Abstract
Objectives
Sprouty (SPRY) 1 is one of the SPRY proteins that inhibits signalling from various growth factors pathways and has also been known as a tumour suppressor in various malignancies. However, no study elucidates the role of SPRY1 in the skin. Our study was conducted to determine the function of SPRY1 in human keratinocytes and the epidermis.
Materials and methods
In vitro primary cultured epidermal keratinocytes were used to investigate the proliferation, differentiation and apoptosis of these cells. We also established overexpression of SPRY1 in vitro and K14‐SPRY1 transgenic mice.
Results
SPRY1 was mainly located in the cytoplasm of the epidermal keratinocytes from the granular epidermal layer of the skin and cultured cells. Overexpressed SPRY1 in keratinocytes resulted in up‐regulation of P21, P27 and down‐regulation of cyclin B1; decrease in MMP3 and integrin α6. SPRY1‐overexpressed primary keratinocytes exhibited a lower proliferation and migration capability and higher rates of apoptosis. Epidermis of SPRY1‐TG mice represented delayed wound healing. Proteomics analysis and GO enrichment showed DEPs of SPRY1 TG mice epidermis is significantly enriched in immune‐ and inflammatory‐associated biological process.
Conclusions
In summary, SPRY1 expression was inversely correlated with cell proliferation, migration and promote cell apoptosis of keratinocytes. SPRY1 maybe a negative feedback regulator in normal human epidermal keratinocytes and cutaneous inflammatory responses. Our study raised the possibility that enhancing expression of SPRY1 may have the potential to promote anti‐inflammatory effects.
1. INTRODUCTION
Sprouty (SPRY) proteins are a highly conserved group of negative feedback loop modulators of the fibroblast growth factor (FGF)‐Ras‐ERK signalling pathway activation and were discovered in a gene screen in Drosophila responsible for shaping the developing trachea.1, 2 Mammalian SPRY proteins comprise of four orthologs: SPRY1, 2, 3 and 4. Their RTK‐induced signalling pathway modulation is growth factor and cell context dependent.1 Subsequent work established the expression of SPRY1, 2, and 4 being widespread in embryonic and adult tissues, whereas expression of SPRY3 was restricted to brain and teses in adult tissues.3, 4 In Drosophila, SPRY proteins play a pivotal role during branching morphogenesis including lung tracheal network formation, nephrogenesis and angiogenesis.5 These proteins act as negative regulators of growth factor signalling pathways. In several malignancies their expression is down‐regulated.6
Loss‐of‐function studies have shown that SPRY1 and SPRY2 play critical roles during early development of multiple‐organ rudiments including kidney, inner ear, teeth, cortical patterning in the brain and eyelid closure.7 Loss of SPRY1 causes specific phenotypes similar to those observed in case of growth factor overdose.8 In addition, SPRY1 may affect tumour cell function through direct interaction with urokinase plasminogen activator receptor and promote its lysosomal degradation.9 SPRY proteins have a crucial role in the regulation of TGF‐β‐induced lens epithelial mesenchymal transition (EMT).10 Overexpression of SPRY1 prevents VEGF secretion and suppresses growth, migration and invasion of human breast cancer cells.11 SPRY1 is downregulated in a variety of cancer types, including breast, prostate, liver, lung and thyroid cancers,12 especially in metastatic malignant stages involving EMT.13 However, it was also reported that SPRY1 mRNA and protein levels were rather increased in the malignant tissue of lung cancer patients.14 Therefore, the role of SPRY1 was controversial in view of the occurrence and progression of malignancies. Although the mechanism by which they function biochemically is still subject of investigation, it is clear that SPRY1 can antagonize RTK signalling. SPRY1 has also diverse cellular functions, such as antiproliferative activity, inhibition of migration, supporting differentiation and survival.1, 15
Despite recent advances in the understanding of roles of SPRY1 on the regulation of FGF receptor signalling in embryogenesis,16 the role of SPRY1 in human skin remains widely unknown. As the inhibitory function of SPRY1 is cell and ligand specific,4 we examined whether SPRY1 affects the differentiation, proliferation, apoptosis and invasion capability of human normal epidermal keratinocytes in vitro and in wound healing, contact hypersensitivity and acute inflammatory responses in K14‐SPRY1 transgenic mouse model in vivo.
2. MATERIALS AND METHODS
2.1. Reagents and antibodies
Antibodies against SPRY1 (1:1000 dilution), β‐actin, E‐cadherin, N‐cadherin and vimentin were purchased from Santa Cruz Bio‐technology, Inc. (Santa Cruz biotechnology, Santa Cruz, CA, USA) and diluted as 1:500. Antibody against K14 and caspase 3 (1:1000 dilution) were purchased from Cell Signaling Technology company. Primary antibody against Ki67 was purchased from Invitrogen (Invitrogen, Auckland, NZ, USA). Dispase, high‐glucose Dulbecco's modified Eagle's medium (DMEM), and foetal bovine serum (FBS) were obtained from Gibco (Paisley, Scotland, UK) and Invitrogen (Invitrogen, Auckland, NZ, USA). EpiGROTM Human Epidermal Keratinocyte Complete Culture Media Kit (EpiGRO) was purchased from Millipore Corporation (Millipore, Bedford, MA, USA). Fluoresce in isothio‐cyanate and Cy3‐conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Cocktail protease inhibitors were purchased from Roche (Roche Diagnostics, Indianapolis, IN, USA).
2.2. Skin sample collection
After obtaining approval by the ethics committee of the Zhejiang University, School of Medicine, 12 donors (mean age 32.6 ± 10.5) undergoing plastic surgery for cosmetic reasons were invited to participate the study. All individuals provided written informed consent. A 1‐2 cm surgical biopsy from donors’ non‐sun‐exposed extremities and/or backs’ skin was taken.
2.3. Immunohistochemistry and Immunofluorescence
For immunohistochemistry (IHC) staining of SPRY1, involucrin and Ki67, OCT embedded sections were used. Slides were then blocked with normal rabbit or goat serum (Vector Laboratories, Burlingame, CA, USA) and incubated with primary antibody followed by incubation with biotinylated rabbit anti‐rat IgG or biotinylated goat anti‐rabbit IgG and treatment of AB complex (Vector Laboratories, Burlingame, CA, USA). Staining was finally visualized with DAB high‐sensitivity substrate chromogen solution (Vector Laboratories, Burlingame, CA, USA) and counterstained with haematoxylin. Immunofluorescence assay was carried out as described in detail before.17 Briefly, skin biopsy specimens (n = 3) were processed, fixed, preheated and blocked using 10% normal FBS for 1 h. Primary monoclonal antibody against human SPRY1 and Cy3‐conjugated secondary antibodies were used. Nuclei were counterstained with DAPI mounting medium. For each case, a negative control incubated with non‐immune mouse IgG (Sigma‐Aldrich, St. Louis, MO, USA) was included. Immunofluorescence cytochemistry on normal human epidermal keratinocytes (NHEK) was carried out according to our previous work.17
2.4. Human embryonic kidney (HEK) 293T cells and primary keratinocytes culture and treatment
HEK293T cells less than 10 passages were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS at 37°C in a humidified atmosphere of 5% CO2 and acted as a tool cell to transfect and produce sufficient lentivirus for further NHEK transfection. NHEK were prepared from skin incubated with dispase as previously described,18 except for the culture medium was replaced by serum‐free medium from Millipore. Second passage with confluence status of NHEK was used in this study except indicated.
