Abstract
Objectives
Long non‐coding RNAs have identified to involve into the tumour cell proliferation, apoptosis and metastasis. We previously found that up‐regulated LncRNA‐SNHG7 (SNHG7) positively correlated to the Fas apoptosis inhibitory molecule 2 (FAIM2) in lung cancer cells with unclear mechanism.
Methods
Non‐small cell lung cancer (NSCLC) and relative normal tissues (n = 25) were collected. The SNHG7 expression and function in NSCLC was determined. The SNHG7‐miR 193b‐FAIM2 network was analysed in vitro and vivo.
Results
We reported that oncogene SNHG7 predicted a poor clinical outcome and functioned as competitive endogenous RNA (ceRNA) antagonized microRNA‐193b (miR‐193b) to up‐regulate the FAIM2 level in NSCLC. Bioinformatic analysis predicted that SNHG7 harboured miR‐193b‐binding sites, and we found decreased miR‐193b levels in NSCLC tissues when compared to relative normal tissues. Luciferase assays indicated that overexpression of miR‐193b inhibited the Ruc expression of plasmid with miR‐193b‐binding sites of SNHG7 in a dose‐dependent manner. Ectopically expressed SNHG7 also as a molecular sponge sequestered endogenous miR‐193b. Besides, FAIM2 was found to be directly targeted by miR‐193b. The restoration of miR‐193b levels in NSCLC cell lines A549 and H125 suppressed the expression of FAIM2 and related tumour proliferation, metastasis and induced apoptosis. However, forced expression of SNHG7 could down‐regulate miR‐193b to elevate the FAIM2 level of tumour cells, leading to impaired miR‐193b/FAIM2‐induced tumour progression. Knockdown of SNHG7 in vivo significantly delayed the tumour growth with decreased tumour volume, which accompanied with enhanced miR‐193b expression and reduced FAIM2 levels.
Conclusion
The results indicated that miR‐193b is indispensible for the ceRNA role of SNHG7 in FAIM2‐supported tumourigenesis of lung cancer.
1. INTRODUCTION
Lung cancer as a global health burden is a leading cause of cancer‐related mortality worldwide with 221 200 estimated new cases.1 Traditionally, lung cancers consists of non‐small cell carcinoma and small cell carcinoma (small cell lung carcinoma, SCLC), with the former accounting for approximately 80% of the cases and the remaining 20% of cases are SCLC. The majority of non‐small cell lung cancer (NSCLC) patients present with locally advanced or metastatic disease, leading to limited therapeutic options and the overall 5‐year survival rates of those patients was approximately 17%.2, 3 Therefore, understanding of tumour biology and pathogenesis of NSCLC is conducive to clarifying favourable response to targeted therapy.
Currently, the majority of transcribed RNAs in mammalian cells are non‐coding RNAs (ncRNAs) with no protein‐coding capacity, which are generally divided into 2 major classes based on the following sizes including microRNAs (miRNAs, <200 nt in length), which mediate post‐transcriptional gene silencing, and long non‐coding RNAs (lncRNAs, >200 nt in length), which act as signals, guides or scaffolds to chromatin to regulate the expression of target genes.4, 5 The aberrant expression of miRNA and LncRNA was found to be involved into the progression of cancer.6, 7, 8, 9 miR‐34 and let‐7 were found to be tumour‐suppressive miRNAs in lung cancer, systemic nano‐delivery of miR‐34 and let‐7 suppressed tumour growth via the inhibition of survival signatures including ITGB3 and MAP4K3, which improved the survival advantage.10, 11 LncRNA‐MALAT‐1 was up‐regulated during the tumourigenesis of lung cancer and demonstrated to be associated with metastasis in NSCLC patients.12 Fas apoptotic inhibitory molecule 2 (FAIM2) could inhibit Fas‐mediated cell death and was found to be overexpressed in SCLC cell lines.13, 14 In our previous study, we identified that the expression of lncRNA‐SNHG7 (SNHG7) was enhanced in lung cancer and positively correlated to FAIM2 to promote tumour proliferation and survival in vitro.15 However, the potential regulation mechanism of SNHG7/FAIM2 is unclear in lung cancer.
