64 ± 2.6 in FaDu‐P group to 9 ± 2.6 in FaDu‐CDDP‐R group, suggesting therapy‐induced stemness in OSCC (Figure 2G,H). To further correlate the expression of CD44, ABCB1 and ADAM17 with chemoresistance, we executed the immunofluorescence analysis of FaDu‐P and FaDu‐CDDP‐R cells and discovered that FaDu‐CDDP‐R had higher level of expression of stemness marker CD44 (Figure 2I,J), drug‐resistant marker ABCB1 (Figure 2K,L) and invasion mediator ADAM17 (Figure 2M,N) than the FaDu‐P cells.
Figure 2.
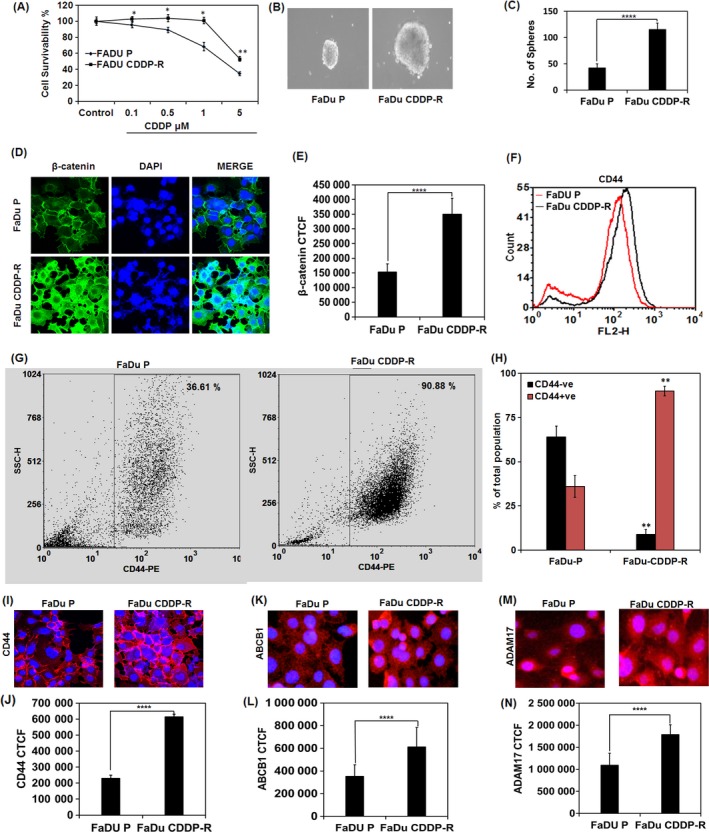
Cisplatin‐resistant cells are endowed with cancer stem‐like features and increased CD44, ABCB1 and ADAM17 expression. The FaDu‐P and FaDu CDDP‐R cells were treated with different concentration of CDDP for 72 h, and the cell viability was measured by MTT dye reduction assay (A). The FaDu P and FaDu CDDP‐R cells were grown in ultralow attachment plate in stem cell media for 10 days, and the number and size of the orosphere produced were quantified (B, C). Expression of β‐catenin in FaDu P and FaDu CDDP‐R cells (D). The corrected total cell fluorescence (CTCF) of β‐catenin expression as analysed by Image J software is depicted (E). Flow cytometric analysis of CD44 expression in FaDu‐P and FaDu‐CDDP‐R cells (F). Flow cytometric analysis of CD44+ population with bar diagram represents the comparison between CD44+ and CD44− population in FaDu‐P and FaDu CDDP‐R cells (G, I). Immunofluorescence image of indicated protein expression and their quantification in FaDu‐P and FaDu CDDP‐R cells (I‐N). **P value ≤0.01 was considered significant when compared between FaDU‐P and Fadu‐CDDP‐R
3.4. CDDP‐mediated autophagy regulates stemness of resistant cancer cells and expression of CD44, ABCB1 and ADAM17
Next, we investigated the autophagic response in FaDu‐P and FaDu‐CDDP‐R cells. From the immunofluorescence analysis, we found augmented formation of punctate structure of LC3, a hallmark of autophagy induction in FaDu‐CDDP‐R cells with FaDu‐P cells showed a diffused LC3 fluorescence with lesser puncta (Figure 3A). Approximately 24 ± 1.8% FaDu‐P cells displayed LC3B‐II puncta per cell, whereas it increased to 57 ± 5.9% in FaDu‐CDDP‐R cells (Figure 3B). Next, we evaluated the extent of autophagosome and autolysosome formation in terms of fluorescent LC3 puncta. Both the numbers of GFP and RFP puncta per cell were found in remarkable amount in FaDu‐CDDP‐R cells (Figure 3C). The number of RFP+GFP+ (yellow) puncta was considerably higher in FaDu‐CDDP‐R cells indicating increased autophagosome formation (Figure 3C,D). Moreover, augmented autolysosome formation and higher autophagic flux in terms of the number of RFP+GFP− (red) puncta per cell also significantly increased in FaDu‐CDDP‐R group as compared with their parental counterparts (Figure 3C,D). Further validating our