2.5. Quantitative real‐time PCR (qRT‐PCR) and PCR analysis
Total RNA was isolated from epidermis of WT and SPRY1‐TG mice by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). qRT‐PCR was done according to previous description.19 CT values were analysed by qBase Plus 2 software (Biogazelle, Zwijnaarde, Belgium).
2.6. Lentiviral infection
The lentiviral vectors were obtained from Applied Biological Materials (ABM) Inc (Richmond, BC, Canada). The amplification, packaging and infection of the lentiviral vectors were carried out according to the manufacturer's protocol. Briefly, lentiviral vectors were firstly amplified by using standard Escherichia coli DH5α strain transformation protocols. Then, the isolated vectors were diluted with ABM's Second Generation (LV003) Packaging Mix in serum‐free medium and mixed with LentiFectin™ Transfection reagent. To produce human SPRY1 lentiviruses, HEK293T cells were transfected with a lentiviral vector plenti‐GIII‐CMV‐hSPRY1‐GFP‐2A‐puro and packaging plasmids. Blank control lentivector, pLenti‐CMV‐GFP‐2A‐Puro‐Blank Vector (LV590), was used as control. Then, the supernatant was harvested and subsequently virus tittering at 106 IU/mL was used to infect NHEK. At 48 h after infection, 2 μg/mL puromycin was added to select virally infected cells for further experiments. Expression of SPRY1 was assayed by qRT‐PCR and Immunoblot.
2.7. Western blot analysis
Cells were lysed with RIPA buffer and lysates were boiled with 4 × Laemmli sample buffer for 5 min. The samples were then separated with SDS‐PAGE gel and immunoblotted with the indicated antibodies. Densitometric analyses were performed using Gel‐Pro Analyzer software V4.0 (Media Cybernetics, Bethesda, MD, USA) and statistical analyses were performed by one‐way analysis of variance followed by an all‐pairwise multiple comparison procedure (Holm‐Sidak method) with an overall significance level of less than 0.05.
2.8. Cell proliferation assay
Lentiviral‐infected NHEK with stable overexpression of SPRY1, along with equal number of blank control lentiviral‐infected keratinocytes were cultured in triplicate (4 × 103 cell/well in 96‐well plates) in DMEM containing 10% FBS at 37°C in 5% CO2 incubator for 48 hours. According to the manufacturer's instruction, CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was performed to analyse the cell viability. The absorbance was measured at 490 nm in Clinibio128 (ASYS‐Hitech, Austria). Three independent experiments were performed.
2.9. Transwell invasion assay
The transwell invasion assay was performed by using the Costar®3422 Cell Invasion Systems (24‐well plates, Corning Corporation, NY, USA) according to the manufacturer's protocol, as previously described.19 Briefly, 1 × 105 NHEK stably expressing SPRY1 Lentivector or the blank control Lentivector was seeded onto the upper surface of a transwell insert precoated with BDMatrigelTMBasement Membrane Matrix. The upper and the low chamber cells were incubated in serum‐free DMEM and DMEM with 10% FBS respectively. After incubation for 24 h, non‐migratory cells on the upper surface were completely removed with a cotton swab. Cells on the lower surface of the insert were fixed by 4% PFA and nuclei were labelled with DAPI. Each cell line was plated in triplicate. The invaded cells were counted in five fields under the fluorescent microscope at 40x magnification.
2.10. Apoptosis assay
Apoptotic events were detected with annexin V PE Apoptosis Detection Kit PE (eBioscience, Inc. San Diego, CA, USA) according to the manufacturer's instruction. Briefly, stable overexpression of SPRY1 and control LV keratinocytes were plated in six‐well plate and incubated at 37°C under normal conditions until 60%‐70% confluence. Then, the cells were collected, washed with PBS and twice with annexin V‐binding buffer, and incubated with annexin V‐FITC at room temperature for 15 min. The cells were also stained with PE to distinguish between live and dead cells. Annexin V‐FITC‐positive cells were determined by flow cytometry under standard conditions.
2.11. Mouse line
K14‐SPRY1 transgenic mice were obtained from Cyagen Biosciences Inc. (Suzhou, Jiangsu, China). All animal experiments were performed in accordance with protocols approved by the Second Affiliated Hospital, Zhejiang University School of Medicine Animal Care Committee. Strains were housed under specific pathogen‐free conditions and investigated at 6‐10 weeks of age. The mice were screened by PCR of genomic DNA for GFP expression, and founder lines validated by genotyping using SPRY1‐specific primers, 5′‐CCTTTCCAATTTACCCGAGCACCT‐3′ (forward) and 5′‐ TTTTCTTGTCTTGGTGCTGTCCGA‐3′ (reverse), which amplifies a 330 bp fragment. Also, the amplified products of PCR were confirmed by direct sequence. The internal mice house‐keeping control gene Rgs7 primers, 5′‐ CAACCACTTACAAGAGACCCGTA‐3′ (forward) and 5′‐ GAGCCCTTAGAAATAACGTTCACC‐3′ (reverse) were designed for PCR. K14–SPRY1 founder lines were identified and crossed with wild‐type C57/BL6 mice. Protein extracted from the mouse skin was determined by Western blot.
2.12. Mouse epidermis acquiring
After sacrifice, mice dorsum skin was removed and cut into pieces. After incubating with 0.5% dispase in 4°C overnight, dermis of skin removed and part of the epidermis were utilized for total epidermis proteins and mRNA extraction as routine.
2.13. Skin wound healing assay in mice
Six SPRY1 TG female mice (7‐week‐old) were used for the study. Age‐ and sex‐matched WT mice (n = 6) were used as controls. Four full‐thickness excisional wounds (6 mm in diameter) on the back skin were generated by sterile disposable biopsy punches (AcuPunch®, Acuderminc. NW, USA). The circle pieces of skin were removed by surgical forceps. The day when the punch biopsy was taken recorded as day 0. Photographs were taken using a digital camera (Nikon Corp., Tokyo, Japan) at day 0, 3, 6, 9 and 12 after the biopsy. Wounds were also monitored and measured by callipers. Length and width of the wounds were used to calculate the wound area in mm2. Healing was defined as the decrease in wound diameter over time and was expressed as the percentage of the day‐0 wound diameter.
2.14. Haematoxylin and eosin (H&E) staining and masson trichrome staining
Mouse skin tissue a specimen was fixed in 10% freshly prepared buffered formalin for 24 h and embedded in paraffin. Sections were cut using a microtome, and collected on superfrost plus slides. The sections were then rehydrated by gradual immersion in 70%, 80%, 95% and 100% ethanol, cleared with xylene and finally stained with H&E staining. Images were taken with an Olympus DP70 camera system (Olympus, Tokyo, Japan). The masson trichrome staining procedure was performed with the operation manual of the masson trichrome staining kit (Solarbio life science, Beijing, China).