Interestingly, the cross‐regulations between miRNAs and lncRNAs have been found in many diseases, including cancer.16, 17 The miRNAs can target lncRNAs to reduce lncRNA stability, whereas lncRNAs can also function as competitive endogenous RNA (ceRNA) to antagonize miRNAs or lncRNAs compete with miRNAs to modulate miRNAs availability to silence target mRNAs.4 During the lung carcinogenesis, an oncogenic gene LncRNA‐UCA1 was highly up‐regulated and competitively spongeing miR‐193a‐3p to increase its target gene ERBB4 levels for the development of NSCLC.18 LncRNA‐NEAT1 was found to accelerate NSCLC cell growth and metastasis, and functioned as a ceRNA for hsa‐miR‐377‐3p to de‐repress its endogenous targets E2F3.19
In this study, we uncovered that high SNHG7 levels predicted a poor clinical outcomes in patients with lung cancer. Besides, SNHG7 functioned as ceRNA to competitively inhibit miR‐193b availability, which down‐regulated the miR‐193b levels to enhance its target gene FAIM2‐induced tumour survival, anti‐apoptosis and metastasis. Knockdown of SNHG7 efficiently inhibits tumour growth with decreased miR‐193b/FAIM2 signature in vivo.
2. MATERIALS AND METHODS
2.1. Tumour samples, cell lines and reagents
In this study, NSCLC tissues (n = 25) and corresponding relative normal tissues (n = 25) were collected from Southern Medical University according to the ethical and legal standards of Southern Medical University. All of the patients diagnosed with primary NSCLC were confirmed by haematoxylin and eosin staining by experienced pathologists. Written informed consent was obtained from all of the patients. The lung cancer cell line H125, 95D, A549 and normal lung epithelial cells Beas‐2B cells were purchased from the cell bank of the Chinese academy of sciences (Shanghai, China) and cultured in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% FBS (Life Technologies, California, Carlsbad, USA), ampicillin and streptomycin at 37°C, 5% CO2 conditions. Negative control and oligonucleotide sequences of miR‐193b mimics, inhibitors or negative control were purchased from RiboBio (Guangzhou, china). The overexpression vector pcDNA3.0‐ SNHG7 and reporter plasmid of full‐length 3′‐UTR (wild type or mutant) of FAIM2, Renilla luciferase reporter vector psiCHECK2‐miR‐193b, psiCHECK2‐SNHG7‐wild or mut were conducted by GenePharma (Shanghai, China). Anti‐GAPDH, E‐cadherin and N‐cadherin antibodies were obtained from Cell Signalling Tech (Denver, MA, USA) and Abcam (Massachusetts, Cambridge, USA).
2.2. Luciferase reporter assay
The A549 and H125 cells were co‐transfected containing psiCHECK2‐miR‐193b, psiCHECK2‐SNHG7‐wild or mut, or psiCHECK2‐3′UTR (wild or mut) of FAIM2, miR‐195 mimics. Luciferase activity was measured using the Dual‐Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase acted as a reporter gene for normalized control.
2.3. RNA isolation and qRT‐PCR
Total RNA from tissues or tumour cells was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the standard RNA isolation protocol. Quantitative real‐time RT‐PCR (qRT‐PCR) was performed, and the expression levels of SNHG7/miR‐193b/FAIM2 were normalized to GAPDH for gene expression. The primers were listed as Table 1.
Table 1.
Primers for qRT‐PCR
| Gene | Primer |
|---|---|
| SNHG7 | F: GTTGGGGTGTTGGCATTCTTGTT |
| R: GCGCCCAATACGACCAAATC | |
| miR‐193b | F: ACACTCCAGCTGGGAACTGGCCCTCAAAGTCC |
| R: CTCAACTGGTGTCGTGGA | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| FAIM2 | F: GGCGTGCTCTTCGTGCTTC |
| R: TGGCGTCGGTACCCATCA | |
| GAPDH | F: ACACCCACTCCTCCACCTTT |
| R: TTACTCCTTGGAGGCCATGT |
2.4. Cell transfection
The A549 and H125 cells were cultured to about 80% confluence in 6/12‐well plates and then, using Lipofectamine 2000 (Invitrogen), the cells were transfected with indicated agents according to the manufacturer's instructions. After transfection for the indicated time, the cells were harvested for further experiments.
2.5. CCK‐8 assay
A549 cells were transfected with negative control, miR‐193b mimics and/or pcDNA3.0‐FAIM2/SNHG7 for 48 hours, and the cells were harvested and washed with PBS and then cell counting kit‐8 (Kumamoto, Japan) mixed with DMEM was used for cell viability assay, and the absorbance was measured at 450 nm by a microplate reader.
2.6. Transwell assay
2 × 104 A549 and H125 cells transfected with negative control, miR‐193b mimics and/or pcDNA3.1‐FAIM2/SNHG7 were in the upper chamber of a non‐coated transwell insert for the migration assay, and for the invasion assay, the upper chamber of the transwell inserts were coated with Matrigel, and tumour cells were plated in the upper chamber of the Matrigel‐coated transwell insert. Cells that did not migrate or invade were removed using a cotton swab and were stained by crystal violet and counted under an inverted microscope. Five random views were selected to count the cells.
