Abstract
Objectives
FBXW7 acts as a tumour suppressor by targeting at various oncoproteins for ubiquitin‐mediated degradation. However, the clinical significance and the involving regulatory mechanisms of FBXW7 manipulation of NSCLC regeneration and therapy response are not clear.
Materials and Methods
Immunohistochemical staining and qRT‐PCR were applied to detect FBXW7 and Snai1 expression in 100 samples of NSCLC and matched tumour‐adjacent tissues. FBXW7 manipulation of cancer biological functions were studied by using MTT assay, immunoblotting, flow cytometry, transwells, wound healing assay, and sphere‐formation assays. Immunofluorescence and co‐immunoprecipitation were used to analyse the possible interaction between Snai1 and FBXW7.
Results
We detected the decreased FBXW7 expression in majority of the NSCLC tissues, and lower FBXW7 level was correlated with advanced TNM stage. Furthermore, those patients with decreased FBXW7 expression tend to have both poorer 5‐year survival outcomes, and shorter disease‐free survival, comparing to those with higher FBXW7 levels. Functionally, we found that FBXW7 enforcement suppressed NSCLC progression by inducing cell growth arrest, increasing chemo‐sensitivity and inhibiting Epithelial‐mesenchymal Transition (EMT) progress. Results further showed that FBXW7 could interact with Snai1 directly to degrade its expression through ubiquitylating alternation in NSCLC, which could be partially abrogated by restoring Snai1 expression.
Conclusions
FBXW7 conduction of tumour suppression was partly through degrading Snai1 directly for ubiquitylating regulation in NSCLC
Keywords: cancer stem‐like cells, chemotherapy resistance, epithelial‐mesenchymal transition, FBXW7, non‐small‐cell lung cancer, snai1
1. INTRODUCTION
In recent years, extensive effort has been made towards the diagnosis and therapeutics of lung cancer, which firmly ranks the first in terms of cancer incidence and cancer‐associated mortality worldwide.1, 2 Non‐small‐cell lung cancer (NSCLC) accounts for over 80% of all lung cancer cases, with the 5‐year survival rate being approximately 15%. Distant metastasis and long‐term recurrence are the major obstacles to improve survival. Previous studies have been performed to dig out metastasis‐associated genetic alterations in NSCLC, however, crucial factors that contribute to lung cancer metastasis are still not determined, and identification of the molecular mechanism of carcinogenesis and metastasis is urgent for developing potential therapeutic targets and strategies.
FBXW7 (F‐box and WD repeat domain‐containing 7, FBXW7, Cdc4, Ago, Sel10) is an evolutionarily conserved F‐box protein, containing two essential functional domains (F‐box and WD), which are necessary for function exertion.3, 4 The F‐box domain mediates Skyp1 binding for SCF complex formation, and the WD repeats as a substrate proteins‐binding domain, form a β‐propeller structure to bind substrates phosphorylated motifs (CPD, Cdc4 phosphodegron).5, 6 Recently, it has been reported that FBXW7 mediated the ubiquitin‐dependent proteolysis of multiple crucial oncoproteins such as Myc, c‐Jun, Cyclin E and Notch1 most of which are involved in the diverse cellular processes, suggesting the suppressive role of degrading these oncoproteins. FBXW7 is commonly mutated in various types of tumours, and the overall mutation rate is approximately 6%. However, the precise mechanism of FBXW7 regulation of tumour initiation and progression is still unknown.
Epithelial‐mesenchymal transition (EMT) is fundamental to malignant progression of cancer,7, 8 which is a developmental process involving loss of apical polarity and obtaining of mesenchymal phenotype, contributing to increased migratory and invasive properties. Also, EMT could help to generate and enrich cancer stem‐like cells (CSC), the small subpopulation of cells with a high tumorigenic and self‐renewal capacity and exist in various human malignancies, including NSCLC.8, 9, 10, 11 To date, CSCs are thought to be responsible for tumour occurrence, recurrence and metastasis. Emerging evidence indicates that FBXW7 plays a pivotal role in EMT, stem cells’ renewal and differentiation. Hui et al. reported that FBXW7 suppressed EMT and stemness potential of cholangiocarcinoma cells through inhibition of mTOR signalling12; Rustighi et al. suggested that FBXW7 decreased the number of breast cancer stem‐like cells and inhibited their self‐renewal capacity by restraining Notch activity.13 A study on gastric cancer showed that FBXW7 induced tumour growth arrest and EMT inhibition in part by targeting RhoA.14 Our group found that miR‐367 could target at FBXW7/Wnt signalling to control the stem cells’ fates of NSCLC.3 The regulatory mechanism of FBXW7 in tumorigenesis and progression is mainly realized via ubiquitin‐mediated degradation of different oncoproteins, as were reported in these studies.
Snai1 is a critical transcription factor for EMT by binding to and sequentially inhibiting E‐cadherin promoter, which reduced cell adhesion and promoted migratory capacity. In addition, current studies have shown that Snai1 is implicated in the regulation of chemo‐resistance and the emergence of cancer stem‐like cell (CSC) phenotype.15, 16, 17 The further elucidation of Snai1 in EMT and CSC provides a critical insight into the development of metastatic cancer and long‐term recurrence. Several F‐box proteins (Fbxw1, Fbxl14, Fbxl5, Fbxo11 and Fbxo45) that targeted Snai1 for degradation have been studied. Furthermore, some studies have reported that FBXW7 inactivation promoted EMT process through regulation of the Snail1 in various human cancers.12, 14 However, the role of FBXW7‐mediated Snai1 degradation remains unclear in NSCLC.
In this study, we planned to explore the role of FBXW7 in NSCLCs’ generation and progression, and hypothesized that FBXW7 is a potent prognostic factor and acts as a tumour suppressor in NSCLC partly by targeting Snai1 for ubiquitination and proteasomal degradation.
2. METHODS AND MATERIALS
2.1. Clinical samples and cell lines
One hundred NSCLC tissue and matched normal tumour‐adjacent tissue specimens were obtained from the First Affiliated Hospital of Xi'an Jiaotong University between 2012 and 2017. All patients have been histopathologically and clinically diagnosed according to the American Joint Committee on Cancer criteria. Detailed demographic features and clinicopathologic data of all patients are shown in Table 1. The study was approved by the Xi'an Jiaotong University Ethics Committee according to the 1975 Helsinki Declaration, and all patients’ signed informed consent was obtained. The NSCLC cell lines (A549, H460, H1299 and PC9), the human bronchial epithelial cells (BEAS‐2B) and 293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), and maintained in Dulbecco's modified Eagle medium (DMEM, Gibco, Grand Island, NY, USA), supplemented with 10% FBS (HyClone, Logan, UT, USA),1% penicillin/streptomycin (Sigma, St‐Louis, MO, USA) and routinely incubated at 37°C in a humidified 5% CO2 incubator. The identity of cell lines was confirmed by short tandem repeats‐polymerase chain reaction (STR‐PCR) genotyping.
Table 1.