2.15. iTRAQ (isobaric tags for relative and absolute quantification)‐based proteomic analysis
Epidermis proteins separated from mouse whole skin (n = 4, Spry1 TG mice and WT mice group) after digestion in 0.25% dispase at 4°C overnight. Peptide preparation with Trypsin Gold (Promega, Madison, WI, USA) at 37°C for 16 h, then desalted with C18 cartridge to remove the high urea, and desalted peptides were dried by vacuum centrifugation. Desalted peptides were labelled with iTRAQ reagents (iTRAQ® Reagent‐8PLEX Multiplex Kit, Sigma), fractionated by HPLC and analysed by LC‐MS/MS with Thermo Scientific™ Q Exactive™ HF‐X. The resulting spectra from each fraction were searched separately against Mus_musculus.fasta database by the search engines: Proteome Discoverer (Thermo Scientific).
The protein quantification results were statistically analysed by Mann‐Whitney Test, the significant ratios, defined as P < .05 and |log 2 FC| >0.585 (ratio >1.5 or ratio <0.67 [fold change, FC]), were used to screen the differentially expressed proteins (DEPs).
The functional analysis of protein and DEPs with Gene Ontology (GO). GO enrichment statistically analysed by Fisher's exact test.
2.16. Statistical analysis
Statistical analyses were performed using SPSS software version 18.0 (IBM SPSS Inc., Chicago, IL, USA) by means of two‐way ANOVA analysis of variance with Bonferroni's post‐test. Grouped data are expressed as mean ± SEM.
3. RESULTS
3.1. Expression of SPRY1 in normal skin and cultured NHEK
To investigate the expression profile of SPRY1 in normal epidermis, immunohistochemistry (IHC) staining against SPRY1 was performed in specimens obtained from three healthy donors, who had signed prior informed consents. According to the results, SPRY1 was mainly detected in epidermis and primarily continuously expressed in the granular layer. In the basal layer, it was only marginal detectable (Figure 1A). Furthermore, we detected the keratinocytes’ terminal differentiation marker of involucrin by IHC and together with SPYR1 by immunofluorescence (IF). As shown in Figure 1A and B, SPRY1 was located in the upper stratum granular compared with involucrin. To further ascertain the localization of SPRY1 in the subcellular compartments, primary cultured NHEK were obtained from healthy donors. As shown in Figure 1C determined by immunofluorescence staining, SPRY1was distributed mainly in the cytoplasm in the cultured NHEK. About 22.2% of the cultured keratinocytes showed bright staining, while less than 77.8% being dim staining (Figure 1C). To further understand that SPRY1 may be involved in the terminal differentiation of keratinocytes, NHEK with different culture time and confluence status were used to detect the expression of SPRY1, together with involucrin and loricrin at mRNA transcriptional and protein levels by qRT‐PCR and western blot. Compared with sub‐confluence NHEK, SPRY1, involucrin and loricrin obviously increased in confluence and post‐confluence NHEK at protein level (Figure 1E). In post‐confluence NHEK, mRNA level of SPRY1 and involucrin up‐regulated as compared with sub‐confluence (Figure 1F). This indicated that SPRY1 may play a role in keratinocytes’ terminal differentiation.
Figure 1.
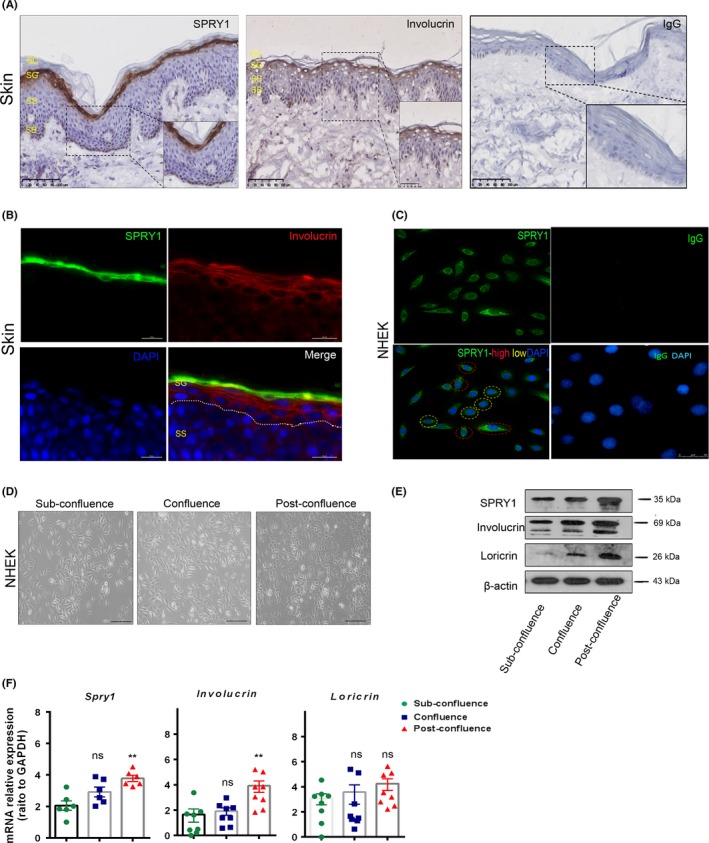
Expression profile of SPRY1 in human normal skin, NHEK and forced overexpression in NHEK. A, Immunohistochemistry staining of SPRY1, involucrin and isotype IgG in normal skin of healthy donors’ skin biopsy. Scale bar = 100 μm, SC: stratum corneum, SG: stratum granular; SS: stratum spinosum; SB: stratum basale; B, Immunofluorescence staining of SPRY1 (green) and Involucrin (red) in normal skin of healthy donors’ skin biopsy. Scale bar = 20 μm. Dotted line indicated the SG and SS. C, Immunofluorescence staining of SPRY1 and isotype IgG in primary NHEK which was obtained from healthy donor. Scale bar = 50 μm. Green: SPRY1 protein; Blue: DAPI; Red dot circle notes SPRY1 specific high fluorescence of NHEK, yellow dot circle notes SPRY1‐specific low fluorescence of NHEK. AC, Primary antibody of mouse IgG was used as negative control compared to SPRY1 antibody. D, Represented photos of cultured NHEK in different culturing status: sub‐confluence, confluence and post‐confluence. Scale bar = 20 μm. E, Western blot of SPRY1, involucrin and loricrin expression in NHEK with different cell confluence status. F, qRT‐PCR analyses SPRY1 and differentiation‐associated genes expression in NHEK. (**P < .01)
3.2. Forced overexpression of SPRY1 established in NHEK
In order to better understand the features of SPRY1 in the epidermis, we established SPRY1‐overexpressed NHEK. We took advantage of the widely used polybrene‐mediated infection of lentivirus for SPRY1 overexpression in NHEK and followed an optimized protocol with slight modifications. We obtained efficient infection in >80% verified by fluorescence microscopy in both SPRY lentivirus (SPRY1 LV) and blank control lentivirus (control LV) groups (Figure 2A). At mRNA level, SPRY1 increased more than 400‐folds in cultured keratinocytes compared to control (Figure 2A). More than 10‐folds increase in SPRY1 at protein level was detected in SPRY1 LV‐transfected keratinocytes compared to control LV‐infected cells (Figure 2A).
Figure 2.
