Abstract
Objectives
Although dramatic improvements of overall survival has achieved in patients with favourable histology Wilms tumour, disease recurrence is still the main cause of cancer‐related death in childhood. Long non‐coding RNAs (lncRNAs) as oncogenes or tumour suppressors are dysregulated during carcinogenesis. However, the role of lncRNAs in the pathogenesis of Wilms tumour is unknown. Here, an lncRNA LINC00473 signature that functioned as oncogene was identified in Wilms tumour.
Methods
Wilms tumour (n = 15) and relative normal tissues were collected. The LINC00473 expression and function in Wilms tumour was determined. The LncRNA‐miRNA network of LINC00473 was analysed in vitro and vivo.
Results
We uncovered that the expression of LINC00473 was elevated in tumour tissues than that in relative normal tissues. Higher LINC00473 levels correlated to higher stage and unfavourable histology Wilms tumour. Mechanistically, knockdown of LINC00473 inhibited cell vitality and induced Bcl‐2‐dependent apoptosis and G1/S arrest via CDK2 and cyclin D1. Moreover, LINC00473 harboured binding sites for miR‐195 and limited miR‐195 availability in a dose‐dependent manner. Forced expression of miR‐195 impaired tumour survival and metastasis, which, however, could be restored by LINC00473. Furthermore, IKKα was the downstream of LINC00473/miR‐195 signals and could be directly targeted by miR‐195 to participate LINC00473‐induced tumour progression. Loss‐of‐function of LINC00473 in vivo effectively promoted the regression of Wilms tumour via miR‐195/IKKα‐mediated growth inhibition.
Conclusion
LINC00473 as an oncogene is up‐regulated to participate into the molecular pathogenesis of Wilms tumour via miR‐195/IKKα.
Keywords: IKKα, LINC00473, miR‐195, Wilms tumour
1. INTRODUCTION
As an embryonal type of renal cancer, Wilms' tumour (nephroblastoma) is the most common primary renal tumour occurring in childhood, which affects one in 10 000 children and accounts for 5% of all childhood cancers.1, 2 In Asia, boys have a greater disease incidence than girls, and approximately 500 cases are identified each year in the United States. Due to the successive clinical trials conducted by the National Wilms Tumor Study Group (NWTSG), now the Children's Oncology Group (COG), the International Society of Pediatric Oncology (SIOP) and other national study groups, the overall survival (OS) rate of patients with Wilms tumour has risen to exceeding 90%, especially for those with favourable histology. However, the survival rate for patient with unfavourable histologic and molecular features (eg, diffuse and focal anaplasia or loss of heterozygosity (LOH) at chromosomes 1p and 16q) remains well below 90%, which is attributed to the increased risk of disease recurrence.3, 4 Therefore, investigations on the molecular pathogenesis of Wilms tumour are conducive to improve the treatment and outcomes of patient with unfavourable histology.
In mammalian cells, the majority of transcribed RNAs are non‐coding, which do not contain protein‐coding sequences. These transcripts are eventually, on one hand, processed into small RNAs including microRNAs (miRNAs), Piwi‐interacting RNAs (piRNAs), tRNA‐derived stress‐induced fragment RNAs and small nucleolar RNAs (snoRNAs), and, on the other hand, processed into long non‐coding RNAs (lncRNAs).5, 6 miRNAs are ~20‐25 nucleotide non‐coding RNAs which involve in post‐transcriptional regulation of gene expression and present in almost all organisms including kidney. Yu et al found that miR‐34a could directly target and impair the CD44 level in renal cancer cells to suppress renal cancer cell growth, tube formation and metastasis in vitro and in vivo.7 miR‐21 was reported to target PTEN in renal cancer cells and control the expression of cyclin D1 to promote proliferation through NF‐κB‐dependent transcription.8 Mounting evidences also demonstrated that lncRNAs were able to participate into a broad range of cellular processes of carcinogenesis, such as tumour cell growth, differentiation, metastasis and chemotherapy resistance.9, 10, 11, 12 However, the bio‐functions and molecular mechanisms of lncRNAs in Wilms' tumour remained largely unknown.
To be noted that the cross‐regulation between microRNAs and lncRNAs is the important way for LncRNA to exert its function.13 LncARSR promoted sunitinib resistance via competitively binding miR‐34/miR‐449 to facilitate AXL and c‐MET expression and predicted poor response of renal cell carcinoma patients.14 In addition, lncRNA MALAT1 was found to be up‐regulated in renal cell carcinoma and interacted with Ezh2 and miR‐205 for E‐cadherin recovery and β‐catenin down‐regulation, leading to the proliferation and invasion of tumour cells.15 In non‐small‐cell lung cancer (NSCLC), LINC00473 as an oncogene was elevated in lung cancer and interacts with NONO, a component of the cAMP signalling pathway, thereby facilitating CREB‐mediated transcription for the growth and survival of LKB1‐inactivated NSCLC cells.16 In this study, 10 human lncRNAs, which were often dysregulated during carcinogenesis, were analysed in our pre‐experiment, including MALAT1, ANRIL, HOTAIR, Xist, H19, MEG3, THRIL, NEAT1, lnc‐ATB and LINC00473. Among these, MALAT1, lnc‐ATB and LINC00473 were up‐regulated but H19 was down‐regulated in Wilms tumour tissues (unpublished data). However, only the LINC00473 level was found to be associated with higher tumour stage and unfavourable histology of Wilms tumour. MiR‐195 was found to be sequestered by LINC00473, which contributed to the inactivation of miR‐195/IKKα signals. Knockdown of LINC00473 was effective strategy for the treatment of Wilms tumour.