Correlation between the expression of Fbxw7 and clinicopathological characteristics of the NSCLC patients
| Clinicopathological characteristics | Cases (100) | FBXW7 expression | P | |
|---|---|---|---|---|
| Low (63) | High (37) | |||
| Age (y) | ||||
| <60 | 50 | 28 | 22 | .147 |
| ≥60 | 50 | 35 | 15 | |
| Gender | ||||
| Male | 83 | 55 | 28 | .853 |
| Female | 17 | 8 | 9 | |
| Tumour size (cm) | ||||
| ≤ 3 | 26 | 13 | 13 | .110 |
| >3 | 74 | 50 | 24 | |
| TNM tumour stage | ||||
| I + II | 66 | 37 | 29 | .045* |
| III + IV | 34 | 26 | 8 | |
| Histology | ||||
| Adenocarcinoma | 62 | 46 | 16 | .004** |
| Squamous cell carcinoma | 27 | 10 | 17 | |
| Other | 11 | 7 | 4 | |
| Degree of differentiation | ||||
| Well | 26 | 18 | 8 | .046* |
| Moderately | 36 | 17 | 19 | |
| Poorly | 38 | 28 | 10 | |
| Lymph node metastasis | ||||
| Yes | 41 | 30 | 11 | .079 |
| No | 59 | 33 | 26 | |
| Smoking history | ||||
| Yes | 68 | 46 | 22 | .161 |
| No | 32 | 17 | 15 | |
*P < .05; **P < .01.
2.2. Immunohistochemistry and immunofluorescence
The immunohistochemistry (IHC) staining was performed as previously described. Briefly, paraformaldehyde‐fixed paraffin sections were deparaffinized and rehydrated. Antigen retrieval was performed in citrate buffer (Beyotime) at 100°C for 10 minutes. The sections were blocked with 3% hydrogen peroxide, followed by incubated with anti‐FBXW7(1:100, Abcam, USA, ab105752) and anti‐Snai1 (1:50, CST, USA, #3879) antibody overnight at 4°C. Slides were then incubated with secondary antibodies for 1 hour at room temperature, developed using 3‐diaminobenzidine (DAB) solution and counterstained with hematoxylin. The immunhistochemical staining of FBXW7 and Snai1 was evaluated by two pathologists in a blinded manner. The percentage of positive cells was divided into five rankings: 0, less than 10%; 1, 10%‐30%; 2, 30%‐50%; 3, more than 50%. The expression of FBXW7 and Snai1 were defined as positive when the score was >3.
For immunofluorescence assay, A549 and HEK‐293T cell lines (2.5 × 104/well) were seeded on glass coverslips in 6‐well plates for 24 hours. These cells were then fixed with 4% paraformaldehyde for 1 hour at room temperature and permeabilized with 0.1% Triton X‐100 for 5 minutes. Next, the fixed cells were blocked with 5% BSA for 30 minutes at 37°C and incubated with primary antibodies against FBXW7 and Snai1 overnight at 4°C. After three washes with PBS for 5 minutes each in the dark, the cells were incubated with secondary antibodies Alexa Fluor‐conjugated anti‐rabbit IgG or anti‐mouse IgG (each, 1:200 dilution, Abbkine, USA) for 1 hour at 37°C.
2.3. Real‐time PCR and Western blot
Total RNA from tissues and cells was extracted by TRIZOL Reagent (Invitrogen, USA) following the manufacturer's instructions. CDNA was synthesized with SYBR PrimeScript RT‐PCR Kit (TaKaRa, Dalian, China). Real‐time PCR was conducted, using SYBR Green qPCR Kit (TaKaRa, Dalian, China) and performed on CFX96 Real‐time PCR detection system (Bio‐Rad). GAPDH was used as an endogenous control and the fold‐difference was the calculated, using the 2‐ΔΔCt method. The primers sequences were list in Table 2. A western bolt (WB) analysis was performed as previously described. Antibodies against EMT markers (including E‐cadherin, N‐cadherin, ZO‐1 and Vimentin) were purchased from Cell signaling technology (CST, USA, #9782) and used according to manufacturer's protocol. The others primary antibodies were used as follows: anti‐FBXW7 (1:500 dilution; Abcam, USA, ab105752), anti‐Snai1 (1:1000 dilution; CST, USA, #3879), anti‐c‐Myc (1:1000 dilution; CST, USA, #13987), anti‐Notch1 (1:1000 dilution; Abnova, USA, 78486), β‐actin (1:1000 dilution; Bioss, China, bs‐0061R), anti‐Sox2 (1:500 dilution; Proteintech, USA, 11064‐1‐AP), anti‐Oct4 (1:500 dilution; Proteintech, USA, 11263‐1‐AP), anti‐Nanog (1:500 dilution; CST, USA, #4903), anti‐TCF4 (1:500 dilution; Santa Cruz, USA, sc‐166699), anti‐Flag (1:1500 dilution; Sigma‐Alorch, USA, F7425); anti‐HA (1:1000 dilution; Proteintech, USA, 51064‐2‐AP); anti‐Ub (1:500 dilution; Santa Cruz, USA, sc‐8017).
Table 2.
Primer sequences of genes
| Gene | Primer sequence of genes |
|---|---|
| FBXW7 | F: 5′‐CACAGGCCTTCAAGAGTGGC‐3′ |
| R: 5′‐TTGCATCATATGCTTCACTTGTGT‐3′ | |
| Snai1 | F: 5′‐ CCT CAA GAT GCA CAT CCG AAG CCA C‐3′ |
| R: 5′‐ CCG GAC ATG GCC TTG TAG CAG C‐3′ | |
| ABCG2 | F: 5′‐CAC AAG GAA ACA CCA ATG GCT‐3′ |
| R: 5′‐ACA GCT CCT TCA GTA AAT GCCTTC‐3′ | |
| Oct4 | F: 5′‐ACA TCA AAG CTC TGC AGA AAG AAC‐3′ |
| R: 5′‐CTG AAT ACC TTC CCA AAT AGA ACC C‐3′ | |
| Sox2 | F: 5′‐GGG AAA TGG GAG GGG TGC AAA AGA‐3′ |
| R: 5′‐TTG CGT GAG TGT GGA TGG GAT TGG‐3′ | |
| Nanog | F: 5′‐AGA AGG CCT CAG CAC CTA‐3′ |
| R: 5′‐GGC CTG ATT GTT CCA GGA TT‐3′ | |
| GAPDH | F: 5′‐GGT GGT CTC CTC TGA CTT CAA CA‐3′ |
| R: 5′‐GTT GCT GTA GCC AAA TTC GTT GT‐3′ |
2.4. Cell viability, migration and invasion
Cell viability assay was evaluated, using a Cell Counting Kit‐8 (CCK‐8, Dojindo Lab, Japan). A549 and H1299 cells were planted in 96‐well plates (3 × 103 cells/well) and then incubated for 0, 24, 48 and 72 hours. After incubation for indicated times, CCK‐8 solution was added per well and plates were incubated at 37°C for 1 hour. The optical density values of each wells was measured by a microplate reader at a wavelength of 450 nm.