hypothesis, the Western blot analysis also displayed increased conversion of LC3B‐I to LC3B‐II in FaDu‐CDDP‐R cells (Figure 3C,D). Moreover, authenticating the occurrence of increased autophagic flux in resistant cells, Western blot analysis showed noteworthy increase in the expression of autophagic signature molecules including ULK1, Beclin 1, ATG5, ATG7 and ATG14 in FaDu‐CDDP‐R cells as compared with FaDu‐P cells (Figure 3E). In addition, enhanced degradation of p62 could be seen in FaDu‐CDDP‐R cells as compared with FaDu‐P cells, confirming FaDu‐CDDP‐R cells exhibit greater autophagic flux (Figure 3E). To further investigate the role of autophagy in chemoresistance, we transiently down regulated autophagy induction by using siATG14 (Figure 3F) and studied the status of autophagy in siATG14 transfected cells and our data showed decreased formation LC3B‐II puncta in ATG14 knocked down FaDu‐CDDP‐R cells as compared with FaDu‐P cells. The percentage of cells positive for LC3B‐II puncta decreased from 33 ± 2.8 to 18 ± 1.3 in parental FaDu cells and 78 ± 7.2 to 0.45 ± 4.1 when autophagy is inhibited (Figure 3G,H).
Figure 3.
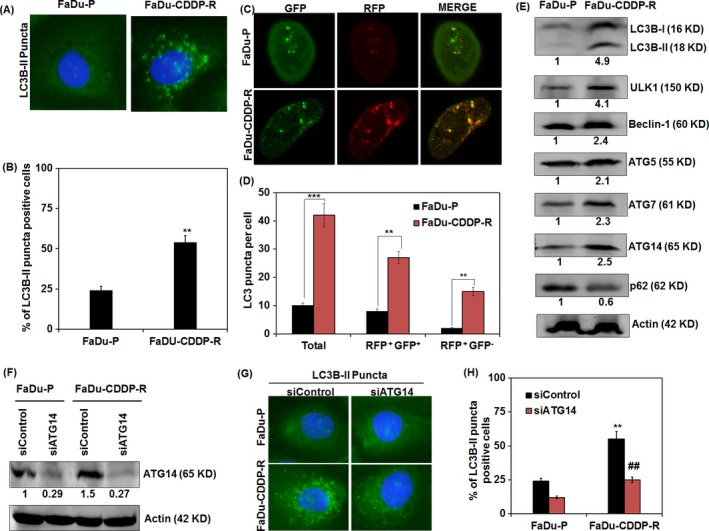
Role of autophagy in chemoresistance in FaDu cells. Fadu‐P and Fadu‐CDDP‐R cells were analysed for the autophagy induction in terms of punctate florescence of LC3B‐II expression (A) and quantification of % of LC3B‐II puncta positive cells (B). Autophagic flux analysis in parental and resistant FaDu cells was done after tF‐LC3 transfection, and the numbers of RFP + GFP + (yellow) and RFP + GFP − (red) puncta per cell were representing autophagosome and autolysosome, respectively, were counted (C and D). Western blot analysis of autophagic protein LC3B, ULK1, Beclin1, ATG5, ATG7, ATG14 and p62 is depicted (E). Western blot image depicts the down regulation of ATG14 with transient transfection of siATG14 in FaDU‐P and FaDu‐CDDP‐R cells (F). Immunofluorescence analysis of LC3B‐II puncta formation in FaDU‐P and FaDu‐CDDP‐R cells in siControl and siATG14 transfected condition were evaluated (G). % of LC3B‐II puncta positive cells in siControl and ATG14‐deficient FaDU‐P and FaDu‐CDDP‐R cells was quantified (H). Densitometry was performed on the original blots; the ratio of protein to actin in control cells was considered as 1. **P value < 0.01 was considered significant when compared between FaDu‐P and FaDu‐CDDP‐R group. ## P value < 0.01 was considered significant when compared between FaDu‐CDDP‐R and siATG14‐ FaDu‐CDDP‐R