2. MATERIALS AND METHODS
2.1. Tumour samples, cell lines and reagents
Wilms tumour tissues (n = 15) and corresponding relative normal tissues (n = 15) were collected from Guangzhou Women and Children's Medical Center. All these retrospective specimens were handled according to the ethical and legal standards of Guangzhou Women and Children's Medical Center. According to WHO classifications, all of the patients diagnosed with primary Wilms tumour were confirmed by haematoxylin and eosin staining by experienced pathologists. Written informed consent was obtained from all of the patients. The Wilms tumour cell line SK‐NEP‐1 was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS (Life Technologies, USA), ampicillin and streptomycin at 37°C, 5% CO2 conditions. siRNA‐LINC00473 (5′‐CCACUUCUGACGUGUCCUU‐3′), siRNA‐IKKα (5′‐ GAGGCUGUGAUAGCUAUAUUU‐3′) or negative control and oligonucleotide sequences of miR‐195 mimics, inhibitors or negative control were purchased from RiboBio (Guangzhou, china). The overexpression vector pcDNA3.1‐LINC00473 and reporter plasmid of full‐length 3′‐UTR (wild‐type or mutant) of IKKα were conducted by GenePharma (Shanghai, China). Renilla luciferase reporter vector psiCHECK2‐miR‐195, psiCHECK2‐LINC00473‐wild or mut, psiCHECK2‐3′UTR (wild or mut) of IKKα were conducted by GenePharma (Shanghai, China). Anti‐GAPDH, CDK2, cyclin D1, caspase‐3 and Bcl‐2 antibodies were obtained from Cell Signaling Tech (Denver, MA) and Abcam (USA).
2.2. Cell transfection
The SK‐NEP‐1 cells were cultured to about 80% confluence in 6/12‐well plates and then, using Lipofectamine 2000 (Invitrogen, USA), the cells were transfected with indicated agents according to the manufacturer's instructions. After transfection for the indicated time, the cells were harvested for further experiments.
2.3. CCK‐8 assay
The treated SK‐NEP‐1 cell lines were harvested and washed with PBS and then cell counting kit‐8 (Kumamoto, Japan) mixed with DMEM was used for cell viability assay, and the absorbance was measured at 450 nm by a microplate reader.
2.4. EdU assay
The treated SK‐NEP‐1 cell lines were treated with 50 μM of EdU for 2 hours at 37°C and harvested to be fixed with 4% formaldehyde in PBS for 10 minutes and rinsed with PBS for 3 times. Then, cells were permeabilized with 100% methanol for 10 min at −20°C. After that, the cells were treated with 1×Apollo reaction cocktail for 30 minutes, and the nuclei of the cells were stained with DAPI and visualized under a fluorescent microscope.
2.5. Flow cytometer assay
For the cell cycle analysis, SK‐NEP‐1 cells were stained with PI staining solution (10 μg mL−1 RNase A and 50 μg mL−1 PI) at 37°C for 30 minutes in dark. The cell cycle distribution was analysed using a flow cytometry provided with the Cell‐Quest software.
2.6. Transwell assay
In the migration assay, 2 × 104 SK‐NEP‐1 cells transfected with indicated agents were in the upper chamber of a non‐coated transwell insert, and in the invasion assay, the upper chamber of the transwell inserts was coated with Matrigel, and tumour cells were plated in the upper chamber of the Matrigel‐coated transwell insert. Cells that did not migrate or invade were removed using a cotton swab and were stained by crystal violet and counted under an inverted microscope. Five random views were selected to count the cells.
2.7. RNA isolation and qRT‐PCR
Total RNA from tissues or tumour cells was extracted using Trizol reagent (Invitrogen) according to the standard RNA isolation protocol. Quantitative real‐time RT‐PCR (qRT‐PCR) was performed, and the expression levels of LINC00473/miR‐195/IKKα were normalized to GAPDH for gene expression. The primers were listed as below:
| Gene | Primer 5′‐3′ |
|---|---|
| LINC00473 | F: AGAGAGGTCTGAGTCCGAAGT |
| R: CAAAAGGCGAGTGTGTGG | |
| miR‐195 | F: ACACTCCAGCTGGGTAGCAGCACAGAAATATT |
| R: CTCAACTGGTGTCGTGGA | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| IKKα | F: ATGAAGAAGTTGAACCATGCCA |
| R: CCTCCAGAACAGTATTCCATTGC | |
| GAPDH | F: ACACCCACTCCTCCACCTTT |
| R: TTACTCCTTGGAGGCCATGT |
2.8. Western blotting
According to the manufacturer's instructions, proteins were separated by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to nitrocellulose membrane (Bio‐Rad, Hercules, CA, USA) and were blocked in 5% non‐fat milk in TBST buffer (Tris Buffer Saline containing 0.1% Tween‐20) for 1 h at room temperature, and subsequently incubated with primary antibodies overnight at 4°C. After washing with TBST buffer, the blots were then incubated with HRP‐conjugated secondary antibody for 1 hour at room temperature. The blots were washed with TBST buffer and visualized using the ECL‐Plus reagent (Millipore, Billerica, MA, USA). GAPDH was used as the loading control in the Western blotting.