Cell migration and invasion assay were carried out, using Boyden chambers containing 8 μm pore filters (Coring Incorporated, NY, USA). After transfected with the indicated virus, 3 × 104 cells were cultured in serum‐free medium for 24 hours and seeded in the upper chamber for cell migration assay. Here, 5 × 104 cells were treated with virus transfection and serum starvation were added to the upper chamber coated with Matrigel (BD Biosciences) for cell invasion assay. Medium containing 10% FBS was added to lower chamber to stimulate migration or invasion. After 36 hour of incubation, migrated cells were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet and counted, using Image‐pro plus software.
2.5. Wound healing assay
For the wound healing assay, cells were cultivated in 6‐well culture plates (4 × 105 cells/well) and grown to 80%‐90% confluence overnight. The confluent monolayer cells were scratched with a sterile 200 μL micropipette tip and washed with PBS buffer to clear cell debris. The scratched cells were incubated in serum‐free medium for 0 h and 48 h at regular culture conditions. Closed area of the wound was determined under an inverted microscopy at 0 and 48 hours.
2.6. Vector construction, transfection and reagents
Human FBXW7 and Snai1 cDNA were amplified by PCR and cloned into pHBLV lentiviral vector to generate the pHBLV‐Flag‐FBXW7 and pHBLV‐HA‐Snai1 recombinant plasmid. The human FBXW7‐targeting shRNA sequences (targeting sequence: 5′‐AACACAAAGCUGGUGUGUGCA‐3′) were cloned into pHBLV‐U6‐Puro vector to generate pHBLV‐U6‐FBXW7‐RNAi. The lentiviral production and transduction were performed as described previously. The expression Snai1‐specific small interfering RNA (siRNA) was purchased from GenePharma Co., Ltd (Shanghai, China) and transfected using the Lipofectamine 2000 reagent (Invitrogen, CA, USA) according to the manufacturer's protocol. MG132 and Cycloheximide (CHX) were purchased from ApexBio (Houston, USA) and dissolved in dimethyl sulfoxide (DMSO).
2.7. Sphere‐formation and flow cytometry assays
Single cells (100 cells and 1000 cells per well) were plated onto 24‐well ultra‐low attachment plate (Corning) with serum‐free DMEM/F12 medium supplemented with 1% B27 (Gibco, 17504‐044), 20 ng/mL EGF (epidermal growth factor) (PeproTech, AF‐100‐15, USA), 10 ng/mL bFGF (basic fibroblast growth factor) (PeproTech, 100‐18B, USA). The primary generation spheres were collected and counted after 7 days of culture. Spheres were resuspended and seeded into fresh medium to promote further generations. After 14 days of culture, the second‐generation spheres were evaluated by an inverted microscope for their number and size.
For flow cytometry assays, the NSCLC cells were routinely cultured, harvested and re‐suspended into single cells in stain buffer, and then stained with PE‐conjugated anti‐CD44 (BioLegend, USA) and FITC‐CD133 (BioLegend, US) for 20 minutes at 4°C in the dark, followed by three additional washes. After staining, cells were subjected to flow cytometry for analysis, using FACS Calibur machine (Becton, Dickinson and Company, USA).
2.8. Co‐immunoprecipitation
For co‐immunoprecipitation assay, beads (Dynabeads® Protein G Immunoprecipitation Kit, Thermo Fisher, cat# 10007D) were mixed with the primary antibodies or lgG and shaken on a rotating shaker at 30 minutes at room temperature. Total protein lysates were extracted from cells, using NP40 cell lysis buffer, and the lysates were incubated with beads‐Ab complex. After 1 hour incubation at room temperature, beads‐Ab‐antigen complex was collected by the magnet and washed 3 times, using 200 μL washing buffer. A quantity of 20 μL elution buffer and 10 μL SDS‐PAGE sample buffer was added to beads‐Ab‐antigen complex before heating for 10 minutes at 70°C. The supernatant was collected and used for western blot analysis.
2.9. Statistical analysis
All statistical analyses were carried out by SPSS 16.0 software (SPSS Inc.). Data were presented as mean±SD. Comparisons between two groups were analysed with the Student's t‐test. A Pearson's correlation coefficient analysis was performed to assess the relationship between FBXW7 and Snai1 expression in NSCLC cancer tissues. The clinical significance of FBXW7 in NSCLC samples was evaluated, using Pearson χ2 test and Kaplan‐Meier survival analysis. P‐values < .05 were considered statistically significant.
3. RESULTS
3.1. Decreased FBXW7 is significantly correlated with poor survival outcomes in patients with NSCLC
We firstly investigated the expression of FBXW7 in 100 pairs of primary NSCLC tumour versus adjacent non‐tumour tissues, using immunohistochemistry (IHC) staining. Representative IHC images of FBXW7 staining were shown in Figure 1A, and FBXW7 expression was significantly decreased in specimens of NSCLC, comparing to adjacent tissues (Figure 1B). This trend was further testified in randomly selected specimens by western blot (Figure 1C). The association between FBXW7 and clinicopathologic features were listed in Table 1 in detail. Chi‐square test indicated that the lower FBXW7 expression in NSCLC was significantly associated with histology, later tumour stage and advanced differentiation degree. Moreover, gene set enrichment analysis (GSEA) on the RNA‐sequencing data from public profiles helped to prove that higher FBXW7 expression was related with better survival in patients with NSCLC (Figure 1D, left), which was also supported by Kaplan‐Meier Plotter analysis (kmplot.com/analysis/) (Figure 1D, right). Kaplan‐Meier analyses and log‐rank tests were also made to reveal the lower FBXW7 expression‐associated poorer overall survival (P = .00241) and lower disease‐free survival rates (P = .0351). These results indicated that FBXW7 may function as an anti‐oncogene involved in the development and progression of human NSCLC (Figure 1E).
Figure 1.