2.7. Flow cytometry assay
For the cell apoptosis, 2 μL of annexin V mixed with 2 μL of propidium iodide (eBioscience, Waltham, Massachusetts) was used to stain cells and analysed using a flow cytometry provided with the Cell‐Quest software.
2.8. Western blotting
The tumour tissues were collected and the protein was abstracted. According to the manufacturer's instructions, proteins were separated by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane (Bio‐Rad, Hercules, CA, USA) and were blocked in 5% non‐fat milk in TBST buffer (tris buffer saline containing 0.1% Tween‐20) for 1 hour at room temperature, and subsequently incubated with anti‐GAPDH, E‐cadherin and N‐cadherin antibodies overnight at 4°C. After washing with TBST buffer, the blots were then incubated with HRP‐conjugated secondary antibody for 1 hour at room temperature. The blots were washed with TBST buffer and visualized using the ECL‐Plus reagent (Millipore, Billerica, MA, USA). GAPDH was used as the loading control in the Western blotting.
2.9. Tumour model
A549 and H125 cells were transfected with lentivirus vector of siRNA‐SNHG7, overexpressed SNHG7 or negative control, 2 × 106 tumour cells were subcutaneously injected in rear flank of nude mice (4 per group). The tumour sizes were measured 1 week apart and the tumour volumes were calculated: V (cm3) = width2 (cm2) × length (cm)/2.
2.10. Statistical analyses
The results are analysed by the Statistical Package for Social Sciences version 16.0 (SPSS 16.0; SPSS Inc., Chicago, IL, USA) and the Prism statistical software package (version 5.0, Graphpad Software Inc., San Diego, California). Unpaired t tests or Mann‐Whitney U tests were used to compare the 2 groups, and multiple group comparisons were analysed with 1‐way ANOVA. P < .05 was considered statistically significant. All experiments were performed at least 3 times.
3. RESULTS
3.1. High expression of SNHG7 correlates to poor clinical outcome
We previously reported that the expression of SNHG7 was elevated in NSCLC, but the correlations between the clinicopathological characteristics of NSCLC patients and SNHG7 expression were unknown. NSCLC tissues (n = 25) were collected to determine the SNHG7 level and the Q‐PCR results showed that the SNHG7 level positively correlated to the high TNM stage (Figure 1A) and lymph node metastases (Figure 1B). Thus, high expression of SNHG7 in clinical was an oncogene during the development of NSCLC.
Figure 1.
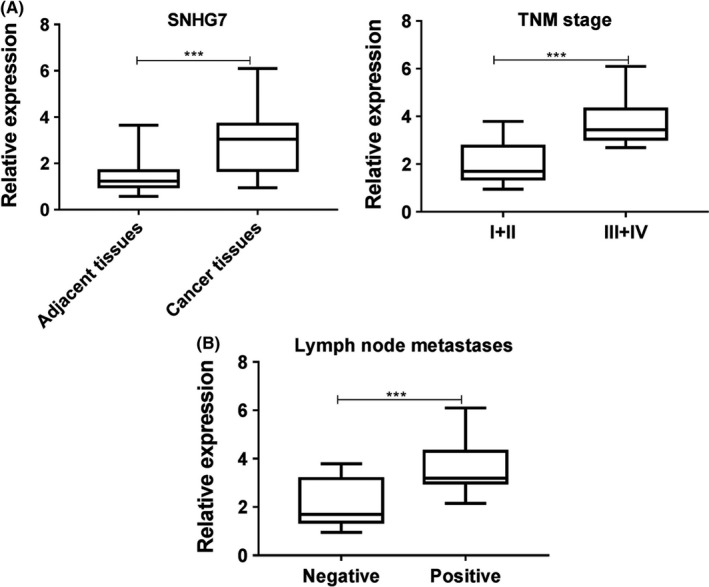
The expression of SNHG7 positively correlates to poor clinical outcome. The expression of SNHG7 in lung cancer tissues was determined by Q‐PCR and the correlation between SNHG7 and (A) TNM or (B) LNM was analysed. ***P < .001
3.2. SNHG7 as ceRNA contains functional miR‐193b interaction sites
Because we have found that the expression of FAIM2 positively correlated to SNHG7 levels in lung cancer with unclear mechanism, we speculated that SNHG7 could modulate miRNA that target FAIM2 to control the FAIM2 levels in NSCLC. Bioinformatic analysis uncovered that human SNHG7 harboured putative complementary sequences for miR‐193b and one predicted miR‐193b‐binding sites (7 bp) were found (Figure 2A).
Figure 2.