2.9. Luciferase reporter assay
The SK‐NEP‐1 cells were co‐transfected containing psiCHECK2‐miR‐195, psiCHECK2‐LINC00473‐wild or mut, psiCHECK2‐3′UTR (wild or mut) of IKKα, pcDNA3.1‐LINC00473 or miR‐195 mimics. Luciferase activity was measured using the Dual‐Luciferase Reporter Assay System (Promega, USA). Firefly luciferase acted as a reporter gene for normalized control.
2.10. Immunohistochemistry
The expression of Ki‐67, caspase‐3 and cycling D1 in Wilms tumour tissues was analysed on 2 μm thick, formalin‐fixed and paraffin‐embedded specimen sections. Slides were incubated in xylene for 5 minutes and followed by 2 washes of 100% ethanol for 10 minutes, 95% ethanol for 10 minutes. Antigen unmasking was performed at room temperature. Then, the primary antibody incubated the FFPE specimen sections at 4°C overnight, and then the EnVision Detection System kit (DAKO, Denmark) was used for the DAB chromogen followed by nuclear staining using haematoxylin.
2.11. Tumour model
To test the role of TFP in vivo, SK‐NEP‐1 cells were transfected with lentivirus vector of siRNA‐LINC00473, pcDNA3.1‐LINC00473 or negative control; 2 × 106 SK‐NEP‐1 cells were subcutaneously injected in rear flank of nude mice (4 per group). The tumour sizes were measured 3 days apart and the tumour volumes were calculated: V (cm3) = width2 (cm2) × length (cm)/2. On day 26, the mice were killed.
2.12. Statistical analyses
The results are analysed by the Statistical Package for Social Sciences version 16.0 (SPSS 16.0, SPSS Inc., Chicago, IL, USA) and the Prism statistical software package (Version 5.0, GraphPad Software Inc., La jolla, CA, USA). Unpaired t test or Mann‐Whitney U test was used to compare the 2 groups, and multiple group comparisons were analysed with one‐way ANOVA. P < .05 was considered statistically significant. All experiments were performed at least 3 times.
3. RESULTS
3.1. Elevated LINC00473 expression correlated to higher stage and unfavourable histology in Wilms tumour
We investigated the clinical expression of LINC00473 in patients of Wilms tumour. Thus, Wilms tumour tissues (n = 15) and corresponding non‐neoplastic tissues (n = 15) and the Q‐PCR results showed that the LINC00473 level was significantly elevated in cancer tissues (Tumour) than that in the corresponding non‐neoplastic tissue (Normal) (Figure 1A). Moreover, we analysed the correlations between the clinicopathological characteristics of Wilms tumour patients and LINC00473 expression. We found that patients with higher tumour stage and unfavourable histology harboured a significant higher LINC00473 level in tumour samples (Figure 1A,B). Thus, up‐regulated LINC00473 expression might participate into the pathogenesis of Wilms tumour.
Figure 1.
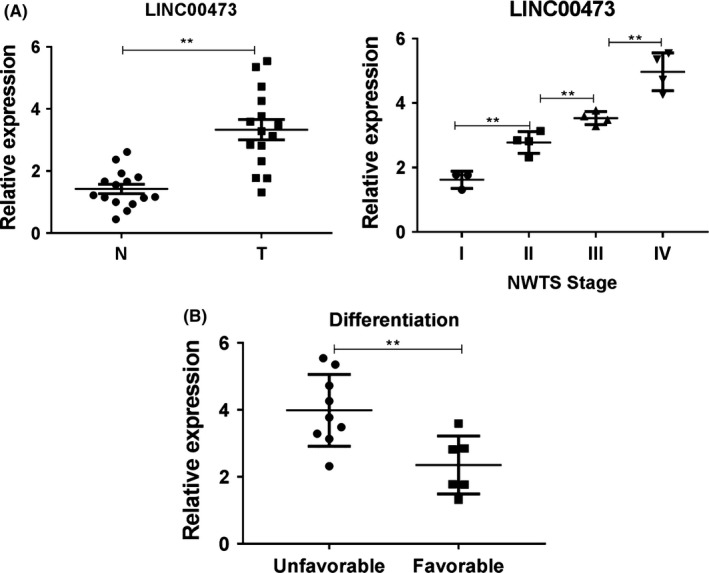
Up‐regulated LINC00473 correlated with the poor clinical outcome of Wilms tumour. (A) The expression of LINC00473 was analysed in Wilms tumour and relatively normal tissues were by Q‐PCR. (B) The clinicopathological characteristics of Wilms tumour patients were analysed according to the expression of LINC00473. **P < .01, data represent the mean ± SD
3.2. LINC00473 promotes tumour growth and metastasis in vitro
High expression of LINC00473 was noticed to be associated with high Wilms tumour stage, then knockdown of LINC00473 in cell line SK‐NEP‐1 of Wilms tumour was performed to investigate its function. Three siRNAs of LINC00473 were designed and transfected into SK‐NEP‐1 cells. We found that siRNA‐2 was the most efficient siRNA to inhibit the LINC00473 level (Figure 2A). The cell vitality was measured in 48 and 72 hours; knockdown of LINC00473 significantly impaired the vitality of SK‐NEP‐1 cells (Figure 2B). The EdU assay indicated that siRNA‐LINC00473 could reduce the ability of proliferation of SK‐NEP‐1 cells (Figure 2C), with increased apoptosis of tumour cells (Figure 2D). Meanwhile, inhibition of LINC00473 was found to induce G1/S arrest, which implicated that the cycle arrest might contribute to the decreased cell vitality and proliferation (Figure 2E). Interestingly, the ability of migration and invasion of SK‐NEP‐1 cells was also decreased by the inhibition of LINC00473 (Figure 2F). Besides, we found that knockdown of LINC00473 was able to repress the anti‐apoptosis protein Bcl‐2 level and promote the cleavage of caspase‐3 for the induction of apoptosis. CDK2 and cyclin D1 were essential for the transition of G1/S cycle, but they were down‐regulated by the siRNA‐LINC00473 (Figure 2G). These results indicated that LINC00473 involved into the cell proliferation, apoptosis and migration in Wilms tumour.