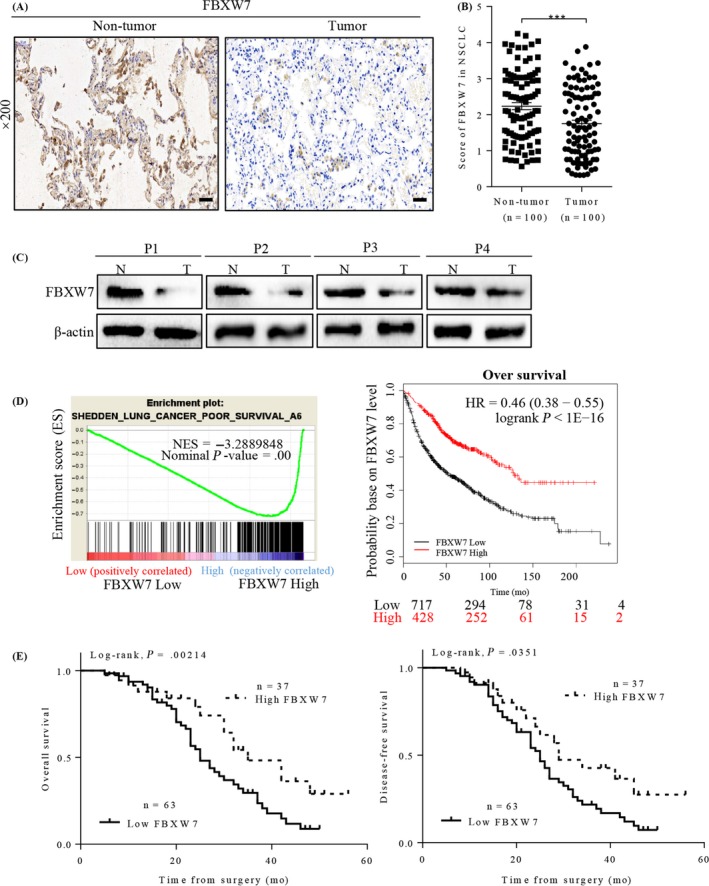
FBXW7 expression and its clinical significance in NSCLC patients. A, Representative images of FBXW7 IHC staining in one pair of matched primary NSCLC samples and their corresponding non‐tumour tissues (Scale bars: 50 μm). B, Corresponding semiquantifcation of FBXW7 expression in 100 pairs NSCLC specimens (***P < .0001). C, Western blotting of FBXW7 in 4 paired samples of NSCLC tissues (T) versus adjacent normal tissues (N). Levels were normalized to β‐actin. D, Performance of GSEA based on publicly available NSCLC patient gene expression profiles (NCBI/GEO/GSE GSE18842; n = 91, left), Survival analysis of FBXW7 in lung cancer was performed at the online website Kaplan‐Meier Plotter, using publicly available datasets (2015 version, Affy ID: 229419_at, right). E, Kaplan‐Meier curves with univariate analysis (log‐rank) for 5‐year overall (P = .00214) and disease‐free survival curves (P = .0351) of NSCLC patients based on their FBXW7 protein expression status
3.2. FBXW7 inhibits proliferation, migration and EMT of NSCLC cells
To better understand the effect of FBXW7 on metastasis‐related cell behaviours in NSCLC cells, we investigated the expression of FBXW7 in normal human bronchial epithelial cells of Beas‐2b, and four paired NSCLC cell lines. Western blot analysis showed that FBXW7 level was lower in NSCLC cells than that in Beas‐2b cells (Figure 2A), to the same as the result shown in specimens study. We constructed A549‐Flag‐FBXW7 cell with consistently FBXW7 expressing, and H1299‐shFBXW7 cells with FBXW7‐knockdown. The results were confirmed by western blot and qRT‐PCR analysis (Figure 2B). The effects of FBXW7 on proliferation and migration of NSCLC cells were shown in Figure 2C. In detail, MTT assay (Figure 2C upper) and colony formation (Figure 2C bottom) demonstrated that FBXW7‐ knockdown cells significantly increased the number of foci formed, and the cell growth rates compared with control cells, while the opposite result was obtained in FBXW7‐overexpressing cells. Furthermore, we observed the morphological changes in two transfected cell lines. As expected, silence of FBXW7 induced, whereas elevated FBXW7 expression reduced a more mesenchymal phenotype as compared with their control cells (Figure S1).
Figure 2.
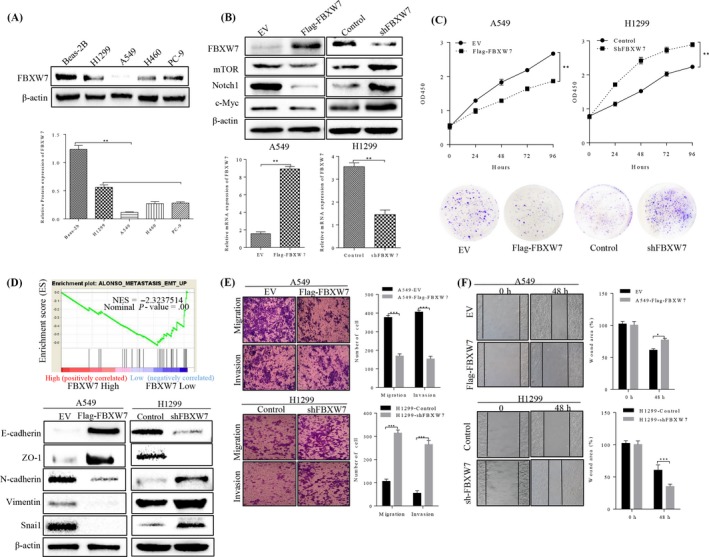
Effect of FBXW7 on cell proliferation, migration, invasion and EMT in NSCLC cells. A, FBXW7 expression in human lung bronchial epithelial (BEAS‐2B) and four NSCLC cell lines was detected by Western blot analysis (**P < .01). B, A549 and H1299 cells were subjected to western blotting for FBXW7 and its downstream target proteins (mTOR, Notch1 and c‐Myc) after transfected with Flag‐FBXW7 and FBXW7‐shRNA, respectively (upper panel). FBXW7 expression was confirmed by qRT‐PCR (lower panel). C, MTT assay for detection of cell proliferation in 24, 48 and 72 hours after transfection in NSCLC cells (upper panel). Representative images of colony forming units induced by FBXW7 in A549 and H1299 cell lines (lower panel). D, Performance of GSEA based on GEO dateset (NCBI/GEO/GSE GSE19188; n = 156, upper panel). E, The migratory and invasion capacities of the FBXW7‐overexpressing A549 cells were decreased compared with empty vector (EV) cells (***P < .01 vs. EV by t test, upper panel). The migratory and invasion capacities of the FBXW7‐silenced H1299 cells were increased compared with negative control (NC) cells (***P < .001 vs. controls by t test, lower panel). n = 3 repeats with similar results. F, wound healing assay showed that cell migration ability was inhibited by FBXW7 overexpression in A549 cells (**P < .01 vs. EV, upper panel) and promoted by FBXW7 knockdown in H1299 cells (***P < .001 vs EV, lower panel). n = 3 repeats with similar results. Values are depicted as Mean ± SD
Migration and invasion are hallmarks for cancer metastasis. Thus, we performed transwell and wound healing assay in stable NSCLC cells to test it. Results indicated that ectopic expression of FBXW7 markedly inhibited migration and invasion of A549 (Figure 3E‐F, upper). Conversely, H1299‐shFBXW7 cells exhibited stronger capacity of migratory and invasion, comparing with the control group respectively (Figure 3E‐F, bottom).
Figure 3.