We further investigated mitophagic activity in CDDP‐resistant FaDu cells through immunostaining of translocase of the outer mitochondrial membrane 20 (TOM20), an integral part of outer mitochondrial membrane. Our data showed that the percentage of cells with less/no mitochondrial TOM20 staining increased from 23 ± 2.2 to 60 ± 3.9 (Figure 4A,B) in FaDu‐CDDP‐R indicating the occurrence of mitophagy. In this connection, we analysed the expression of another mitochondrial marker COX‐IV (cytochrome c oxidase subunit IV) which is an integral part of inner mitochondrial membrane in CDDP‐resistant FaDu cells. From the Western blot analysis, it is clear that FaDu‐CDDP‐R cells have significantly lesser expression of COX‐IV as compared with its parental counterpart (Figure 4C). To further substantiate our hypothesis that mitophagy plays a significant role in chemoresistance, we assayed functional mitophagy by using mitochondria‐targeted mKeima‐Red‐Mito7, a lysosomal proteases resistant probe whose fluorescence shifts from green to red in acidic pH as an indicative of the mitochondrial delivery into the maturing lysosome. As expected, a considerable increase in red mKeima‐Red‐Mito7 puncta per cell was found in FaDu‐CDDP‐R cells as compared with FaDu‐P cells (Figure 4D,E). To further validate the induction of CDDP‐mediated mitophagy in resistant cells, we investigated the expression of TOM20 in ATG14‐deficient cells. The percentage of cells with less/no mitochondrial TOM20 in FaDu‐P and FaDu‐CDDP‐R groups were 33 ± 2.8 and 78 ± 7.2 in siControl group. Intriguingly, the CDDP induced reduction in mitochondrial mass was reversed in siATG14 transfected group and the percentage of cells with less/no mitochondrial TOM20 in FaDu‐P and FaDu‐CDDP‐R reduced to 18 ± 1.3 and 45 ± 4.1 (Figure 4F,G). Similar to the TOM20 expression, the immunoblotting showed efficient restoration COX‐IV expression in ATG14‐deficient FaDu‐CDDP‐R cells indicating the potential association of mitophagy in chemoresistance of oral cancer (Figure 4H).
Figure 4.
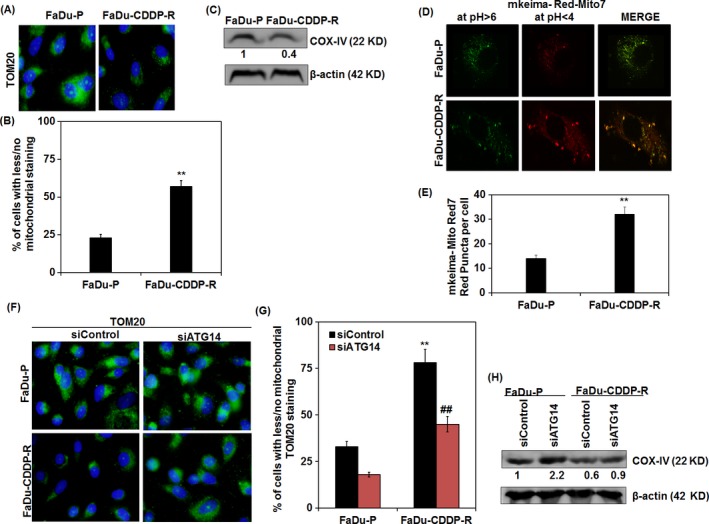
Mitophagy regulates chemoresistance in FaDu cells. Immunofluorescence expression (A) and quantification of outer mitochondrial marker protein TOM20 in FaDU‐P and FaDu‐CDDP‐R (B) were analysed. Western blot analysis of inner mitochondrial marker protein COX‐IV was investigated (C). The mitochondrial delivery to lysosome in order to measure the mitophagic flux (in terms of increased red puncta) was assayed by employing mKeima‐Red‐Mito7 (D and E). Immunofluorescence expression of TOM20 with quantification of % of cells with less/no mitochondrial TOM20 staining in FaDU‐P and FaDu‐CDDP‐R cells after transient transfection of siControl and siATG14 are depicted (F and G). Western blot analysis of COX‐IV expression in ATG14‐deficient FaDU‐P and FaDu‐CDDP‐R cells is represented (H). Densitometry was performed on the original blots, the ratio of protein to actin in control cells was considered as 1. **P value < 0.01 was considered significant when compared between FaDu‐P and FaDu‐CDDP‐R groups. ## P value < 0.01 was considered significant when compared between FaDu‐CDDP‐R and siATG14‐ FaDu‐CDDP‐R