Figure 2.
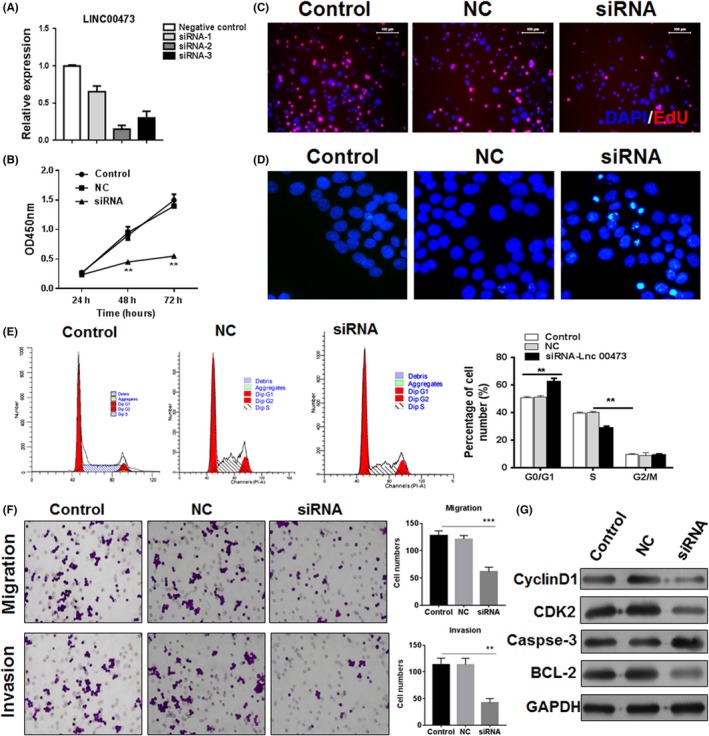
Knockdown of LINC00473 inhibits the tumour growth. (A) The efficiency of siRNA transfection in SK‐NEP‐1 cells was determined by Q‐PCR. (B) After the knockdown of LINC00473, the cell vitality was estimated by CCK‐8. (C and D) The proliferation and apoptosis were determined by EdU assay and Hoechst assay. (E) The cell cycle was also analysed by flow cytometry. (F) The ability of migration and invasion was assessed by transwell assay. (G) The expression of Bcl‐2, caspase‐3, CDK2 and cyclin D1. *P < .05, **P < .01, ***P < .001, data represent the mean ±SD
3.3. LINC00473 contains functional miR‐195 interaction sites and inhibits miR‐195 availability
We next analysed the molecular mechanism of LINC00473‐related tumour progression. In SK‐NEP‐1 cells, we analysed the micro‐location of LINC00473 and the Q‐PCR results indicated that transcript levels of LINC00473 were found both in the nuclear (Nuc) and cytoplasmic (Cyt) fractions obtained from SK‐NEP‐1 cells, which provided the potential of LncRNA‐miRNA interaction model (Figure 3A). Bioinformatic analysis revealed putative complementary sequences for miR‐195 in human LINC00473 and a predicted miR‐195 binding sites were found (Figure 3B). The plasmid psiCHECK2‐LINC00473 was conducted which harboured the miR‐195 binding site in the 3′UTR of a Renilla luciferase (Rluc) gene and also contained a downstream constitutively expressed firefly luciferase gene as an internal control for normalization (Figure 3C). The results showed that co‐transfected different concentrations of miR‐195 with psiCHECK2‐miR‐195 and miR‐195 significantly inhibited Rluc expression in a dose‐dependent manner in SK‐NEP‐1 cells, which was also observed with psiCHECK2‐LINC00473‐wild containing a human LINC00473 fragment at the 3′UTR of the Rluc in place of miR‐195 binding sites. However, psiCHECK2‐LINC00473‐mut with deleted sequences complementary to the seed region of miR‐195 could not responded to miR‐195‐indeced inhibition of Rluc expression in SK‐NEP‐1 cells, which suggested that inhibition was dependent on the predicted miR‐195 sites (Figure 3D). Besides, when miR‐16‐1 (not predicted to bind LINC00473) was tested on psiCHECK2‐LINC00473, a dose‐dependent inhibition was not observed (Figure 3D). Of importance, to determine that LINC00473 specifically sequestered endogenous miR‐195, a different dose of pcDNA3.1‐LINC00473 was transfected into SK‐NEP‐1 cells. We found that the Rluc activity of psiCHECK2‐miR‐195 increased in response to pcDNA3.1‐LINC00473 in a dose‐dependent manner, which could be explained by the down‐regulation of endogenous miR‐195 specifically sequestered by LINC00473 (Figure 3E). Therefore, LINC00473 was able to sequester miR‐195 to inhibit its function to increase Rluc expression.
Figure 3.