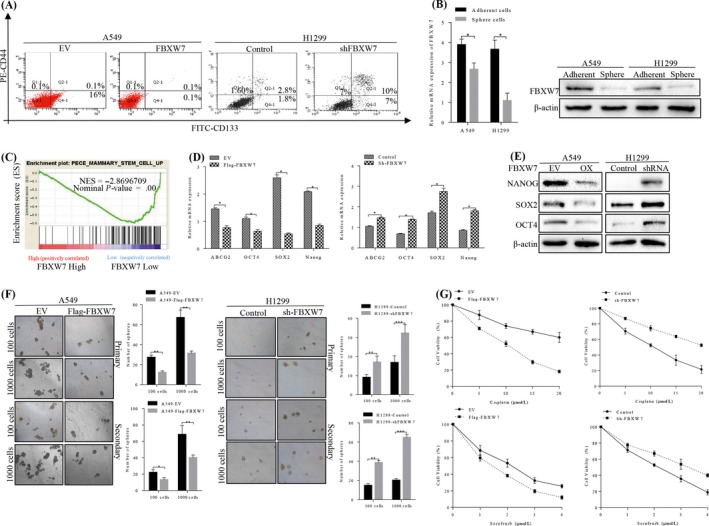
Effect of FBXW7 on CSC and chemo‐resistance in NSCLC cells. A, Representative FACS profiles of A549 cell transduced with Flag‐FBXW7 or empty control vector and H1299 cell transduced with sh‐FBXW7 or control. Quantification of CSC and non‐CSC populations stained with CD44‐PE and CD133‐FITC. B, qRT‐PCR and immunoblot showing decreased FBXW7 expression in NSCLC cells spheres relative to adherent cells (*P < .05). D‐E, Real‐time PCR (D) and western blot analysis (E) of CSC‐associated markers in the indicated cells (*P < .05). F, Representative images of spheres formed by FBXW7‐knockdown H1299 cells and FBXW7‐overexpressing A549 cells. Number of spheres per well was quantified on primary spheroid formation assay and secondary spheroid formation assay for the two stable cells (**P < .05, *** P < .01 vs controls by T test). G, The sensitivity of FBXW7‐knockdown H1299 cells and FBXW7‐overexpressing A549 cells to cisplatin and sorafenib were determined by MTT, respectively. n = 3 repeats with similar results. Values are depicted as Mean ± SD
As to the effects of FBXW7 on EMT process, the EMT markers and EMT‐related transcription factors were qualified by western blot, which proved that FBXW7 could upregulate epithelial markers of E‐cadherin and ZO‐1, and decrease mesenchymal markers of N‐cadherin and vimentin, as well as the EMT‐related transcription factor of Snai1. The opposite results were observed when slicing FBXW7 in H1299 cells (Figure 3D), demonstrating the functional cascade of FBXW7 in suppressing lung cancer cell proliferation, migration and EMT.
3.3. FBXW7 restrains the acquisition of stem cell‐like phenotypes of NSCLC cells
Emerging evidence has clarified that EMT is directly relevant to maintenance of stem‐like cells’ renewal ability. Using FACS sorting, we found that enforced FBXW7 expression caused a significant reduction in the subpopulation of CD44−/CD133+ cells, and on the contrary, knocking down FBXW7 in H1299 cells increased the percentage of CD44+/CD133+ population (Figure 3A). Furthermore, the RNA and protein levels of FBXW7 were found to be strikingly down‐regulated in sphere cells, comparing to the parental adherent cells (Figure 3B). Moreover, we performed GSEA analysis by using RNA‐seq data of NSCLC tissues to elucidate the possible association between FBXW7 and ratio of CSCs, and GSEA results showed that increased ratio of stem cell was increased greatly in FBXW7 lowly expressed tissues (Figure 3C). To further validate the GSEA results, we detected the RNA levels of stem cell‐associated markers of Oct4, Sox2, Nanog and ABCG2, results revealing highly expressed status of these markers in H1299‐shFBXW7 cells (Figure 3D), which were consistent with the results in western blot (Figure 3E). The tumour sphere formation assay was used to investigate the effects of FBXW7 on the regulation of stem cells’ renewal. Notably, FBXW7‐silenced H1299 cells formed more spheres with larger size, than that of vector control cells. When reintroducing FBXW7 into A549 cells, the putative stem cells formed fewer spheres with smaller size, comparing to control cells in primary and secondary spheroid formation assays (Figure 3F).
Chemoresistance was considered to be the key stem cells’ signature, and we chose two commonly used chemotherapy agents of cisplatin and sorafenib for sensitivity testing of two stable NSCLC cells. As expected, FBXW7 overexpressed cells were more sensitive to cisplatin and sorafenib induction of proliferation inhibition, and FBXW7 inhibition increased the resistance to cisplatin and sorafenib in a dose‐dependent manner (Figure 3G).
3.4. FBXW7 is inversely related to snai1 in NSCLC tissues
Previous studies have identified that multiple members of the F‐box family were involved in the regulation of ubiquitination degradation of Snai1, while Snai1 has been reported to play important roles in EMT and stem cell phenotype of NSCLC. However, whether FBXW7 restrained these biological functions by inducing Snai1 ubiquitination and proteasomal degradation is still unknown. Thus, we firstly evaluated the relationship between FBXW7 and Snai1 in NSCLC tissues. The correlation between FBXW7 and Snai1 was examined in serial sections of 100 NSCLC cases. IHC staining showed that Snai1 overexpression was detected in NSCLC samples compared with the corresponding non‐tumorous tissues (Figure 4A). Besides, FBXW7 high expression was inverse correlation with Snai1 low expression in Case 3(Figure 4B, left). Inversely, FBXW7 low expression was associated with high Snai1 levels in Case 4 (Figure 4B, left). Although the protein expression of FBXW7 was positively correlated with its mRNA level in NSCLC tissues (Figure S2), spearman rank correlation analysis further confirmed that FBXW7 expression was negatively associated with Snai1 expression in protein levels (Figure 4B, right) but not in mRNA (Figure 4C). These results demonstrated that increased FBXW7 expression was closely correlated with reduced Snai1 levels in NSCLC samples, supporting our results in vitro data.
Figure 4.