In order to justify our hypothesis, we inhibited autophagy and performed the spheroid formation assay. Interestingly, the numbers of orospheres produced were remarkably lower in ATG14‐deficient FaDu‐CDDP‐R cells (86 ± 7.3) as compared to siControl group (266 ± 18.3) (Figure 5A). We then further analysed the self‐renewal ability of capacity of FaDu‐CDDP‐R cells in terms of β‐catenin expression. Our immunofluorescence data showed a considerable decrease in the β‐catenin expression in ATG14‐deficient‐resistant FaDu cells against the siControl group, indicating the potential involvement of autophagy in maintaining self‐renewal ability which is an indispensable characteristic of CSCs (Figure 5B,C). The inhibition of autophagy in FaDu‐CDDP‐R cells was found to effectively down regulate the stemness surface marker CD44 (Figure 5D,E) along with the down regulation of drug resistance marker ABCB1 (Figure 5F,G) and invasion mediator ADAM17 (Figure 5H,I), suggesting that autophagy regulates the expression of these proteins to induce stemness and chemoresistance. However, further study is required to understand how autophagy modulates the expression CD44, ABCB1 and ADAM17 during chemoresistance.
Figure 5.
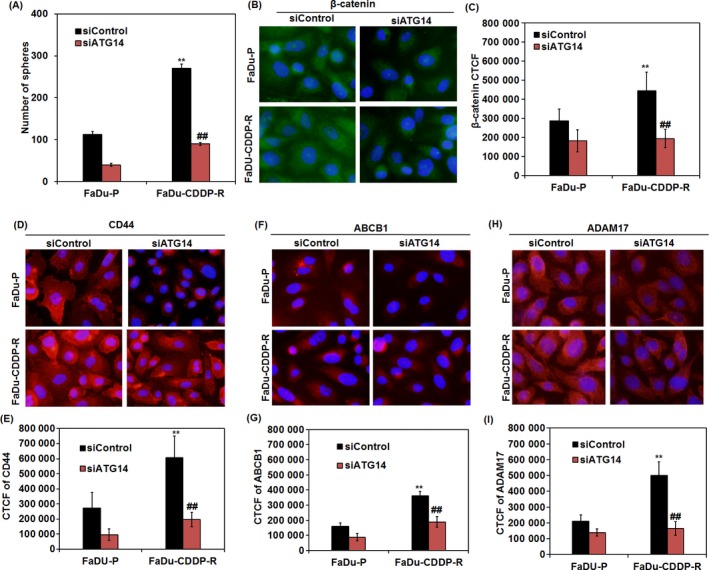
Autophagy regulates stemness and expression of CD44, ABCB1 and ADAM17 in CDDP‐resistant FaDu cells. After knock down of ATG14, FaDu P and FaDu CDDP‐R cells were grown in ultralow attachment plate in stem cell media for 10 days and the number orospheres produced were quantified (A). Immunofluorescence analysis of β‐catenin, CD44, ABCB1 and ADAM17 in ATG14‐deficient FaDU‐P and FaDu‐CDDP‐R cells and their corresponding quantification was analysed (B‐I). **P value < 0.01 was considered significant when compared between FaDu‐P and FaDu‐CDDP‐R group. ## P value < 0.01 was considered significant when compared between FaDu‐CDDP‐R and siATG14‐ FaDu‐CDDP‐R
4. DISCUSSION
Most of the cotemporary therapeutic regimens target only the bulk tumour mass leaving behind the assassin CSCs that serve as a reservoir for tumour repopulation post‐therapy. Likely, CSCs in oral cancer were reported to impose a great therapeutic implication in cancer relapse and patient survival.14 A correlation between CD44 expression and poor prognosis has been reported in many cancers.15, 16 Moreover, CD44 may also symbolize a potential marker of OSCC stem cells.17, 18 This study confirms the expression of stem cell surface marker CD44 expression in OSCC tissue samples. Moreover, CD44 expression in OSCC was found to be associated with tumour grade and tumour differentiation. Again, CSCs have been postulated to be drug resistance phenotypes by virtue of the expression of several drug efflux transporters.19, 20 Previous study showed that expression of ABCB1 marker correlates with the degree of differentiation in gastric cancer.8 Similarly, in our study, the expression of ABCB1 highly correlated with the degree of differentiation of OSCC tissue. Moreover, the upregulated expression of ADAM17 was shown to be associated with poor prognosis of cancers.21, 22 Likewise, we found that ADAM17 expression in OSCC significantly correlated with degree of cancer progression and aggressiveness.