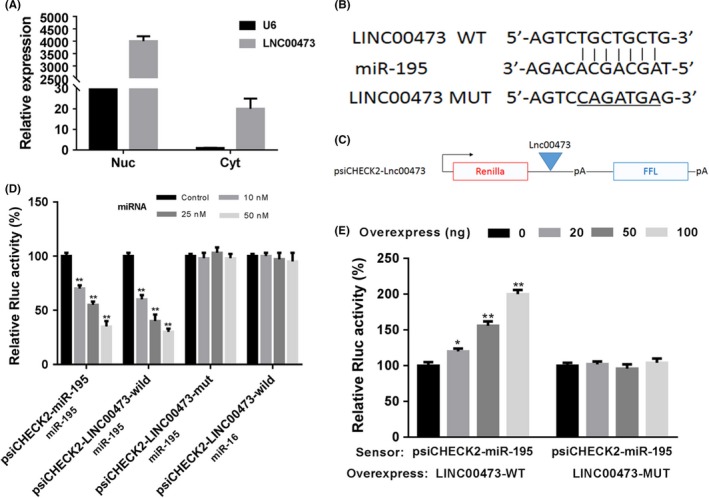
LINC00473 contains functional miR‐195 interaction sites. (A) The micro‐location of LINC00473 in SK‐NEP‐1 cells was analysed. (B) The plasmid psiCHECK2‐LINC00473 was conducted as indicated. (C) The predicted miR‐195 binding sites for LINC00473. (D) The SK‐NEP‐1 cells were co‐transfected with miR‐195 and psiCHECK2‐miR‐195 or psiCHECK2‐LINC00473 and the Rluc expression was determined. (E) the Rluc activity of psiCHECK2‐miR‐195 was impaired by siRNA‐LINC00473. *P < .05, **P < .01, ***P < .001, data represent the mean ± SD
3.4. The anti‐tumour role of miR‐195 could be inhibited by up‐regulated LINC00473
Due to the inhibition of LINC00473 to miR‐195, we assessed the expression of miR‐195 in Wilms tumour tissues and cell lines. In SKNEP1 cell, the concentration of LINC00473 and miR‐195 were about 24600 pmol L−1 and 250 pmol L−1 by RT‐PCR (date show in Data S1), and the miR‐195 level was remarkably decreased in cancer tissues, which negatively correlated to the LINC00473 level in clinical (Figure 4A), and a predicted miR‐195 binding sites in LINC00473 further emphasized the possibility of LINC00473/miR‐195 interaction. Thus, we suspected that the function of miR‐195 could be inhibited by up‐regulated LINC00473 in Wilms tumour. Ectopically expressed miR‐195 mimics inhibited the cell vitality and proliferation of SK‐NEP‐1 cells, which, but, was restored by overexpression of LINC00473 (Figure 4B,C). In addition, miR‐195‐induced apoptosis and cell cycle arrest could also be abrogated by LINC00473 (Figure 4D,E), and the ability of migration and invasion showed the similar effects (Figure 4F). Correspondingly, apoptosis‐related Bcl‐2 and caspase‐3 were inhibited by miR‐195 but rescued by LINC00473. LINC00473 could also impair miR‐195‐induced inhibition of CDK2 and cyclin D1 (Figure 4G). These data indicated that LINC00473 inhibited miR‐195 to participate the growth of Wilms tumour.
Figure 4.
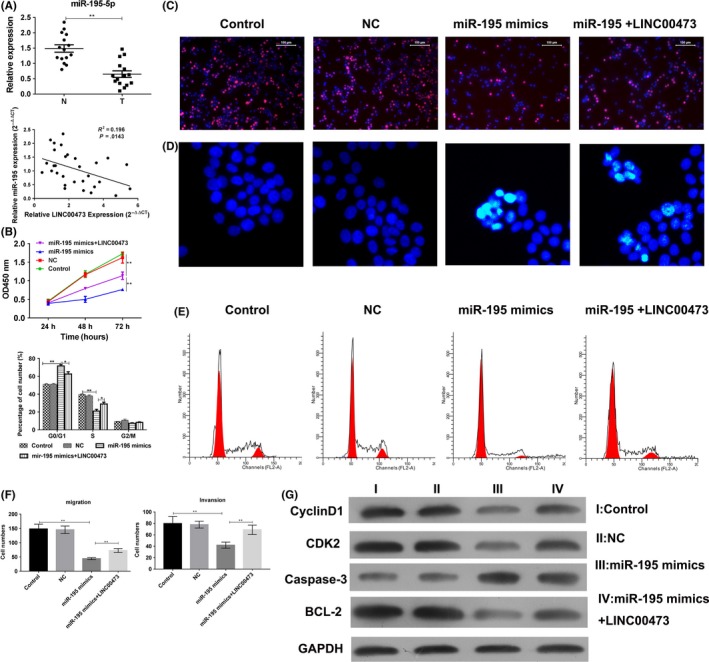
The anti‐tumour effect of miR‐195 was impaired by LINC00473. (A) The expression of miR‐195 and its correlation to LINC00473 were analysed in Wilms tumour and relatively normal tissues were by Q‐PCR. (B) After the transfection of miR‐195 mimics and/or pcDNA 3.1‐LINC00473 in SK‐NEP‐1 cells, the cell vitality was estimated by CCK‐8. (C and D) the proliferation and apoptosis were determined by EdU assay and Hoechst assay. (E) The cell cycle was also analysed by flow cytometry. (F) The ability of migration and invasion was assessed by transwell assay. (G) The expression of Bcl‐2, caspase‐3, CDK2 and cyclin D1. *P < .05, **P < .01, ***P < .001, data represent the mean ± SD
3.5. IKKα is a direct target of LINC00473/miR‐195 signal axis
Amounting evidences indicated that IKKα was high expressed during the progression of cancer and involved into tumour growth and metastasis.17, 18 MiR‐195 was reported to target and inhibit the IKKα level in cancer,19 thus we proposed a hypothesis that LINC00473 was able to regulate the activity of IKKα by inhibition of miR‐195. The results showed that overexpression of miR‐195 mimics reduced IKKα level which could be impaired by LINC00473 (Figure 5A). Luciferase reporter assay also confirmed that LINC00473 decreased the expression of miR‐195 to increase the Luciferase activity (Figure 5B). To confirm the role of miR‐195/IKKα in Wilms tumour, knockdown of IKKα combined with miR‐195 inhibitor was performed in SK‐NEP‐1 cells. We found that miR‐195/IKKα signal suppressed the features of tumour including cell vitality (Figure 5C), proliferation (Figure 5D), apoptosis (Figure 5E) and metastasis (Figure 5F) via regulation of the expression of Bcl‐2, caspase‐3, CDK2 and cyclin D1 (Figure 5G). These results demonstrated that LINC00473 could up‐regulate IKKα via miR‐195 inhibition in Wilms tumour.