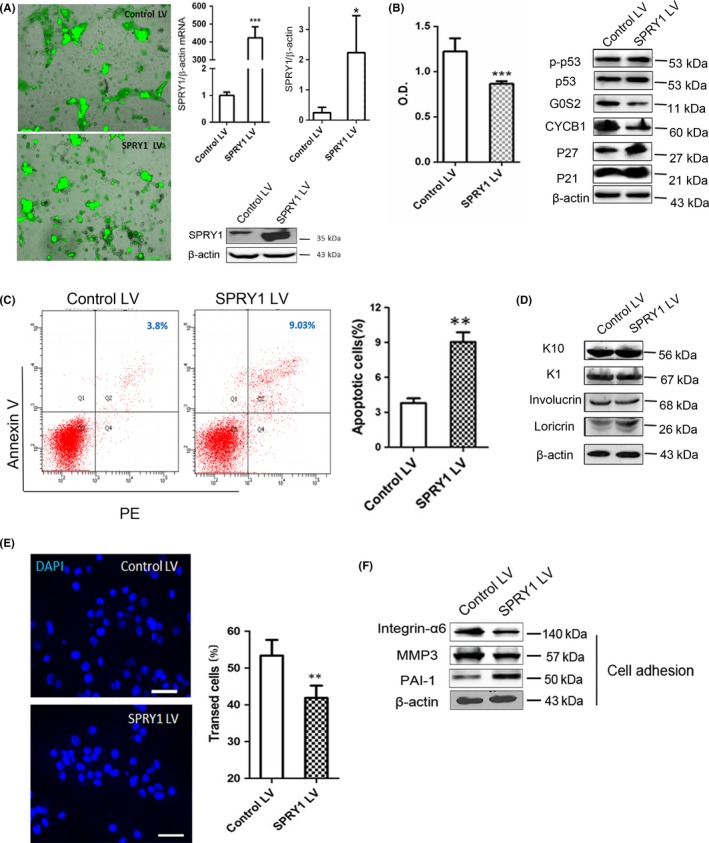
Overexpression of SPRY1 inhibited cell grow and proliferation while promoted apoptosis in NHEK. A, Images of NHEK cells transfected with SPRY1 LV and control after 24 h observed by fluorescence microscopy. The transfection rate was over 80% in all experiments as demonstrated by GFP production. Scale bar = 200 μm. D, mRNA relative expression levels of SPRY1 in NHEK transfected with control vector and SPRY1‐overexpressed vector by qRT‐PCR. Protein levels of SPRY1 LV and control LV transfected NHEK by western blot analysis. β‐actin was used as internal control. The histogram showed quantification of SPRY1 protein levels in two groups. Ctl: control; LV: lentivirus; In histograms, data were showed as mean±SEM and student t test was used. *P < .05, ***P < .001. B, Comparison of cell proliferation in SPRY1 LV and control LV‐transfected primary keratinocytes after 24 hours by MTT assay. Optical density value (O.D) at 490 nm was used for quantitative (n > 3 in each group, ***P < .001). Cell cycle‐related proteins (P21, P27, CYCB1, G0S2, P53 and p‐P53) were detected in SPRY1 LV and control LV group by western blot and β‐actin used as internal control. Alteration of differentiation‐associated proteins of keratinocyte (keratin1, keratin10, involucrin and loricrin) expression was measured by western blot with β‐actin used as internal control. C, Representative images of overexpressed SPRY1 in primary keratinocytes on the cell apoptosis rate determined by FACS. The annexin V and PI both positive cells are the cells undergoing apoptosis and are represented in the upper right quadrant. The mean apoptosis rate of SPRY1 LV group is 9.03% vs 3.8% in the controls and the right histogram shows quantification of apoptosis cell rate between the two groups (**P < .01). D, The expression of differentiation markers K1, K10, loricrin and involucrin in SPRY1 LV and control determined by Western blot. E, Immunofluorescence images of transwell assay indicated migration capability was applied in keratinocytes with forced overexpression of SPRY1. The right histogram showed quantification of at least 10 randomly selected fields after 24 h for rates of transfected cells in transwell assay. Scale bar = 100 μm. Blue: DAPI; **P < .01.) F, Western blot analyse the effect of up‐regulated SPRY1 in primary keratinocytes on the levels of cell adhesion‐related proteins (integrin‐α6, MMP3 and PAI‐1). β‐actin was used as internal control. Representative data from three independent experiments are shown
3.3. Overexpressed SPRY1 inhibited cell proliferation and promoted apoptosis of NHEK
After SPRY1‐overexpressed primary cultured keratinocytes being established, we further examined the role of SPRY1 in cell function of NHEK. As shown in Figure 2B, MTT assay was used to investigate cell proliferation of SPRY1 overexpressed NHEK and increased SPRY1 inhibited primary keratinocytes’ proliferation compared to controls (P < .001). In addition, the mitosis‐regulation proteins cyclin B1 (CYCB1) and G0/G1 switch gene 2 (G0S2) were remarkably decreased in SPRY1‐overexpressed NHEK at protein level (Figure 2B). Cell cycle‐related proteins such as p21, p27, and p53 exert an important role in the cell cycle or apoptosis. In our study, p21 and p27 were slightly increased under SPRY1 overexpression in NHEK, while phospho‐p53 and p53 did not change notably (Figure 2B).
We further analysed the apoptosis of SPRY1‐overexpressed keratinocytes through FACS. Our results showed that the population of annexin V single‐positive apoptotic cells was markedly increased in the SPRY1 LV group as compared to the control LV group (9.03% vs 3.8%, P < .01) (Figure 2C). Up‐regulation of SPRY1 resulted in an increased number of cells undergoing apoptosis, suggesting that SPRY1 contributes to apoptosis of keratinocytes.
3.4. SPRY1 and differentiation of NHEK
As SPRY1 is primarily expressed in the granular layer, we assumed that SPRY1 may be associated with the differentiation of keratinocytes. As shown in Figure 2D, protein levels of the differentiation markers of the epidermal keratinocytes, keratin 10 (K10) and 1 (K1), were not affected by overexpression of SPRY1. Involucrin is a minor component of cornified envelope (CE) and expressed in stratified squamous epithelia in the upper spinous and granular layer.20, 21 In this study, involucrin was not changed upon overexpression of SPRY1 in NHEK. Interestingly, overexpressed SPRY1 up‐regulated loricrin in NHEK (Figure 2D), which is considered as a later differentiation marker of NHEK.20 Therefore, our results suggested that overexpressed SPRY1 may be associated with later differentiation of NHEK which was indicated by the fact that SPRY1 was located primarily in granular layer of epidermis.
3.5. Overexpression of SPRY1 in NHEK‐inhibited cell migration
To further evaluate the role of SPRY1 in keratinocytes, we conducted a transwell assay to examine the migration of SPRY1‐overexpressed epidermal keratinocytes. Our results indicated that migrated SPRY1‐overexpressed cells were much less than control counterparts, which was significantly reduced about by 29.9% (P < .01) in 24 hours (Figure 2E). PAI‐1, a secreted protein that belongs to the serine proteinase inhibitor superfamily and interacts with the extracellular matrix (ECM) components, was up‐regulated; but integrin α6 being decreased, under the condition of SPRY1 overexpression in NHEK. In addition, protein levels of matrix metalloproteinase 3 (MMP3), a family member of MMPs degrading proteoglycan in ECM, were also decreased somewhat in the SPRY1 LV group (Figure 2F).