While CD44 populations may indeed encompass a subpopulation of CSCs, by itself it does not appear to be an adequate stem cell marker for OSCC. Hence, the histological or clinical implications of CD44+/ABCB1+/ADAM17+ cells as putative CSCs in OSCC and their role in chemoresistance have been elucidated in this study. Higher tumour grades and poor tumour differentiation are one of the basic indicative features of cancer cell aggressiveness.23, 24 A remarkable expression of CD44+/ABCB1+/ADAM17+ was obvious in higher OSCC tumour grade. Our data advocate that tumours with higher grades are more likely to encompass CSCs than the lower grade tumours. Likewise, marked CD44+/ABCB1+/ADAM17+ expression was evident in the component of OSCC showing poor differentiation. This study also elucidates that morphologically, CSCs may show poorly differentiated (dedifferentiated) characteristics. This is consistent with a report by Ohara et al. indicating that poorly differentiated colon cancers contained more CSCs than well‐differentiated tumours.25 Therefore, our data imply that poor differentiation might be common characteristics of CSCs in clinical specimens.
Our study showed that the resistant FaDu cells had more stem characteristics as compared to parental cells as evident with the higher sphere forming capacity, CD44 and β‐catenin expression. Accordingly, Nor et al. reported that stemness attributes of cancer cells is highly influenced by chemotherapeutic drugs and anticancer therapies and elucidated that cisplatin could induce the stem cell fraction in UM‐SCC‐22B HNSCC cells.26 Moreover, the increased expression of CD44, ABCB1 and ADAM17 in resistant FaDu cells emphasized the role of these proteins in chemoresistance. Likely, chemoresistance in colon carcinoma cells was reported to be mediated by CD44 survival signalling via Lyn Kinase and phosphoinositide 3‐kinase/Akt.27 In addition, it reported that CD44 expression was associated with chemoresistance in T‐cell acute lymphoblastic leukaemia mediated in part through enhanced drug efflux system.28 The chemoresistance in CSCs is mainly alleged to the higher expression of ABC drug efflux transporters.2 The expression of ABCB1 in colorectal cancer has been shown to promote stemness and chemoresistance.29 In consistent with the previous report,9 ADAM17 expression was also reported to be involved in stemness and chemoresistance in OSCC.