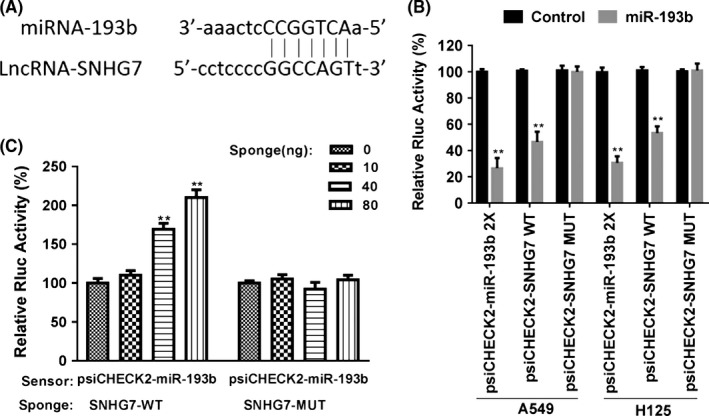
SNHG7 as a molecular sponge antagonizes miR‐193b availability. (A) The predicted miR‐193b‐binding sites for SNHG7. (B) miR‐193 and psiCHECK2‐miR‐193 or psiCHECK2‐SNHG7 were co‐transfected into A549 and H125 cells and the Rluc expression was determined. (C) The Rluc activity of psiCHECK2‐miR‐193b was analysed after overexpression of SNHG7 in Beas‐2B cells. **P < .01, data represent the means ± SD
Then, we conducted the plasmid psiCHECK2‐miR‐193b which harboured 2 copies of miR‐193b‐binding sites in the 3′UTR of a Renilla luciferase (Rluc) gene, downstream constitutively expressed firefly luciferase gene was also contained as an internal control for normalization. miR‐193b was co‐transfected with psiCHECK2‐miR‐193b into NSCLC cell lines A549 and H125, the results showed that miR‐193b significantly inhibited Rluc expression in a dose‐dependent manner in 2 cell lines, which was also observed with psiCHECK2‐SNHG7‐wild containing a human SNHG7 fragment at the 3′UTR of the Rluc in place of miR‐193b‐binding sites (Figure 2B). To confirm that the observed inhibition was dependent on the predicted miR‐193b sites, psiCHECK2‐SNHG7‐mut with deleted sequences complementary to the seed region of miR‐193b could not respond to miR‐193‐indeced inhibition of Rluc expression in a dose‐dependent manner, which suggested that inhibition was dependent on the predicted miR‐193b sites (Figure 2B). Human normal lung epithelial cells Beas‐2B express low expression of SNHG7 but appreciable levels of miR‐193b. Overexpression of full‐length human SNHG7 (sponge) in Beas‐2B cells was able to increase the Rluc activity of psiCHECK2‐miR‐193b, which could be explained by the down‐regulation of endogenous miR‐193b specifically sequestered by SNHG7 (Figure 2C). These data indicated the ceRNA role of SNHG7 to antagonize miR‐193b availability in lung cancer cells.
3.3. FAIM2 is directly targeted by miR‐193b
We have demonstrated that SNHG7 could inhibit the miR‐193b function, thus we next analysed the relationship between miR‐193b and FAIM2. The expression of miR‐193b was decreased in NSCLC tissues and lung cancer cell lines (Figure 3A), which was negatively associated with the FAIM2 level (Figure 3B), indicating that miR‐193b might negatively regulate the expression of FAIM2.
Figure 3.