3.6. Establishment of K14‐SPRY1 transgenic mice
In order to study the function of SPRY1 in vivo, we established skin specifically overexpressed SPRY1 transgenic (SPRY1‐TG) mice with the gene profile K14‐SPRY1. The SPRY1‐TG mice expressed SPRY1 in the tissue or cells with keratin 14 expression, such as epidermis. Firstly, we determined SPRY1 at DNA and protein levels. As shown in Figure 3A, the genomic DNA of SPRY1 was indicated at 330 bp in SPRY1 TG by PCR compared to wild‐type and Rsg7 (632 bp) selected as internal control gene. Western blot result further confirmed overexpressed SPRY1 in the epidermis of SPRY1 TG mice (Figure 3A). Immunohistochemistry and immunofluorescence were performed in SPRY1 TG skin (samples gained from the dorsum). As shown in Figure 3B, SPRY1 was continuously expressed in the epidermis (especially under the stratum corneum) of SPRY1 TG mice. In the zoomed image, it is recognizable that SPRY1 protein was located in cytoplasm, just the same as in NHEK. These results demonstrated that K14‐SPRY1 transgenic mice were successfully established. Both male and female transgenic mice were fertile.
Figure 3.
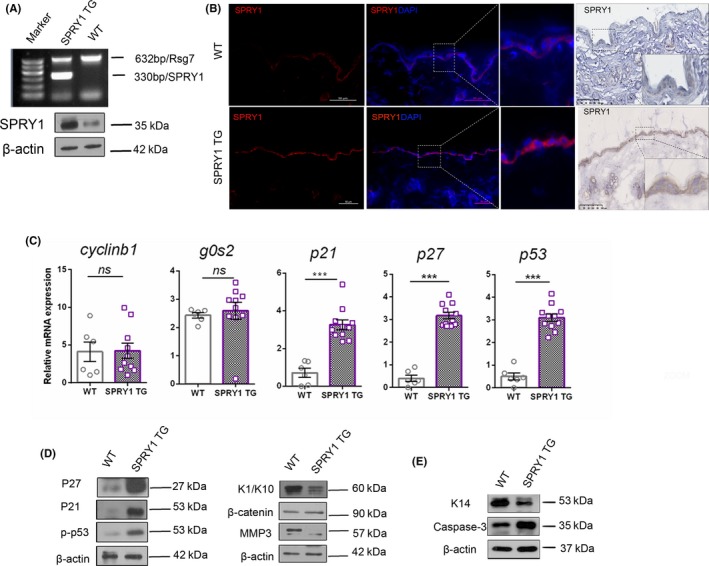
Confirmation of overexpressed SPRY1 protein in the transgenic mice. A, Genotyping was performed by PCR products electrophoresis. The left line was DNA markers, 330 bp band was SPRY1 and Rsg7 (632 bp) was selected as internal control. Western blot analysis of tissue samples from epidermis of SPRY1‐TG and WT. β‐actin was used as internal control. B, SPRY1 expression by immunofluorescence and immunochemistry staining of mice skin from SPRY1‐TG and WT mice respectively. The tissue sections were also stained with DAPI to visualize the nuclei. Scale bar = 100 μm for all panels. C, qRT‐PCR analysis relative mRNA expression of cell cycle‐associated genes (p21, p27, cyclinb1, g0s2, p53) in SPRY1‐TG and WT mice skin. Vertical axis indicated relative expression of mRNA by GAPDH as control. n.s: no statistically significant. ***P < .001. Symbols indicate mean mean ± SEM of six mice per group with student t test. D,E, Images of western blot showed alteration of cell cycle‐related proteins (P27, P21, p‐P53), EMT‐related proteins (K1/K10, β‐catenin, MMP3 and K14 and caspase‐3. β‐actin was used as internal control (Lysate was obtained from epidermis of SPRY1 TG and wild‐type mice)
3.7. Cell cycle and differentiation‐associated proteins of epidermis altered in SPRY1 transgenic mice
To confirm the role of SPRY1 in cell cycle and differentiation in vivo, expression of associated proteins was detected in the epidermis of SPRY1 transgenic mice and wild‐type mice. Epidermis‐derived sample tissues were obtained from both SPRY1 TG and WT mouse skin, then cell cycle, differentiation‐associated proteins were detected by western blot.
Results in Figure 3C demonstrated epidermis‐derived cell cycle‐related proteins, p27, p53 and p‐p53, were notably overexpressed in SPRY TG that consistent with mRNA level by qRT‐PCR. However, cyclinB1 and G0S2 did not change at mRNA level. Terminal differentiation makers, k1 and k10, which are localized in spinous and granular layers, were decreased in the epidermis of SPRY1 TG mice (Figure 3D). Furthermore, the proliferation marker K14 decreased in SPRY1 TG while apoptosis marker of caspase 3 increased (Figure 3E). These results indicated that SPRY1 might play a role in the terminal epidermal differentiation and has the similar role in vivo as in cultured human primary keratinocytes. Overexpression of SPRY1 in the epidermis or keratinocytes suppresses cell proliferation, cell migration, promote cell apoptosis and may contribute to later differentiation of keratinocytes.
3.8. SPRY1 TG mice exhibited delayed skin wound healing
To elucidate whether SPRY1 could act as a suppressor of the epithelial growth, we performed wounding experiments in WT and SPRY1 TG mice. Specifically, four wounds diameter six millimetre (mm) were punched on the shaved dorsum of each mouse (Figure 4A). Wounded skin was harvested at 3, 6 and 9 days after the punch (Figure 4B). The average wound area in the experimental mice of WT (SPRY1 TG) was 13.9 (18.7), 6.9 (12.7) and 2.7 (5.4) mm2, at day 3, 6 and 9 after wound injury. Skin healing delayed at these days in the SPRY1 TG group compared to the WT group with significant statistical difference at day 3 (P < .01), 6 (P < .001) and 9 (P < .01) (Figure 4B). In addition, masson staining of wound tissue sections revealed that SPRY1 TG mice had more severe haemorrhages and more delayed wound healing than WT mice (Figure 4C). Especially at day 3, blood vessels in the granulation tissue of SPRY1 TG mice were much more than those in WT mice (P < .05) under microscopy (Figure 4C). To illustrate the role of SPRY1 in wound healing, we further performed IHC analysis of proliferation marker Ki67 and western blot of myofibroblast differentiation marker α‐SMA in the wound area. As showed in Figure 4D, less Ki67‐positive cells were detected in SPRY1 TG group while α‐SMA upregulated at day 3 and day 6 after punching. This result indicated that SPRY1 may affect the proliferation of epidermis and the maturity of granulation tissue after skin wounded.
Figure 4.