Many reports claim that autophagy in cancer cells promotes chemoresistance. Likely, our data show that resistant FaDu cells had higher autophagic flux as compared with parental counterpart which is in accordance with a report by Wang et al. which document that miRNA‐25 regulates chemoresistance‐associated autophagy in breast cancer cells.30 Moreover, Song et al. also reported that hypoxia‐induced autophagy is responsible for chemoresistance in hepatocellular carcinoma cells.31 Mitophagy is a selective form of autophagy where mitochondria are selectively degraded in autophagolysosomes. Though there are substantial reports on the involvement of mitophagy in cancer and many neurodegenerative diseases, there is no report on the association of mitophagy and chemoresistance.5 We, for the first time, showed that mitophagy is responsible for chemoresistance in oral cancer. However, the potential of specific targeting of mitophagy as opposed to bulk autophagy in general as a therapeutic strategy remains to be elucidated. Furthermore, previous studies have demonstrated that autophagy acts as a protective mechanism in CSCs in many cancers including osteosarcoma, urothelial carcinoma, colorectal cancer, bladder cancer, brain cancer and breast cancer offering a greater survival and chemoresistance advantage.6, 32, 33, 34, 35, 36 Recently, it has been shown that JAK‐mediated autophagy is responsible for maintenance of stemness in cisplatin‐resistant bladder cancer cells.6 Moreover, in urothelial carcinoma cells, gemcitabine‐ and mitomycin‐induced autophagy was also found to regulate cancer stem cell pool.33 In accordance with previous studies and validating our hypothesis, the present investigation documents the autophagic inhibition in CCDP resistance in FaDu cells is associated with the down regulation of stem properties. Previous reports have demonstrated that osteoponin (OPN) triggers autophagy activation via the integrin/CD44 and p38 MAPK‐mediated pathways.37 Moreover, Whelan et al. reported the potential implication of mitophagy in regulation of tumour cells with high CD44 expression.38 Likely, our study has demonstrated that inhibition of autophagy leads to the declined expression of CD44 in CDDP resistant FaDu cells indicating a potential link between CD44 expression and autophagy mediated chemoresistance. Interestingly, it was also reported that ABCB1 expression is positively correlated with LC3, Beclin1, Rictor expression and negatively correlated with the expression of Raptor in colorectal cancer patients, suggesting a strong association of autophagy in cancer progression and multidrug resistance which is in agreement with this study involving the autophagic regulation of ABCB1‐mediated drug resistance in oral cancer. However, paradoxically, it has been found that both autophagy inhibitor and inducer enhanced the activity of α secretases like ADAM10 and ADAM17.39 Moreover, it has also been described that ADAM17 inhibitor, TIMP3 reduction causes a concomitant STAT1‐dependent loss of FoxO1 activity, which consequentially diminishes protective autophagy genes to fuel glomeruli damage suggesting the prospective association of ADAM17 and autophagy process.40 Accordingly, autophagy was shown to regulate the expression of ADAM17 in resistant FaDu cells which is a key molecule in the process of carcinogenesis and metastasis. As reported earlier, CDDP induces Mitochondrial‐ROS Response that leads to mitochondrial dysfunction as a part of its cytotoxicity.41 Dysfunctional mitochondria are a hub of ROS generation which is encountered by selective clearance of reactive mitochondria through mitophagy.42 The basal level of mitophagy is higher in resistant cells as compared with the parental cells, suggesting mitophagy as a protective response in chemoresistant cells. Moreover, stem cells are reported to keep their basal redox status at a low level through inducing mitophagy,43 which is in consistent with our investigation that higher mitophagy in resistant cell might be a driving force in the acquisition of stemness in oral cancer.
In conclusion, we have established that CD44+/ABCB1+/ADAM17+ cells have histologically poor differentiation, indicating that they are responsible for tumour aggressiveness. This simple eccentricity may manage CD44+/ABCB1+/ADAM17+ expression as a useful biomarker. We expect that our present results designating the clinical significance of CSC markers will contribute to new insight in targeting CSCs as well as address a number of problems associated with CSC biology. Further, we have established that selective mitochondrial autophagy (mitophagy) might be responsible for inducing chemoresistance in oral cancer. Moreover, our finding indicates that autophagy promotes stemness and drug tolerance in OSCC.
CONFLICT OF INTEREST
The authors disclose no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
Research support was partly provided by the Science and Engineering Research Board (SERB) [number: SR/SO/BB‐0101/2012], Department of Science and Technology (DST), Government of India. Research infrastructure was partly provided by Fund for Improvement of S&T Infrastructure in Universities & Higher Educational Institutions (FIST) [number: SR/FST/LSI‐025/2014], Department of Science and Technology, Government of India. PPN is obliged to DST‐SERB for providing fellowship. Authors sincerely thank Sushanta Pradhan for assisting in confocal facility for this research work.