Figure 5.
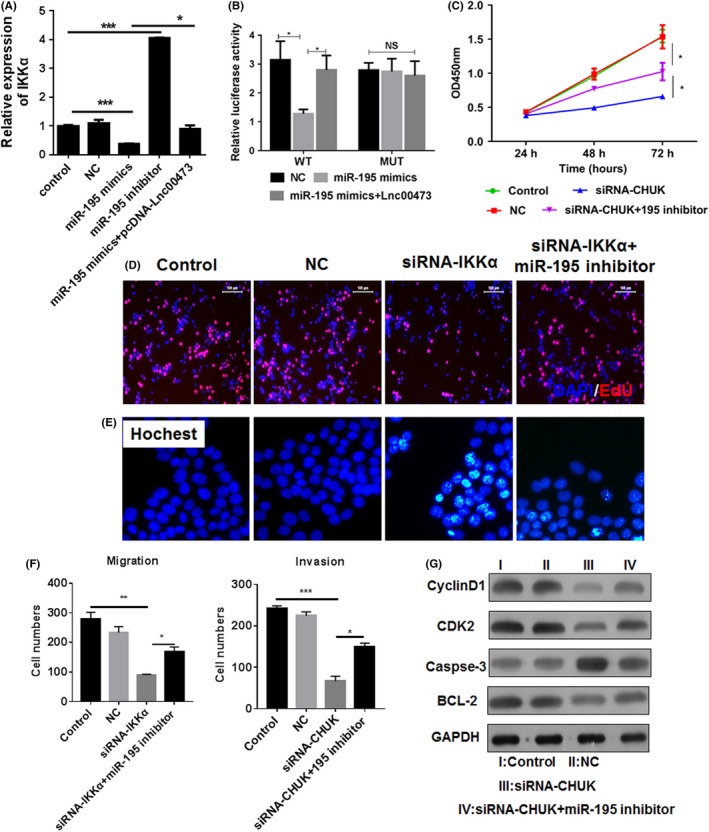
LINC00473 regulates the miR‐195/IKKα signals in Wilms tumour. (A) The expression of IKKα was regulated by miR‐195 mimics and pcDNA 3.1‐LINC00473 in SK‐NEP‐1 cells. (B) A luciferase reporter vector with full‐length promoter construct of IKKα containing mutation at miR‐195 binding sites was transfected into SK‐NEP‐1 cells and the luciferase activity was determined. (C) The cell vitality was estimated by CCK‐8. (D and E) The proliferation and apoptosis were determined by EdU assay and Hoechst assay. (F) The ability of migration and invasion was assessed by transwell assay. (G) The expression of Bcl‐2, caspase‐3, CDK2 and cyclin D1. *P < .05, **P < .01, ***P < .001, data represent the mean ± SD
3.6. Knockdown of LINC00473 effectively promotes Wilms tumour regression in vivo
The pro‐tumour effects of LINC00473 in vivo were determined, the xenograft model of human SK‐NEP‐1 was established and the nude mice were treated with conditional tumour cells with LINC00473 overexpression or knockdown. The results indicated that knockdown of LINC00473 significantly delayed the tumour growth with small tumour volume, whereas the mice treated with highly expressed LINC00473 bared the largest tumour (Figure 6A). The expressions of LINC00473, miR‐195 and IKKα were determined and found that LINC00473 in vivo could also inhibit the miR‐195 level to up‐regulate IKKα (Figure 6B). Besides, the expression of proliferation index Ki‐67 was increased in mice with LINC00473 overexpression but decreased in mice with LINC00473 knockdown (Figure 6C). Similarly, LINC00473 knockdown promoted the cleavage of caspase‐3 to inhibit the caspase‐3 level which was accompanied with decreased cyclinD1 expression in vivo (Figure 6C).
Figure 6.