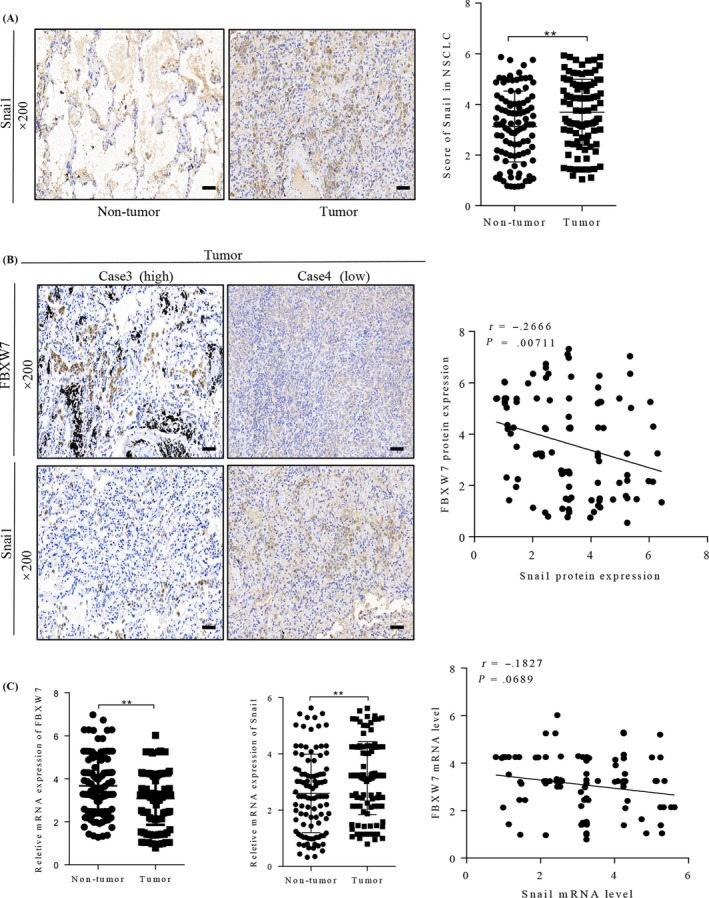
Relationship between the expression of FBXW7 and Snai1 in NSCLC tissues. A, Immunohistochemical (IHC) staining for Snai1 in normal tumour adjacent tissues and NSCLC tissues (left panel). Corresponding semiquantifcation of Snai1 expression was shown in right panel (**P < .01). B, Representive staining of FBXW7 and Snai1 in two NSCLC samples (left panel). Linear regression analyses of IHC scores between FBXW7 and Snai1 in NSCLC tissues (r = −.2666, P = .00711). Case3 represent FBXW7 high expression sample, Case 4 represent FBXW7 low expression sample. C, The mRNA level of Snai1 and FBXW7 were detected in NSCLC tissues and matched normal tumour adjacent tissues, qRT‐PCR assay (left and middle panel, ** P < .01). Linear regression analyses of mRNA level between FBXW7 and Snai1 in NSCLC tissues (right, r = −.1827, P = .0689)
3.5. FBXW7 directly binds to and ubiquitylates snai1 for proteasomal degradation
To further investigate the regulatory relationship between FBXW7 and Snai1, we examined whether Snai1 is a potential recognizable substrate of FBXW7. Substrate recognition of FBXW7 is mainly accomplished by binding to phosphorylated threonine and serine in the CPD motif of FBXW7 substrates. Scansite software (UbiBrower) analysis revealed that the transcription factor of Snai1 contains one potential CPD, which includes the evolutionary conserved phosphoamino acids (Figure 5A). Subsequently, we used a protease inhibitor (MG132) to confirm the FBXW7‐mediated Snai1 degradation via the proteasome‐dependent pathway, and the protease inhibitor MG132 blocked the Snai1 degradation, suggesting that Snai1 might be degraded through a proteasome‐dependent manner (Figure 5B). To explore whether Snai1 level is regulated at post‐translational level, qRT‐PCR assay was applied and showed that no changes in Snai1 mRNA levels were observed after exogenous FBXW7 expression in A549 and 293T cells were conducted, consistent with the post‐translational regulatory model (Figure 5C). Immunofluorescence staining revealed that Snai1 was mainly positioned in the nucleus that co‐localized with FBXW7. Moreover, ectopic expression of Flag‐FBXW7 notably reduced the half‐life of Snai1 by using the CHX chase assay in HEK239T and A549 cells.
Figure 5.
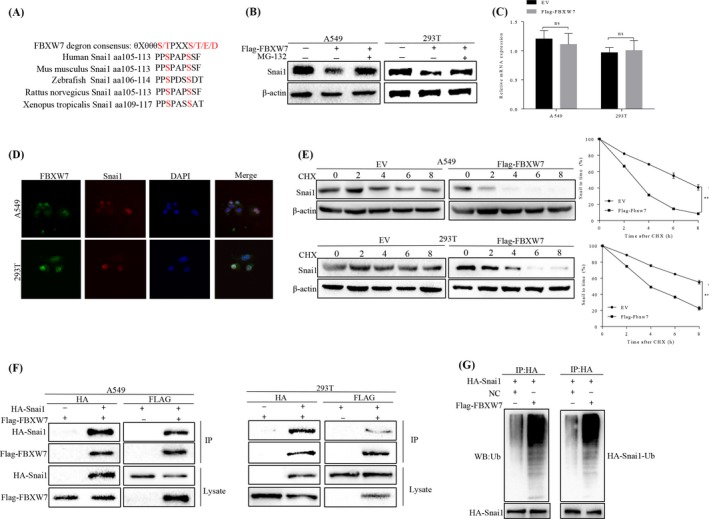
FBXW7 binded to Snai1 and induced its ubiquitination and proteasomal degradation. A, Sequence alignment of Snai1 with FBXW7 degron motif (θXθθθS/TPXXS/T/E/D), θ, hydrophobic amino acid; X, any amino acid. B, Western blot assay for Snai1 expression in FBXW7‐overexpressing A549 and FBXW7‐overexpressing 293T after treatment with MG‐132 for 6 h. C, qRT‐PCR assay to detect the mRNA level of Snai1 in A549 and 293T after transfected with Flag‐FBXW7 plasmids (ns: no significance). D, Immunofluorescence staining of A549 and 293T showing nuclear co‐localization of FBXW7 (Alexa Fluor 488; green) and Snai1 (Alexa Fluor 568; red). DAPI (blue) was used for nucleic acid staining. E, The protein half‐life of Snai1 was analysed following treatment with cycloheximide. The graphs show quantitative analysis of the CHX chase data (**P < .01 vs EV by Two‐way ANOVA). F, Flag‐FBXW7 and HA‐Snai1 were transfected into A549 or HEK293 cells. Cell lysates were immunoprecipitated with anti‐Flag or anti‐HA to detect the specific proteins indicated on the left side of each panel, respectively. G, HA‐Snai1 was immunoprecipitated from HA‐Snai1‐overexpressing A549 or 293T cells using an anti‐HA antibody; then, using western blot to detect Snai1 ubiquitination. Snail 1 ubiquitination was markedly promoted by FBXW7 overexpression. All results are representative of three independent experiments
We next sought to determine whether FBXW7 binds to Snai1 and leads to its ubiquitination. To this end, we tested whether Snai1 interacted with FBXW7 using reciprocal coimmunoprecipitation (co‐IP). Flag‐FBXW7 and HA‐Snai1 were co‐expressed in HEK293T and A549 cells. Co‐immunoprecipitation analysis showed that Snai1 co‐immunoprecipitated with Flag‐FBXW7 by anti‐Flag antibody. Similarly, immunoprecipitation of HA‐Snail by anti‐HA antibody led to co‐immunoprecipitation of Flag‐FBXW7. As a component of E3 ubiquitin ligase, FBXW7 promotes the degradation of target proteins through ubiquitination. Thus, using in vitro ubiquitination assay, we have identified that FBXW7 promotes Snai1 ubiquitination. HA‐Snai1 was precipitated from HEK293T and A549 cells expressing HA‐Snai1, and Snai1 ubiquitination was detected by an ubiquitin western blot. As shown in Figure 5G, FBXW7 overexpression markedly increased the ubiquitination of Snai1 in HEK293T and A549 cells. In conclusion, FBXW7 binds to Snai1 and induces the down‐regulation of Snai1 via its ubiquitination and proteasomal degradation.