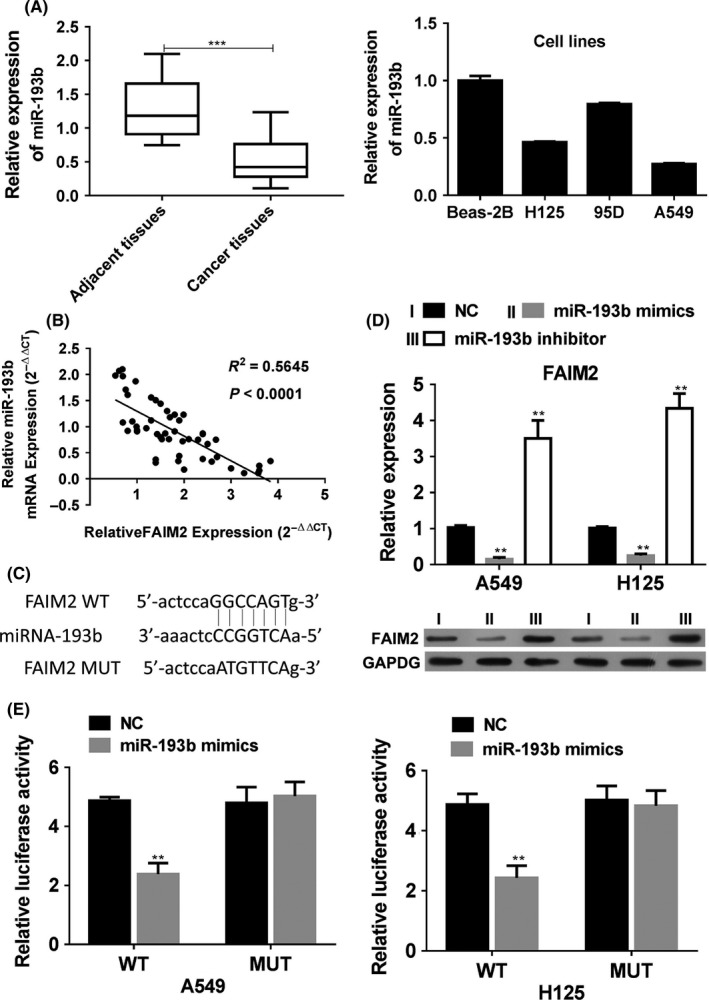
FAIM2 is a direct target of miR‐193b. (A) The expressions of miR‐193b in non‐small cell lung cancer tissues and in H‐125, 95D, A549 and Beas‐2B cells were determined by Q‐PCR and its relationship with FAIM2 was also assessed (B). (C) Sequence alignment of putative miR‐193b‐binding sites in the FAIM2 3ʹ‐UTRs. (D) After the transfection of miR‐193b mimics, inhibitors or negative control in A549 and H125 cells, the expression of FAIM2 was determined by Q‐PCR and WB. (E) A549 and H125 cells were transfected with full‐length 3′‐UTR (wild type or mutant) of FAIM2, and the luciferase reporter was performed to confirm the direct target sites. **P < .01, ***P < .001, data represent the means ± SD
According to miRWalk (http://www.targetscan.org/, we found that FAIM2 harboured a binding site of miR‐193b which indicated that FAIM2 could be targeted by miR‐193b (Figure 3C). Besides, overexpression of miR‐193b mimics in A549 and H125 cells inhibited the expression of FAIM2 in mRNA and protein level, which, but, could be restored by the miR‐143b inhibitors (Figure 3D). To conform the directly target relationship, the luciferase reporter assays were performed. A luciferase reporter vector with full‐length 3′‐UTR (wild‐type or mutant) of FAIM2 was co‐transfected with miR‐193b mimics into A549 and H125 cells (Figure 3E). The results showed that miR‐193b mimics could significantly impair the luciferase activity of wild‐type FAIM2‐3′UTR, but not the mutant‐type, indicating that FAIM2 is direct target of miR‐193b in lung cancer.
3.4. SNHG7 promotes tumour growth and metastasis via miR‐193b/FAIM2 signals
In our previous study, we found that FAIM2 promoted the proliferation and inhibited the apoptosis of A594 cells. Thus, we next investigated the impacts of miR‐193b on FAIM2 function in A549 and H125 cells. The results showed that overexpression of FAIM2 improved the proliferation, migration and invasion of tumour cells. However, restoration of miR‐193b in tumour cells significantly repressed FAIM2‐induced proliferation, migration and invasion (Figure 4A,B). Moreover, forced expressed miR‐193b was able to increase tumour cell apoptosis, despite of the pro‐survival effects of FAIM2 in NSCLC (Figure 4C). Because SNHG7 could antagonize miR‐193b availability in lung cancer cells, we found that overexpression of SNHG7 reduced the miR‐193b levels but up‐regulated the FAIM2 (Figure 4D), which contributed to FAIM2‐induced proliferation, migration and invasion of A549 and H125 cells via miR‐193b (Figure 4E‐G).
Figure 4.