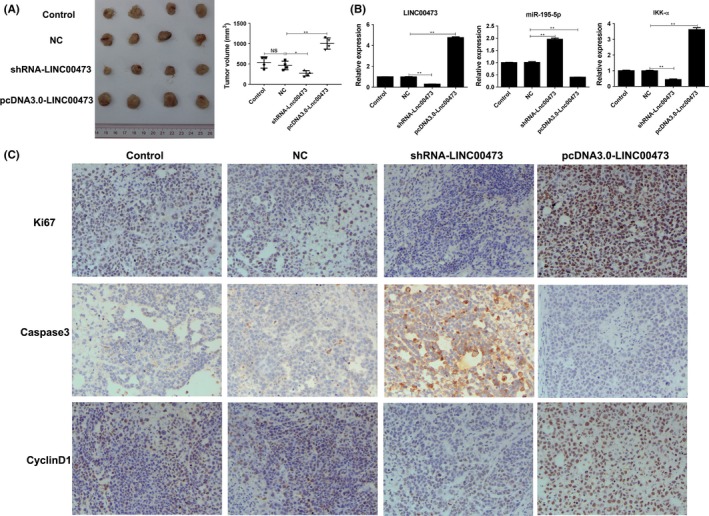
Knockdown of LINC00473 inhibits the growth of Wilms tumour in vivo. 2 × 106 conditional SK‐NEP‐1 cells (knockdown or overexpression of LINC00473) were subcutaneously injected in rear flank of nude mice (4 per group). (A) The mean tumour size (mm3) was analysed. (B) The expressions of LINC00473/miR‐195/IKKα were estimated by Q‐PCR. (C) The expressions of Ki‐67, caspase‐3 and cycling D1 were determined by immunohistochemistry. *P < .05, **P < .01, data represent the mean ± SD
4. DISCUSSION
Wilms tumour accounts for approximately 90% of all paediatric tumours of the kidney, and currently, the overall survival rate of Wilms tumour has been improved to exceeding 90% in patients with favourable histology, which is attributed to the risk‐stratification system based on patient age, tumour stage, tumour weight, histology and tumour‐specific loss of heterozygosity (LOH) at chromosomes 1p and 16q2. However, high‐risk Wilms tumour such as unfavourable histology or post‐chemotherapy blastemal‐type Wilms tumours still needs to identify novel molecules and therapies for stratification of treatment intensity.20 In this study, we found that the LINC00473 level was elevated in higher stage and unfavourable histology Wilms tumour. We found that LINC00473 sequestered miR‐195 to limited miR‐195 availability, leading to the dysregulation of miR‐195/IKKα signals to participate LINC00473‐induced tumour progression, suggesting that LINC00473 is an oncogene in the molecular pathogenesis of unfavourable histological Wilms tumour.
Actually, non‐coding RNA, especially miRNA, had been identified to be diagnostic or prognostic factors for Wilms tumour. Ludwig et al demonstrated that high expression of cell‐free circulating 2 miRNA miRs‐100, miR‐130b could be potentially used as diagnostic marker for Wilms tumour to differentiate patients with Wilms tumour from healthy controls.21 miR‐204 was down‐regulated in Wilms tumour to activate the oncogene meis homeobox 1 (MEIS1).22 We here found that the expression of miR‐195 was also decreased during the development of Wilms tumour, which was a tumour suppressor to target IKKα and induce tumour apoptosis and cell cycle arrest. Cao et al reported that the expression level of miR‐370 was also elevated in Wilms tumour tissues compared with adjacent non‐tumour tissues, which was transcriptionally activated by STAT3 to promote proliferation and enhances tumorigenicity.23 Although the role of miRNA in Wilms tumour had been clarified by different researches, the clinical evidence of LncRNA in Wilms tumour was unknown. In this study, we identified the pro‐tumour role of LINC00473 in the progression of Wilms tumour, higher LINC00473 levels correlated to higher stage and unfavourable histology Wilms tumour. This finding provides a support for the oncogene role of LncRNA in Wilms tumour.
Mounting researches had found that LncRNA functioned as tumour suppressors, oncogenes to affect cell division, senescence, stress response, apoptosis and chemotherapy resistance in many types of cancer, including hepatocellular carcinoma, lung cancer, colorectal cancer, breast cancer, oral cancer, etc.9 CCAT1‐L, a novel CRC‐specific lncRNA in human colorectal cancers, is abundantly transcribed from a locus 515 kb upstream of MYC. Overexpression of CCAT1‐L enhances MYC expression via interaction between the MYC promoter and its upstream regulatory elements to promote tumorigenesis.24 Lnc‐ATB was the most remarkably up‐regulated lncRNA in the tissues of breast cancer patients with trastuzumab resistance and could induce invasion‐metastasis cascade in breast cancer.25 We here in Wilms tumour found that LINC00473 promoted the cell proliferation, and its knockdown promoted Bcl‐2‐dependent apoptosis and G1/S arrest via CDK2 and cyclin D1. The interaction among mammalian lncRNAs and miRNAs was the important mechanism for lncRNA to exert its anti‐ or pro‐tumour function. Zhuang et al found that H19 are precursors for the generation of miR‐675 to target RUNX1, a well‐known tumour suppressor, and involved into gastric cell carcinogenesis.26 But, in turn, let‐7 family functioned as miRNA‐triggered lncRNA decay could target LincRNA‐p21, HOTAIR and H19 to degradation.13 LncRNA as miRNA sponges was also an essential regulation way.27 During the gastric carcinogenesis, the expression of HOTAIR was elevated and negatively correlated with miR‐331. HOTAIR acts as a competing endogenous RNA (ceRNA) by binding miR‐331 to abolish the miR‐331‐induced inhibition on HER2, leading to tumour growth through the Akt pathway.28 Similarly, we here demonstrated that LINC00473 was able to sequester miR‐195 to inhibit its function and decrease the miR‐195 level; thus, the anti‐tumour role of miR‐195 was abrogated and activated IKKα‐dependent cell proliferation and survival, which highlighted the miRNA regulation role of LncRNA in cancer.