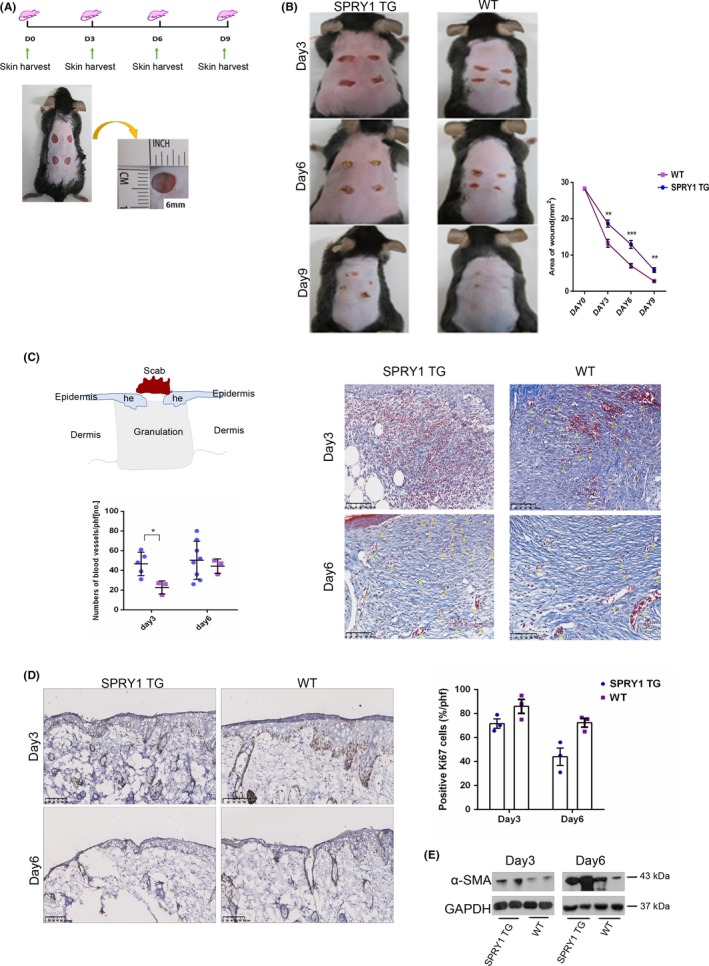
Delayed wound healing process was observed in SPRY1 transgenic mice. A, The graph showed establishment profile of wound healing models, wound tissue sections were harvested at day 0, 3, 6 and 9 post‐injury; Lower picture demonstrated diameter with 6 mm2 punches on shaved back of SPRY1‐TG and WT groups. B, Representative pictures of wound closure process in both groups at day 3, 6, 9 post‐injury. Right histogram showed quantification of wound area at day 0, 3, 6 and 9 after biopsy. C, Diagram demonstrated the histology of excisional wound. Histogram showed quantification blood vessel numbers after injury at day 3 and 6. Manifestation of relevant microscope expression within Masson's trichrome staining in thus two groups. Arrows indicated the blood vessels in granulation. D, Left panel: IHC staining of Ki67 at day 3 and 6. Right panel: Histogram showed quantification of positive Ki67 basal layer cells of left panel. E, Images of α‐SMA protein expression detected by Western Blot. he: hyperproliferative epithelium; hpf: high power field; no.: numbers (data indicated mean score ± SEM, n = 6 at least six mice per group with student t test. *P < .05; **P < .01; ***P < .001)
3.9. Proteomic analysis reveals SPRY1 related to epidermal immune‐ and inflammatory‐associated biological processes
iTRAQ was used to detect the differentially expressed proteins (DEPs) in SPRY1 TG mice’ epidermis compared to WT (n = 4 in each group). According to the experimental data, 2751 proteins were detected and 129 of them were DEPs. As showed in Figure 5A, 82 DEPs were up‐regulated in SPRY1 TG mice compared to WT and 47 DEPs were down‐regulated. Furthermore, based on the GO‐enrichment analysis of biological process of DEPs in SPRY1 TG mice, we found that DEPs are significantly enriched in immune‐ and inflammatory‐associated biological process such as immune response (GO:0006955, P = .0421 P < .05), innate immune response (GO:0045087, P = .0468 P < .05), metabolic processes as well, especially retinoid metabolic process (GO:0001523, P = .0004 P < .001) and steroid metabolic process (GO:0008202, P = .0289 P < .005)(Figure 5B). Additionally, heatmap in Figure 5C presented that pro‐inflammatory cytokines and receptors IL‐1α (Il1a), Complement C3 (C3), IL‐36RA (Il36rn), COX‐1(Ptgs1) and IL‐1RA(Il1rn) were down‐regulated in SPRY1 TG mice while IL‐36β(Il1f8) was a DEP significantly up‐regulated. Thus, indicated SPRY1 may play an anti‐inflammatory role in the immune pathways of skin.
Figure 5.
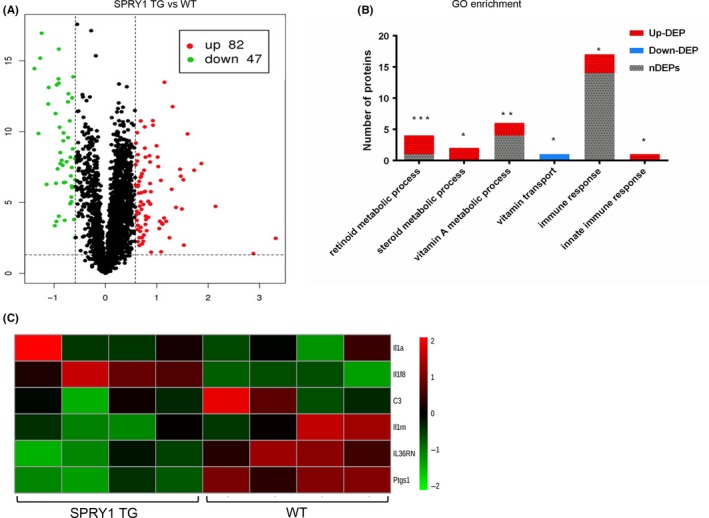
The epidermal proteomics in SPRY1 TG mouse. A, Volcano map demonstrated the up‐ and down‐regulated proteins in SPRY1 TG mice epidermis compared to WT. The green dots represented down‐regulated proteins (n = 42) and red represented up‐regulated proteins (n = 82). B, The histogram shows GO‐enrichment analysis DEPs in Biological Process. Every column shows all detected proteins enriched to GO term, including up‐regulated DEPs (red), down‐regulated DEPs (blue) and non‐DEPs (grey). GO term: immune response (GO:0006955, P = .0421), innate immune response (GO:0045087, P = .0468, retinoid metabolic process (GO:0001523, P = .0004) and steroid metabolic process (GO:0008202, P = .0289). (*P < .05, **P < .01, ***P < .001). C, Heatmap depicting the relative abundance of six detected proteins, which enrich to Biological Process: inflammatory response (GO:0006954) identified from epidermis of Spry1 TG and WT (n = 4). On the right of heatmap showed encoded genes of proteins. The colour key indicates the relative abundance of each protein (−2.0 to 2.0) across eight samples
4. DISCUSSION
It is now well established that SPRY proteins inhibit signalling from various growth factor receptors, including epidermal growth factor (EGF), fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF), platelet‐derived growth factor (PDGF) and glial cell‐derived neurotrophic factor (GDNF).22, 23 As during development, expression of SPRY proteins is mainly observed at sites of known growth factor signalling, a negative feedback loop was also postulated.24 SRPY1 has also been proposed to function as a tumour‐suppressor gene in various malignancies, including prostate cancer,25 hepatocellular carcinoma and thyroid carcinoma.26 However, controversial results evidenced that the level of SPRY1 altered inconsistently in different experiments based on various contexts.24
The concise functions of SPRY1 in epidermal keratinocytes were not determined so far. Therefore, we investigated the role of SPRY1 in the epidermal biology preliminarily. Firstly, we found SPRY1 in normal human skin mainly localized at the granular layer of epidermis and slightly detected at stratum basal. In cultured primary keratinocytes, SPRY1 was expressed in the cytoplasm. Interestingly, SPRY1 displayed high and low stain in cultured epidermal keratinocytes, which may indicate that different levels of SPRY1 may be associated with differentiation or proliferation of keratinocytes differently. Subsequently, forced overexpression of SPRY1 in NHEK was used as a cell model to investigate its functions in vitro. Upon overexpression of SPRY1 in NHEK, the expression profile, including important components in signalling pathways and downstream pertinent molecules were examined. Unexpectedly, in this study, the level of phospho‐ERK/ERK, phospho‐AKT/AKT, phospho‐PTEN/PTEN and phospho‐Smad1, 3/Smad1, 3, 6, 7 were relatively stable and no obvious fluctuation was observed (data not shown). These results indicated that SPRY1‐mediated cell functions might not be driven by these signalling pathways in keratinocytes.