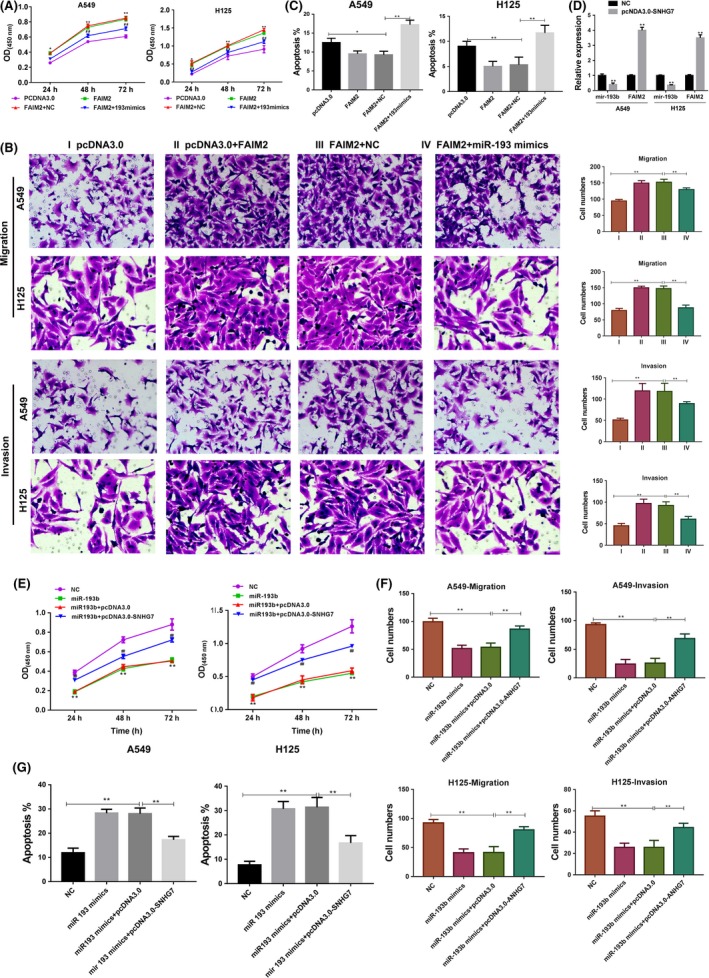
SNHG7 regulates FAIM2‐induced tumour growth and metastasis via miR‐193b. (A) The A549 and H125 cells were transfected with negative control, miR‐193b mimics and/or pcDNA3.0‐FAIM2, the proliferation was estimated by CCK‐8, (B) the migrations and invasions of 2 tumour cells were determined by transwell assay and (C) the apoptosis was estimated by flow cytometry. (D) Overexpression of SNHG7 in A549 and H125 cells, the miR‐193b and FAIM2 levels was estimated. (E‐G) The A549 and H125 cells were transfected with negative control, miR‐193b mimics and/or pcDNA3.1‐SNHG7, the proliferation, migrations and invasions, and apoptosis were analysed. *P < .05, **P < .01, ***P < .001, data represent the means ± SD
3.5. Knockdown of SNHG7 restricts the development of lung cancer in vivo
To confirm the oncogene role of SNHG7 in vivo, the xenograft model of human A549 and H125 cells was established and the nude mice were administrated of conditional tumour cells with SNHG7 overexpression or knockdown. Knockdown of SNHG7 significantly restricted the tumour growth with small tumour volume, whereas the mice treated with overexpressed SNHG7 bared the largest tumour (Figure 5A). The expression of SNHG7, miR‐193b and FAIM2 was determined and found that SNHG7 in vivo inhibits the miR‐193b level to up‐regulate FAIM2 (Figure 5B). Besides, the expression of epithelial marker E‐cadherin was dramatically reduced in mice with SNHG7 overexpression, but expression of mesenchymal marker N‐cadherin was significantly up‐regulated. (Figure 5C,D).
Figure 5.
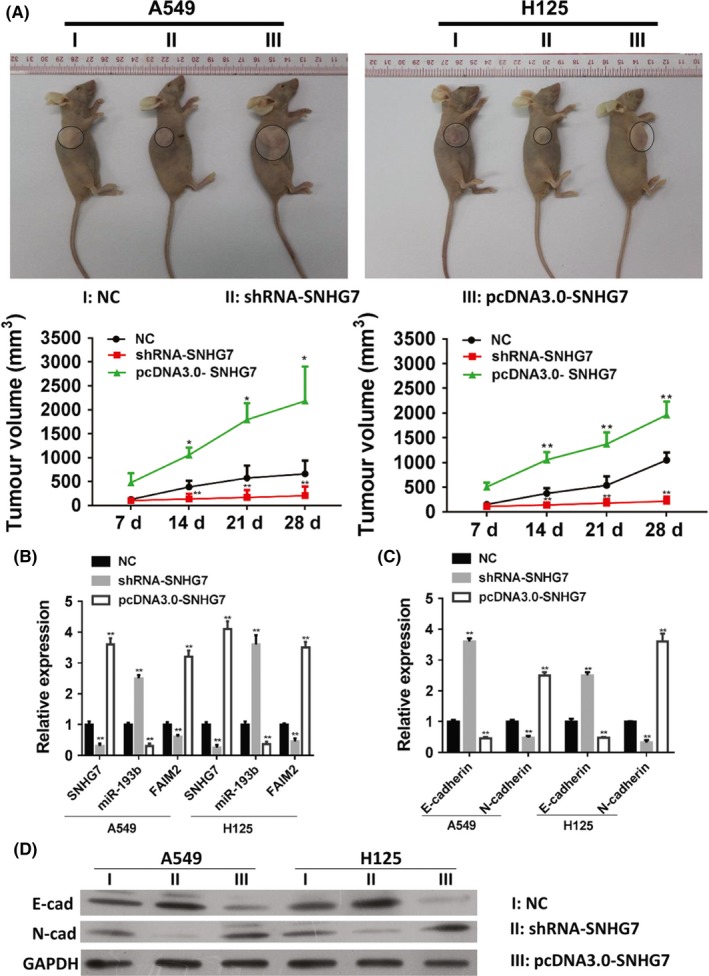
Knockdown of SNHG7 restricts the growth of non‐small cell lung cancer in vivo. 2 × 106 conditional A549 and H125 cells (knockdown or overexpression of SNHG7) were subcutaneously injected in rear flank of nude mice (4 per group). (A) The mean tumour size (mm3) was analysed. (B) The expressions of SNHG7/miR‐193b/ FAIM2 were estimated by Q‐PCR. The expression of EMT markers (E‐cadherin and N‐cadherin) was assessed by Q‐PCR (C) and WB (D). *P < .05, **P < .01, data represent the means ± SD
4. DISCUSSION
Lung cancer is being the most common incident cancer in china, and the leading cause of cancer death in 2015.20 Retrospective data showed that the prognosis for advanced‐stage lung cancer is very poor, with a 5‐year overall survival rate of 17%. Besides, the 5‐year overall survival rate for widely disseminated disease only has 4.2%.3 Therefore, understanding the mechanism of proliferation and metastasis of lung cancer favoured to find new target of therapy. Non‐coding RNAs (ncRNAs) have been proposed to be effective targets of cancer therapy.21 We here found that we uncovered that high SNHG7 levels predicted a poor clinical outcomes in patients