In summary, we uncovered a dysregulated lncRNA signature LINC00473/miR‐195/IKKα that functioned as pro‐tumour pathogenesis in Wilms tumour and knockdown of LINC00473 could be an alternative therapy for the treatment of Wilms tumour.
CONFLICT OF INTEREST
No competing financial interests exist.
Supporting information
ACKNOWLEDGEMENTS
This study was supported by the Guangdong Provincial Department of Science and Technology Foundation, China (grant no. 2016A020215009).
Zhu S, Fu W, Zhang L, et al. LINC00473 antagonizes the tumour suppressor miR‐195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif. 2018;51:e12416 10.1111/cpr.12416
Contributor Information
Wei Jia, Email: jiawei198044@hotmail.com.
Guochang Liu, Email: starbless2003@126.com.
REFERENCES
- 1. Gratias EJ, Dome JS. Current and emerging chemotherapy treatment strategies for Wilms tumor in North America. Paediatr Drugs. 2008;10:115‐124. [DOI] [PubMed] [Google Scholar]
- 2. Davidoff AM. Wilms' tumor. Curr Opin Pediatr. 2009;21:357‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dome JS, Graf N, Geller JI, et al. Advances in Wilms Tumor treatment and biology: progress through International Collaboration. J Clin Oncol. 2015;33:2999‐3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szychot E, Apps J, Pritchard‐Jones K. Wilms' tumor: biology, diagnosis and treatment. Transl Pediatr. 2014;3:12‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126‐139. [DOI] [PubMed] [Google Scholar]
- 6. Li CH, Chen Y. Targeting long non‐coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895‐1910. [DOI] [PubMed] [Google Scholar]
- 7. Yu G, Li H, Wang J, et al. miRNA‐34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J Urol. 2014;192:1229‐1237. [DOI] [PubMed] [Google Scholar]
- 8. Bera A, Ghosh‐Choudhury N, Dey N, et al. NFkappaB‐mediated cyclin D1 expression by microRNA‐21 influences renal cancer cell proliferation. Cell Signal. 2013;25:2575‐2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253‐1261. [DOI] [PubMed] [Google Scholar]
- 10. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097‐1109. [DOI] [PubMed] [Google Scholar]
- 11. Liu S, Mitra R, Zhao MM, et al. The potential roles of long noncoding RNAs (lncRNA) in glioblastoma development. Mol Cancer Ther. 2016;15:2977‐2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding L, Ren J, Zhang D, et al. The TLR3 agonist inhibit drug efflux and sequentially consolidates low‐dose cisplatin‐based chemoimmunotherapy while reducing side effects. Mol Cancer Ther. 2017;16:1068‐1079. [DOI] [PubMed] [Google Scholar]
- 13. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu L, Ding J, Chen C, et al. Exosome‐transmitted lncARSR promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653‐668. [DOI] [PubMed] [Google Scholar]
- 15. Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR‐205. Cancer Res. 2015;75:1322‐1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z, Li JL, Lin S, et al. cAMP/CREB‐regulated LINC00473 marks LKB1‐inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267‐2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bennett L, Quinn J, McCall P, et al. High IKKalpha expression is associated with reduced time to recurrence and cancer specific survival in oestrogen receptor (ER)‐positive breast cancer. Int J Cancer. 2017;140:1633‐1644. [DOI] [PubMed] [Google Scholar]
- 18. Alameda JP, Navarro M, Ramirez A, et al. IKKalpha regulates the stratification and differentiation of the epidermis: implications for skin cancer development. Oncotarget. 2016;7:76779‐76792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding J, Huang S, Wang Y, et al. Genome‐wide screening reveals that miR‐195 targets the TNF‐alpha/NF‐kappaB pathway by down‐regulating IkappaB kinase alpha and TAB 3 in hepatocellular carcinoma. Hepatology. 2013;58:654‐666. [DOI] [PubMed] [Google Scholar]
- 20. van den Heuvel‐Eibrink MM, van Tinteren H, Bergeron C, et al. Outcome of localised blastemal‐type Wilms tumour patients treated according to intensified treatment in the SIOP WT 2001 protocol, a report of the SIOP Renal Tumour Study Group (SIOP‐RTSG). Eur J Cancer. 2015;51:498‐506. [DOI] [PubMed] [Google Scholar]
- 21. Ludwig N, Nourkami‐Tutdibi N, Backes C, et al. Circulating serum miRNAs as potential biomarkers for nephroblastoma. Pediatr Blood Cancer. 2015;62:1360‐1367. [DOI] [PubMed] [Google Scholar]
- 22. Koller K, Pichler M, Koch K, et al. Nephroblastomas show low expression of microR‐204 and high expression of its target, the oncogenic transcription factor MEIS1. Pediatr Dev Pathol. 2014;17:169‐175. [DOI] [PubMed] [Google Scholar]
- 23. Cao X, Liu D, Yan X, et al. Stat3 inhibits WTX expression through up‐regulation of microRNA‐370 in Wilms tumor. FEBS Lett. 2013;587:639‐644. [DOI] [PubMed] [Google Scholar]
- 24. Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer‐specific CCAT1‐L lncRNA regulates long‐range chromatin interactions at the MYC locus. Cell Res. 2014;24:513‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA‐ATB promotes trastuzumab resistance and invasion‐metastasis cascade in breast cancer. Oncotarget. 2015;6:11652‐11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non‐coding RNA H19‐derived miR‐675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315‐322. [DOI] [PubMed] [Google Scholar]
- 27. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272‐283. [DOI] [PubMed] [Google Scholar]
- 28. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