Naik PP, Mukhopadhyay S, Panda PK, et al. Autophagy regulates cisplatin‐induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 2018;51:e12411 10.1111/cpr.12411
REFERENCES
- 1. Costea D, Tsinkalovsky O, Vintermyr O, Johannessen A, Mackenzie I. Cancer stem cells—new and potentially important targets for the therapy of oral squamous cell carcinoma. Oral Dis. 2006;12:443‐454. [DOI] [PubMed] [Google Scholar]
- 2. Naik PP, Das DN, Panda PK, et al. Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncol. 2016;62:122‐135. [DOI] [PubMed] [Google Scholar]
- 3. Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431‐3434. [DOI] [PubMed] [Google Scholar]
- 5. Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ojha R, Singh S, Bhattacharyya S. JAK‐mediated autophagy regulates stemness and cell survival in cisplatin resistant bladder cancer cells. Biochim Biophys Acta 2016;1860:2484‐2497. [DOI] [PubMed] [Google Scholar]
- 7. Kokko L‐L, Hurme S, Maula SM, et al. Significance of site‐specific prognosis of cancer stem cell marker CD44 in head and neck squamous‐cell carcinoma. Oral Oncol. 2011;47:510‐516. [DOI] [PubMed] [Google Scholar]
- 8. Jiang Y, He Y, Li H, et al. Expressions of putative cancer stem cell markers ABCB1, ABCG2, and CD133 are correlated with the degree of differentiation of gastric cancer. Gastric Cancer. 2012;15:440‐450. [DOI] [PubMed] [Google Scholar]
- 9. Kamarajan P, Shin JM, Qian X, Matte B, Zhu JY, Kapila YL. ADAM17‐mediated CD44 cleavage promotes orasphere formation or stemness and tumorigenesis in HNSCC. Cancer Med. 2013;2:793‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng Z, Niu G, Cai L, Wei R, Zhao X. The prognostic significance of CD44V6, CDH11, and‐catenin expression in patients with osteosarcoma. Biomed Res Int. 2013; 2013:496193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimm M, Krimmel M, Polligkeit J, et al. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eu J Cancer. 2012;48:3186‐3197. [DOI] [PubMed] [Google Scholar]
- 12. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pisco AO, Huang S. Non‐genetic cancer cell plasticity and therapy‐induced stemness in tumour relapse: ‘What does not kill me strengthens me'. Br J Cancer. 2015;112:1725‐1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sinha N, Mukhopadhyay S, Das DN, Panda PK, Bhutia SK. Relevance of cancer initiating/stem cells in carcinogenesis and therapy resistance in oral cancer. Oral Oncol. 2013;49:854‐862. [DOI] [PubMed] [Google Scholar]
- 15. Jung WY, Kang Y, Lee H, et al. Expression of moesin and CD44 is associated with poor prognosis in gastric adenocarcinoma. Histopathology. 2013;63:474‐481. [DOI] [PubMed] [Google Scholar]
- 16. Washington K, Gottfried MR, Telen MJ. Expression of the cell adhesion molecule CD44 in gastric adenocarcinomas. Human Pathol. 1994;25:1043‐1049. [DOI] [PubMed] [Google Scholar]
- 17. Prince M, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973‐978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliveira LR, Oliveira‐Costa JP, Araujo IM, et al. Cancer stem cell immunophenotypes in oral squamous cell carcinoma. J Oral Pathol Med. 2011;40:135‐142. [DOI] [PubMed] [Google Scholar]
- 19. Moitra K, Lou H, Dean M. Multidrug efflux pumps and cancer stem cells: insights into multidrug resistance and therapeutic development. Clinical Pharmacol Ther. 2011;89:491. [DOI] [PubMed] [Google Scholar]
- 20. Chiba T, Kita K, Zheng YW, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell–like properties. Hepatology. 2006;44:240‐251. [DOI] [PubMed] [Google Scholar]
- 21. Shou ZX, Jin X, Zhao ZS. Upregulated expression of ADAM17 is a prognostic marker for patients with gastric cancer. Ann Surg. 2012;256:1014‐1022. [DOI] [PubMed] [Google Scholar]
- 22. Zhai X, Wang H, Wang J, Xu P, Zhao Y, Qiu J. Correlation between ADAM17 protein expression level and prognosis of neuroglioma. Int J Clin Exp Pathol. 2016;9:1764‐1769. [Google Scholar]