with lung cancer. Knockdown of SNHG7 efficiently inhibited tumour proliferation, metastasis and promoted apoptosis of lung cancer cells. Similarly, Lu et al22 reported that the inhibition of a new LncRNA linc00673 significantly attenuated the tumourigenesis ability of A549 cells in vivo, indicating the oncogene role of linc00673. Overexpression of LncRNA MEG3 decreased NSCLC cells proliferation and induced apoptosis in vitro and impeded tumourigenesis in vivo, which suggested the anti‐tumour role of MEG3.23 High expression of MEG3 was associated with advanced pathologic stage, and tumour size of patients with NSCLC. Therefore, SNHG7 and linc00673 were oncogene but MEG3 was a tumour suppressor in lung cancer.
Amounting evidences over the past decade have also begun to uncover the interaction among mammalian lncRNAs and miRNAs. The interaction among mammalian lncRNAs and miRNAs was the important mechanism for lncRNA to exert its anti‐ or pro‐tumour function.4 Zhuang et al24 found that H19 was precursors for the generation of miR‐675 to target RUNX1, a well‐known tumour suppressor, and involved into gastric cell carcinogenesis. The abundance of numerous lncRNAs could be controlled by miRNAs. Liu et al25 reported that miR‐211 level was enhanced in colon cancer to target and decrease the tumour suppressor transcript LOC285196. The ceRNA hypothesis proposes that transcripts with shared miRNA‐binding sites compete for post‐transcriptional control. LncRNAs as ceRNAs have been demonstrated in different types of cancer. In MCF‐7 breast cancer cells, TUG1 knockdown was significantly associated with decreased cell proliferation and it promoted apoptosis of breast cancer cells through the regulation of miR‐9.26 LncRNA CRNDE in colorectal cancer could bind to miR‐181a‐5p and represses its expression, leading to the up‐regulation of Wnt/β‐catenin signalling and cell proliferation and chemoresistance.27 H19 functioned as ceRNA for miR‐138 and miR‐200a, antagonized their functions and increased endogenous targets Vimentin, ZEB1 and ZEB2 for epithelial to mesenchymal transition of colorectal cancer cells.28 In this study, SNHG7 modulated miR‐193b availability by acting as a molecular sponge, which down‐regulated the miR‐193b levels to enhance its target gene FAIM2‐induced tumour survival, anti‐apoptosis and metastasis.
In conclusion, we found that miR‐193b was required for SNHG7 to induce FAIM2‐supported proliferation, anti‐apoptosis and metastasis of lung cancer. We highlighted the ceRNA role of SNHG7 during FAIM2‐related carcinogenesis. Knockdown of SNHG7 in vivo could be a promising target therapy for lung cancer.
DISCLOSURE STATEMENT
No competing financial interests exist.
ACKNOWLEDGEMENTS
This study was supported by Southern Medical University.