3.6. Introduction of snai1 reverses the suppressive effects of FBXW7 on oncogenic natures of NSCLC cells
To determine whether the Snai1 protein participates in FBXW7‐induced tumour suppression in NSCLC cells. We overexpressed Snai1 protein in A549‐FBXW7, using recombinant plasmid, and results showed that Snai1 overexpression obviously decreased epithelial markers of E‐cadherin and ZO‐1, and increased mesothelial markers of N‐cadherin and Vimentin of A549‐FBXW7 cells in protein levels. Nevertheless, down‐regulated Snai1 in H1299‐shFBXW7 cells caused the opposite results (Figure 6B).
Figure 6.
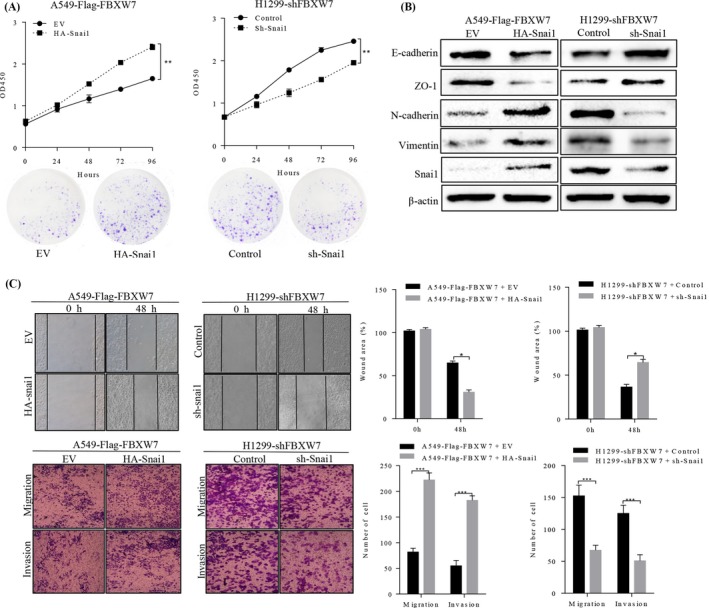
FBXW7‐induced suppression of NSCLC cell growth, migration and invasion, and EMT were partially reverted by Snai1. FBXW7‐silenced H1299 cell or FBXW7‐overexpressing A549 cell was transfected with Sh‐Snai1 or HA‐Snai1, respectively. These transfected cells were then subject to MTT assay and plate clone formation (A), Western blotting (B), wound healing assay (C), Transwell migration assay (D, upper two panels), Matrigel invasion assays (D, lower two panels). * P < .05, ** P < .01, *** P < .0001; n = 3 repeats with similar results. Values are depicted as the Mean ± SD
When restoring Snai1 expression in A549 cells, the effect of exogenous FBXW7 overexpression was partially reverted, leading to a significant induction in both cell migration and proliferation (Figure 6A,C).
Several studies reported that Snai1‐induced EMT promoted self‐renewal capabilities and the stem‐like phenotype, and we therefore hypothesized that Snai1 may also participated in FBXW7 regulation of stemness of NSCLC cells. The effect of Snai1 on CSC‐associated markers, mammosphere‐forming ability and chemoresistance were tested after transfecting sh‐Snai1 or HA‐Snai1 plasmids in two stable NSCLC cells. As shown in Figure 7A, restoring Snai1 in A549‐FBXW7 cells significantly increased expression of Nanog, Sox‐2 and Oct‐4 relative to their control cells, while the protein level of these the stemness markers were partially decreased in FBXW7‐silenced H1299 cells following Snai1 inhibition. Consistently, self‐renewal ability of FBXW7‐silenced H1299 cells and FBXW7‐overexpressing A549 cells were partially decreased or increased following Snai1 inhibition or re‐expression (Figure 7B), accompanied by enhanced or attenuate sensitivity to cisplatin and sorafenib (Figure 7C), respectively. Taken together, these data indicate that Snai1 may function as a downstream factor by which FBXW7 regulates epithelial‐mesenchymal transition, stemness and metastatic potential of NSCLC cells.
Figure 7.
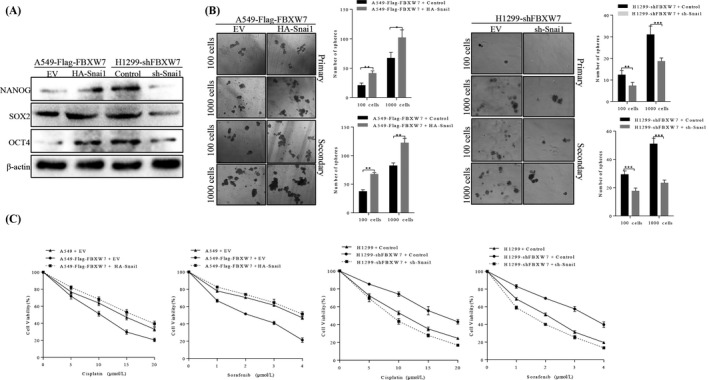
FBXW7‐induced suppression of NSCLC stemness and chemoresistance were partially reverted by Snai1. A, A549‐Flag‐FBXW7 cells and H1299‐shFBXW7 cells that were transfected with HA‐ Snai1 and shSnai1, respectively, and subjected to Western blotting for NANOG, SOX2 and OCT4. B, Number of spheres per well were quantified on primary spheroid formation assay and secondary spheroid formation assay for A549‐Flag‐FBXW7 cells and H1299‐shFBXW7 cells after transfected with HA‐Snai1 and shSnai1, respectively (right panels). Left panels showed representative spheres. C, After restoring Snai1 expression, the sensitive of A549‐Flag‐FBXW7 cells and H1299‐shFBXW7 cells to cisplatin and sorafenib was determined by MTT assay (*P < .05, ** P < .01)
4. DISCUSSION
FBXW7 is frequently mutated or depleted in various human cancers, and its dysfunctional mutation is firmly correlated with tumour progression. In this study, we initially detected FBXW7 expression in 100 pairs of NSCLC tissues and adjacent non‐cancerous tissues, using immunohistochemistry and qRT‐PCR, and results showing that FBXW7 was significantly lower in NSCLC tissues than that in paired non‐cancerous tissues, which are similar to previous study.18 Moreover, public GSEA data based clinical analyses showed that FBXW7 expression was significantly associated with later TNM stage and advanced histology. Lower FBXW7 expression was correlated with poorer patients’ overall survival, and tended to have low disease‐free survival rate.