MTT assay has shown that increased SPRY1 inhibited proliferation of primary cultured keratinocytes. Furthermore, SPRY1 LV‐treated NHEK showed a trend of higher rates of cells in S phase and increased annexin V‐positive apoptotic cells determined by FACS. Therefore, it seems that SPRY1 may inhibit cell proliferation but promote apoptosis in the epidermal keratinocytes.
To further define the underlying mechanism behind the role of SPRY1 in keratinocyte proliferation, cell cycle‐associated markers were investigated next. P21 and P27 are well‐known cell cycle inhibitors. In our study, we detected an increase in the expression of the P27 and P21 in SPRY1 LV cells. Cyclin B1 is a family member of proteins that activate specific cyclin‐dependent kinases required for progression through the cell cycle. Decreased cyclin B1 together with increased P21 and P27 would affect the cell cycle progression in the SPRY1 overexpressed NHEK. G0S2 is a basic protein that was identified as an early activating gene in a screen of lectin‐activated human lymphocytes.27 Results from a limited number of studies have implied that G0S2 is a multifaceted protein involved in proliferation, apoptosis and carcinogenesis.27 In human primary fibroblasts, G0S2 interacts with Bcl‐2, promoting apoptosis by preventing the formation of Bcl‐2/Bax heterodimers.17 In our study, forced overexpression of SPRY1 induced a decrease in G0S2. Furthermore, we detected apoptosis marker of caspase 3 and proliferation marker of K14 in epidermis of SPRY1 transgenic mice and caspase 3 increased while K14 increased when compared to WT. Therefore, based on these data, we suggest that SPRY1 may inhibit proliferation and promote apoptosis through regulating cell cycle‐associated protein.
Transwell results showed less migration in SPYR1‐overexpressed NHEK. Till now, little is known about SPRY1 and migration. Here, we investigated the association between SPRY1‐ and ECM‐associated components, including MMP3, PAI‐1 and integrin α6. MMP3 is a family member of peptidase enzymes responsible for the degradation of ECM components. PAI‐1 is transcriptionally regulated by TGF‐β and mediates TGF‐β‐induced inhibition of cell migration and invasion in cancer cells.28 Integrins play a crucial role in epithelial cell‐cell adhesion and cell‐matrix interaction.29, 30 In the SPRY1 LV keratinocytes, MMP3 and integrin α6 were decreased while PAI‐1 was up‐regulated. That is, SPRY1 may regulate migration of keratinocytes through modulating production of ECM and alteration of cell adhesion as well.
As SPRY1 in normal epidermis was mainly in the granular layer, and most of the differentiation markers are localized in this layer, we hypothesized that SPRY1 may also be associated with differentiation of keratinocytes. To confirm this hypothesis, we examined the link between SPRY1 and established differentiation molecular markers, including K1, K10, involucrin and loricrin. Unexpectedly, the expression of K1, K10 and involucrin did not change, except loricrin, a marker for later differentiation,17 which was up‐regulated in SPRY1‐overexpressed NHEK. Moreover, in cultured NHEK, cells with post‐confluence status which contains cells within differentiation phase expressed much more SPRY1 than in sub‐confluence cells. These results suggested that SPRY1 may regulate the later differentiation, but not early differentiation of keratinocytes.
K14‐SPRY1 mice were generated to further clarify the role of SPRY1 in the epidermal biology in vivo. By detecting associated markers in epidermis, the in vivo results were in accordance with the in vitro, which showed decreased proliferation and promoted differentiation by SPRY1 overexpression. An orchestrated cascade of biochemical events will be set into motion to repair skin damage. This process includes haemostasis, inflammation, tissue growth (cell proliferation) and maturation. In our study, we performed wound healing in mice and results showed that SPRY1 TG mice demonstrated a delayed healing process, which may be due to the functions of SPRY1 in inhibiting cell proliferation, migration and promoting apoptosis in keratinocytes. As Ki‐67‐positive basal layer keratinocytes in SPRY1 TG after wounded were much less than WT, while the myofibroblast differentiation marker α‐SMA was up‐regulated, the epidermal migration and granulation tissue maturity were delayed in SPRY1 TG mice. Furthermore, proteomic analysis was conducted through iTRAQ, we compared the DEPs in the epidermis between SPRY1 TG and WT mice. Some pro‐inflammatory cytokines and receptors were down‐regulated. On the basis of GO‐enrichment analysis, the DEPs are significantly enriched in immune‐ and inflammatory‐associated biological process such as immune response, innate immune response, metabolic processes as well, especially retinoid metabolic process and steroid metabolic process. As known that immune pathways play a significant role in inflammatory‐associated diseases including psoriasis. Moreover, retinoid and steroid metabolic processes also involved in the systemic treatments of psoriasis. This indicated that SPRY1 may be associated with the pathogenesis of psoriasis and was worthy of further investigation. These results suggested that SPRY1 was an important negative feedback regulator of cutaneous inflammatory responses. Our study also raised the possibility that enhancing expression of SPRY1 may have the potential to promote anti‐inflammatory effects.
In conclusion, based on the evidence that comes from in vitro overexpression‐derived data in keratinocytes, and epidermal expressed SPRY1 TG mouse in vivo, we suggest that SPRY1 possesses potential characteristics of anti‐proliferative, anti‐migrate, anti‐inflammatory effects and pro‐apoptotic effects. These effects may be associated with SPRY1‐regulated cell cycle protein, integrin α6, MMP3 and PAI‐1. Similar effects on epidermal proliferation, differentiation and wound healing have been observed for transcription factors such as p63, KLF4, CTIP2 and CTIP1, which might regulate cell signalling in a similar manner as SPRY1.31, 32, 33, 34, 35, 36 The role of SPRY1 in the pathogenesis of wound healing and proliferative skin diseases, such as psoriasis, and its interaction with the above transcription factor, deserves extensive research in the next future.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
ACKNOWLEDGEMENTS
We thank all healthy donors for their enthusiastic participation in this study. This study was supported by grants from the National Natural Science Foundation of China (No. 91542124, 81371740, 81371741, 81472019 and 81630082).
Wang P, Zhou Y, Yang J‐Q, et al. The role of Sprouty1 in the proliferation, differentiation and apoptosis of epidermal keratinocytes. Cell Prolif. 2018;51:e12477 10.1111/cpr.12477
Ping Wang, Yuan Zhou and Jian‐Qiang Yang contributed equally to this work.