She K, Yan H, Huang J, Zhou H, He J. miR‐193b availability is antagonized by LncRNA‐SNHG7 for FAIM2‐induced tumour progression in non‐small cell lung cancer. Cell Prolif. 2018;51:e12406 10.1111/cpr.12406
REFERENCES
- 1. Mery B, Guy JB, Swalduz A, et al. The evolving locally‐advanced non‐small cell lung cancer landscape: building on past evidence and experience. Crit Rev Oncol Hematol. 2015;96:319‐327. [DOI] [PubMed] [Google Scholar]
- 2. Antoni D, Mornex F. Chemoradiotherapy of locally advanced nonsmall cell lung cancer: state of the art and perspectives. Curr Opin Oncol. 2016;28:104‐109. [DOI] [PubMed] [Google Scholar]
- 3. Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11:1411‐1422. [DOI] [PubMed] [Google Scholar]
- 4. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272‐283. [DOI] [PubMed] [Google Scholar]
- 6. Ding L, Ren J, Zhang D, et al. The TLR3 agonist inhibit drug efflux and sequentially consolidates low‐dose cisplatin‐based chemoimmunotherapy while reducing side effects. Mol Cancer Ther. 2017;16:1068‐1079. [DOI] [PubMed] [Google Scholar]
- 7. Chen D, Dang BL, Huang JZ, et al. MiR‐373 drives the epithelial‐to‐mesenchymal transition and metastasis via the miR‐373‐TXNIP‐HIF1alpha‐TWIST signaling axis in breast cancer. Oncotarget. 2015;6:32701‐32712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Augoff K, McCue B, Plow EF, Sossey‐Alaoui K. miR‐31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple‐negative breast cancer. Mol Cancer. 2012;11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liang D, Xiao‐Feng H, Guan‐Jun D, et al. Activated STING enhances Tregs infiltration in the HPV‐related carcinogenesis of tongue squamous cells via the c‐jun/CCL22 signal. Biochim Biophys Acta. 2015;1852:2494‐2503. [DOI] [PubMed] [Google Scholar]
- 10. Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non‐small cell lung cancer. Oncogene. 2015;34:3547‐3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao B, Han H, Chen J, et al. MicroRNA let‐7c inhibits migration and invasion of human non‐small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43‐51. [DOI] [PubMed] [Google Scholar]
- 12. Ji P, Diederichs S, Wang W, et al. MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22:8031‐8041. [DOI] [PubMed] [Google Scholar]
- 13. Planells‐Ferrer L, Urresti J, Coccia E, et al. Fas apoptosis inhibitory molecules: more than death‐receptor antagonists in the nervous system. J Neurochem. 2016;139:11‐21. [DOI] [PubMed] [Google Scholar]
- 14. Kang HC, Kim JI, Chang HK, et al. FAIM2, as a novel diagnostic maker and a potential therapeutic target for small‐cell lung cancer and atypical carcinoid. Sci Rep. 2016;6:34022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA‐SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673‐2680. [DOI] [PubMed] [Google Scholar]
- 16. Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let‐7 microRNAs. Mol Cell. 2013;52:101‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kataoka M, Wang DZ. Non‐coding RNAs including miRNAs and lncRNAs in cardiovascular biology and disease. Cells. 2014;3:883‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nie W, Ge HJ, Yang XQ, et al. LncRNA‐UCA1 exerts oncogenic functions in non‐small cell lung cancer by targeting miR‐193a‐3p. Cancer Lett. 2016;371:99‐106. [DOI] [PubMed] [Google Scholar]
- 19. Sun C, Li S, Zhang F, et al. Long non‐coding RNA NEAT1 promotes non‐small cell lung cancer progression through regulation of miR‐377‐3p‐E2F3 pathway. Oncotarget. 2016;7:51784‐51814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271‐289. [DOI] [PubMed] [Google Scholar]
- 21. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460‐469. [DOI] [PubMed] [Google Scholar]
- 22. Lu W, Zhang H, Niu Y, et al. Long non‐coding RNA linc00673 regulated non‐small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR‐150‐5p. Mol Cancer. 2017;16:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu KH, Li W, Liu XH, et al. Long non‐coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non‐coding RNA H19‐derived miR‐675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315‐322. [DOI] [PubMed] [Google Scholar]
- 25. Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53‐regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976‐4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao XB, Ren GS. LncRNA taurine‐upregulated gene 1 promotes cell proliferation by inhibiting MicroRNA‐9 in MCF‐7 cells. J Breast Cancer. 2016;19:349‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR‐181a‐5p‐mediated regulation of Wnt/beta‐catenin signaling. Mol Cancer. 2017;16:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513‐22525. [DOI] [PMC free article] [PubMed] [Google Scholar]