- 23. Jögi A, Vaapil M, Johansson M, Påhlman S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups J Med Sci. 2012;117:217‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kademani D, Bell RB, Bagheri S, et al. Prognostic factors in intraoral squamous cell carcinoma: the influence of histologic grade. J Oral Maxillofacial Surg. 2005;63:1599‐1605. [DOI] [PubMed] [Google Scholar]
- 25. Ohara Y, Oda T, Sugano M, et al. Histological and prognostic importance of CD44+/CD24+/EpCAM+ expression in clinical pancreatic cancer. Cancer Sci. 2013;104:1127‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nör C, Zhang Z, Warner KA, et al. Cisplatin induces Bmi‐1 and enhances the stem cell fraction in head and neck cancer. Neoplasia. 2014;16:137‐W138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bates RC, Edwards NS, Burns GF, Fisher DE. A CD44 survival pathway triggers chemoresistance via lyn kinase and phosphoinositide 3‐kinase/Akt in colon carcinoma cells. Cancer Res. 2001;61:5275‐5283. [PubMed] [Google Scholar]
- 28. Hoofd C, Wang X, Lam S, et al. CD44 promotes chemoresistance in T‐ALL by increased drug efflux. Exp Hematol. 2016;44(3):166‐171.e117. [DOI] [PubMed] [Google Scholar]
- 29. Liu YS, Hsu HC, Tseng KC, Chen HC, Chen SJ. Lgr5 promotes cancer stemness and confers chemoresistance through ABCB1 in colorectal cancer. Biomed Pharmacother. 2013;67:791‐799. [DOI] [PubMed] [Google Scholar]
- 30. Wang Z, Wang N, Liu P, et al. MicroRNA‐25 regulates chemoresistance‐associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013‐7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song J, Qu Z, Guo X, et al. Hypoxia‐induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131‐1144. [DOI] [PubMed] [Google Scholar]
- 32. Zhang D, Zhao Q, Sun H, et al. Defective autophagy leads to the suppression of stem‐like features of CD271+ osteosarcoma cells. J Biomed Sci. 2016;23:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ojha R, Jha V, Singh S. Gemcitabine and mitomycin induced autophagy regulates cancer stem cell pool in urothelial carcinoma cells. Biochim Biophys Acta. 2016;1863:347‐359. [DOI] [PubMed] [Google Scholar]
- 34. Yang HZ, Ma Y, Zhou Y, et al. Autophagy contributes to the enrichment and survival of colorectal cancer stem cells under oxaliplatin treatment. Cancer Lett. 2015;361:128‐136. [DOI] [PubMed] [Google Scholar]
- 35. Lomonaco SL, Finniss S, Xiang C, et al. The induction of autophagy by γ‐radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717‐722. [DOI] [PubMed] [Google Scholar]
- 36. Gong C, Bauvy C, Tonelli G, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem‐like/progenitor cells. Oncogene. 2013;32:2261‐2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng YH, Tian C, Meng Y, et al. Osteopontin stimulates autophagy via integrin/CD44 and p38 MAPK signaling pathways in vascular smooth muscle cells. J Cellular Physiol. 2012;227:127‐135. [DOI] [PubMed] [Google Scholar]
- 38. Whelan K, Chandramouleeswaran P, Tanaka K, et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin‐mediated mitochondrial clearance. Oncogene. 2017;36:4843‐4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cai Z, Zhou Y, Liu Z, Ke Z, Zhao B. autophagy dysfunction upregulates beta‐amyloid peptides via enhancing the activity of γ‐secretase complex. Neuropsychiatr Dis Treat. 2015;11:2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fiorentino L, Cavalera M, Menini S, et al. Loss of TIMP3 underlies diabetic nephropathy via FoxO1/STAT1 interplay. EMBO Mol Med. 2013;5(3):441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marullo R, Werner E, Degtyareva N, et al. Cisplatin induces a mitochondrial‐ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS ONE. 2013;8:e81162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal. 2011;14:1919‐1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joshi A, Kundu M. Mitophagy in hematopoietic stem cells: the case for exploration. Autophagy. 2013;9:1737‐1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