Contributor Information
Min Zheng, Email: minz@zju.edu.cn.
Xiao‐Yong Man, Email: manxy@zju.edu.cn.
REFERENCES
- 1. Cabrita MA, Christofori G. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis. 2008;11:53‐62. [DOI] [PubMed] [Google Scholar]
- 2. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253‐263. [DOI] [PubMed] [Google Scholar]
- 3. Minowada G, Jarvis LA, Chi CL, et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465‐4475. [DOI] [PubMed] [Google Scholar]
- 4. Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol. 2009;203:191‐202. [DOI] [PubMed] [Google Scholar]
- 5. Edwin F, Anderson K, Ying C, Patel TB. Intermolecular interactions of Sprouty proteins and their implications in development and disease. Mol Pharmacol. 2009;76:679‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma B, Joshi S, Sassano A, et al. Sprouty proteins are negative regulators of interferon (IFN) signaling and IFN‐inducible biological responses. J Biol Chem. 2012;287:42352‐42360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuracha MR, Siefker E, Licht JD, Govindarajan V. Spry1 and Spry2 are necessary for eyelid closure. Dev Biol. 2013;383:227‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Urs S, Henderson T, Le P, Rosen CJ, Liaw L. Tissue‐specific expression of Sprouty1 in mice protects against high‐fat diet‐induced fat accumulation, bone loss and metabolic dysfunction. Br J Nutr. 2012;108:1025‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu X, Lan Y, Zhang D, Wang K, Wang Y, Hua ZC. SPRY1 promotes the degradation of uPAR and inhibits uPAR‐mediated cell adhesion and proliferation. Am J Cancer Res. 2014;4:683‐697. [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao G, Wojciechowski MC, Jee S, Boros J, McAvoy JW, Lovicu FJ. Negative regulation of TGFbeta‐induced lens epithelial to mesenchymal transition (EMT) by RTK antagonists. Exp Eye Res. 2015;132C:9‐16. [DOI] [PubMed] [Google Scholar]
- 11. Mekkawy AH, Pourgholami MH, Morris DL. Human Sprouty1 suppresses growth, migration, and invasion in human breast cancer cells. Tumour Biol. 2014;35:5037‐5048. [DOI] [PubMed] [Google Scholar]
- 12. Macia A, Vaquero M, Gou‐Fabregas M, et al. Sprouty1 induces a senescence‐associated secretory phenotype by regulating NFkappaB activity: implications for tumorigenesis. Cell Death Differ. 2014;21:333‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shin EH, Basson MA, Robinson ML, McAvoy JW, Lovicu FJ. Sprouty is a negative regulator of transforming growth factor beta‐induced epithelial‐to‐mesenchymal transition and cataract. Mol Med. 2012;18:861‐873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rathmanner N, Haigl B, Vanas V, Doriguzzi A, Gsur A, Sutterluty‐Fall H. Sprouty2 but not Sprouty4 is a potent inhibitor of cell proliferation and migration of osteosarcoma cells. FEBS Lett. 2013;587:2597‐2605. [DOI] [PubMed] [Google Scholar]
- 15. Felfly H, Klein OD. Sprouty genes regulate proliferation and survival of human embryonic stem cells. Sci Rep. 2013;3:2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang X, Harkins LK, Zubanova O, et al. Overexpression of Spry1 in chondrocytes causes attenuated FGFR ubiquitination and sustained ERK activation resulting in chondrodysplasia. Dev Biol. 2008;321:64‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochim Biophys Acta. 2013;1831:276‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Man XY, Li W, Chen JQ, et al. Impaired nuclear translocation of glucocorticoid receptors: novel findings from psoriatic epidermal keratinocytes. Cell Mol Life Sci. 2013;70:2205‐2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cai SQ, Dou TT, Li W, et al. Involvement of pituitary tumor transforming gene 1 in psoriasis, seborrheic keratosis, and skin tumors. Discov Med. 2014;18:289‐299. [PubMed] [Google Scholar]
- 20. Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp Dermatol. 2012;21:643‐649. [DOI] [PubMed] [Google Scholar]
- 21. Henry J, Toulza E, Hsu CY, et al. Update on the epidermal differentiation complex. Front Biosci (Landmark Ed). 2012;17:1517‐1532. [DOI] [PubMed] [Google Scholar]
- 22. Faedo A, Borello U, Rubenstein JL. Repression of Fgf signaling by sprouty1‐2 regulates cortical patterning in two distinct regions and times. J Neurosci. 2010;30:4015‐4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S, Bui Nguyen TM, Kovalenko D, et al. Sprouty1 inhibits angiogenesis in association with up‐regulation of p21 and p27. Mol Cell Biochem. 2010;338:255‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kral RM, Mayer CE, Vanas V, Gsur A, Sutterluty‐Fall H. In non‐small cell lung cancer mitogenic signaling leaves Sprouty1 protein levels unaffected. Cell Biochem Funct. 2014;32:96‐100. [DOI] [PubMed] [Google Scholar]
- 25. Wang J, Thompson B, Ren C, Ittmann M, Kwabi‐Addo B. Sprouty4, a suppressor of tumor cell motility, is down regulated by DNA methylation in human prostate cancer. Prostate. 2006;66:613‐624. [DOI] [PubMed] [Google Scholar]
- 26. Macia A, Gallel P, Vaquero M, et al. Sprouty1 is a candidate tumor‐suppressor gene in medullary thyroid carcinoma. Oncogene. 2012;31:3961‐3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Payne KJ, Dovat S. G0S2–a new player in leukemia. Leuk Res. 2014;38:147‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Humbert L, Lebrun JJ. TGF‐beta inhibits human cutaneous melanoma cell migration and invasion through regulation of the plasminogen activator system. Cell Signal. 2013;25:490‐500. [DOI] [PubMed] [Google Scholar]
- 29. Tjin MS, Chua AW, Ma DR, Lee ST, Fong E. Human epidermal keratinocyte cell response on integrin‐specific artificial extracellular matrix proteins. Macromol Biosci. 2014;14:1125‐1134. [DOI] [PubMed] [Google Scholar]
- 30. Stemmler S, Parwez Q, Petrasch‐Parwez E, Epplen JT, Hoffjan S. Association of variation in the LAMA3 gene, encoding the alpha‐chain of laminin 5, with atopic dermatitis in a German case‐control cohort. BMC Dermatol. 2014;14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356‐360. [DOI] [PubMed] [Google Scholar]
- 32. Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang LJ, Bhattacharya S, Leid M, Ganguli‐Indra G, Indra AK. Ctip2 is a dynamic regulator of epidermal proliferation and differentiation by integrating EGFR and Notch signaling. J Cell Sci. 2012;125:5733‐5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang X, Bhattacharya S, Bajaj G, et al. Delayed cutaneous wound healing and aberrant expression of hair follicle stem cell markers in mice selectively lacking Ctip2 in epidermis. PLoS ONE. 2012;7:e29999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S, Teegarden A, Bauer EM, et al. Transcription factor CTIP1/BCL11A regulates epidermal differentiation and lipid metabolism during skin development. Sci Rep. 2017;7:13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koster MI, Dai D, Marinari B, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3255‐3260. [DOI] [PMC free article] [PubMed] [Google Scholar]