Our previous studies showed that FBXW7 expression was associated with cell proliferation, cell cycle and apoptosis in NSCLC cells.19 Nevertheless, the detailed mechanisms of FBXW7 in EMT and characteristics of NSCLC stem cells are rarely reported. Mechanistically, we found that silencing FBXW7 in NSCLC cells promoted the EMT procedure and CSCs’ self‐renewal, which both contributed to the chemoresistance, and conversely, FBXW7 overexpression did the opposite, suggesting the pivotal role of FBXW7 in NSCLCs’ EMT, and the renewal ability of subgroup of stem cell. Importantly, re‐sensitize the NSCLC to chemotherapeutics strongly proved that FBXW7 could be a potential biomarker for NSCLC therapy and prognostic prediction.
In vitro studies, we identified the existence of a physical interaction between FBXW7 and Snai1 in A549 and 293T cells by using immunofluorescent staining and co‐immunoprecipitation. Furthermore, our results indicated that FBXW7 targeted Snai1 and mediated ubiquitination of Snai1 in A549 and 293T cells, and that MG132 treatment could block the FBXW7 overexpression‐induced Snai1 downregulation. According to these data, we confirmed that FBXW7 facilitates the ubiquitination of Snai1, and thus its subsequent proteasomal degradation. In most case, FBXW7 was reported to target substrates in a GSK3β‐dependent manner.20 Therefore, further investigation is required to confirm whether FBXW7 targets Snai1 also in a GSK3β‐dependent manner.
We previously identified that loss of FBXW7 contributed to Wnt signalling activation through a LIN28B/Let‐7‐dependent manner in NSCLC stem cells. Meanwhile, it has been well established that Wnt/β‐catenin activation induced both Snai1 mRNA and Snai1 protein expression via promoting GSK‐3β inhibition and β‐catenin stability,21 implying that apart from FBXW7 induced‐Snai1 degradation by ubiquitylation process, FBXW7 may also inhibit the expression of snail partly by inactivating Wnt signalling pathway.
In conclusion, our study identified that FBXW7 was lowly expressed in cancer tissues, compared to non‐cancerous tissues, and reduced FBXW7 expression indicated poorer prognosis of patients with NSCLC. In vitro studies found that elevated FBXW7 expression inhibited EMT, CSCs’ renewal ability, and re‐sensitizes NSCLC cells to chemotherapeutics by targeting Snai1 for proteasomal degradation. Taken together, our study strongly suggested that FBXW7 may be a novel therapeutic and prognostic target of NSCLC, paving the way for possible pre‐clinical application.
CONFLICT OF INTEREST
All co‐authors implicated in this research approved this article to be published. The authors declare that they have no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
The authors acknowledge assistants in Center for Translational Medicine of the First Affiliated Hospital of Xi'an Jiaotong University, and the staff of the Key Laboratory of Environment and Genes Related to Disease, Ministry of Education, for their technical assistance. This experiment was supported by Natural Science Foundation of China, Grant No. 81272418 (H.R), Key Research and Development Program of Shaanxi Province Grant No.2017KW‐061(H.R), and National Science Foundation for Young Scientists of China, Grant No. 81602597 (X.S).
Xiao G, Li Y, Wang M, et al. FBXW7 suppresses epithelial‐mesenchymal transition and chemo‐resistance of non‐small‐cell lung cancer cells by targeting snai1 for ubiquitin‐dependent degradation. Cell Prolif. 2018;51:e12473 10.1111/cpr.12473
Guodong Xiao, Yuan Li, Meng Wang and Xiang Li are Co‐First authors.
Contributor Information
Hong Ren, Email: medicinedr.ren@gmail.com.
Dapeng Liu, Email: prof_ldp_oncology@126.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5‐29. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐32. [DOI] [PubMed] [Google Scholar]
- 3. Xiao G, Zhang B, Meng J, et al. miR‐367 stimulates Wnt cascade activation through degrading FBXW7 in NSCLC stem cells. Cell Cycle. 2017;16:2374‐2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu Z, Xu Y, Zhao J, et al. miR‐367 promotes epithelial‐to‐mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7‐TGF‐beta signalling pathway. Br J Cancer. 2015;112:1367‐1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang SL, Yang M, Herrlinger S, Liang C, Lai F, Chen JF. MiR‐302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation. Dev Biol. 2015;408:140‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaid C, Silva PB, Cortez BA, Rodini CO, Semedo‐Kuriki P, Okamoto OK. miR‐367 promotes proliferation and stem‐like traits in medulloblastoma cells. Cancer Sci. 2015;106:1188‐1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun X, Liu J, Xu C, Tang SC, Ren H. The insights of Let‐7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. 2016;20:1779‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun X, Jiao X, Pestell TG, et al. MicroRNAs and cancer stem cells: the sword and the shield. Oncogene. 2014;33:4967‐4977. [DOI] [PubMed] [Google Scholar]
- 9. Sun X, Xu C, Tang SC, et al. Let‐7c blocks estrogen‐activated Wnt signaling in induction of self‐renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83‐89. [DOI] [PubMed] [Google Scholar]
- 10. Zakaria N, Yusoff NM, Zakaria Z, et al. Human non‐small‐cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer. 2015;15:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang H, Lu X, Liu Z, et al. FBXW7 suppresses epithelial‐mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget. 2015;6:6310‐6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rustighi A, Zannini A, Tiberi L, et al. Prolyl‐isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Wang Z, Zhang W, Qian K, Xu W, Zhang SR. Fbxw7 regulates tumor apoptosis, growth arrest and the epithelial‐to‐mesenchymal transition in part through the RhoA signaling pathway in gastric cancer. Cancer Lett. 2016;370:39‐55. [DOI] [PubMed] [Google Scholar]
- 15. Xiao G, Li X, Li G, et al., et al. MiR‐129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem‐like cells. Oncotarget. 2017;8:103261‐103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pang Y, Liu J, Li X, et al. Nano Let7b sensitization of eliminating esophageal cancer stemlike cells is dependent on blockade of Wnt activation of symmetric division. Int J Oncol. 2017;51:1077‐1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun X, Qin S, Fan C, Xu C, Du N, Ren H. Let‐7: a regulator of the ERalpha signaling pathway in human breast tumors and breast cancer stem cells. Oncol Rep. 2013;29:2079‐2087. [DOI] [PubMed] [Google Scholar]
- 18. Yokobori T, Yokoyama Y, Mogi A, et al. FBXW7 mediates chemotherapeutic sensitivity and prognosis in NSCLCs. Mol Cancer Res. 2014;12:32‐37. [DOI] [PubMed] [Google Scholar]
- 19. Xiao G, Gao X, Sun X, et al. miR‐367 promotes tumor growth by inhibiting FBXW7 in NSCLC. Oncol Rep. 2017;38:1190. [DOI] [PubMed] [Google Scholar]
- 20. Akhoondi S, Sun D, von der Lehr N, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Can Res. 2007;67:9006‐9012. [DOI] [PubMed] [Google Scholar]
- 21. Horvay K, Casagranda F, Gany A, Hime GR, Abud HE. Wnt signaling regulates Snai1 expression and cellular localization in the mouse intestinal epithelial stem cell niche. Stem Cells Dev. 2011;20:737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


