Abstract
Objectives
The aim of this study was to investigate whether sclerostin (SOST) regulates the osteogenic differentiation of rat ectomesenchymal stem cells (EMSCs) and whether SOST and low‐affinity nerve growth factor receptor (LNGFR) regulate the osteogenic differentiation of EMSCs.
Materials and methods
EMSCs were isolated from embryonic facial processes from an embryonic 12.5‐day (E12.5d) pregnant Sprague‐Dawley rat. LNGFR + EMSCs and LNGFR − EMSCs were obtained by fluorescence‐activated cell sorting and were subsequently induced to undergo osteogenic differentiation in vitro. SOST/LNGFR small‐interfering RNAs and SOST/LNGFR overexpression plasmids were used to transfect EMSCs.
Results
LNGFR + EMSCs displayed a higher osteogenic capacity and lower SOST levels compared with LNGFR − EMSCs. SOST silencing enhanced the osteogenic differentiation of LNGFR − EMSCs, while SOST overexpression attenuated the osteogenic differentiation of LNGFR + EMSCs. Moreover, LNGFR was present upstream of SOST and strengthened the osteogenic differentiation of EMSCs by decreasing SOST.
Conclusions
SOST alleviated the osteogenic differentiation of EMSCs, and LNGFR enhanced the osteogenic differentiation of EMSCs by decreasing SOST, suggesting that the LNGFR/SOST pathway may be a novel target for promoting dental tissue regeneration and engineering.
Keywords: ectomesenchymal stem cells, LNGFR, osteogenic differentiation, SOST
1. INTRODUCTION
Ectomesenchymal stem cells (EMSCs), which originate from the cranial neural crest (CNC), are considered to be the progenitors of almost all tooth tissues, except for enamel and maxillofacial mineralized tissues.1 After migrating to the maxillary and mandibular processes at an early stage of embryogenesis, CNC cells are defined as EMSCs.2 Then, the EMSCs settle down and interact with dental epithelium, ultimately differentiating into multiple dental tissue‐derived cells. EMSCs serve as the primary source of differentiation, which occurs during dentin, cementum and alveolar bone formation in tooth development.1 Therefore, there is an urgent need to investigate the mechanisms of EMSC differentiation, as they may provide potential strategies for dental tissue regeneration and engineering.
Sclerostin (SOST), which is produced by osteocytes, was originally considered to be non‐classical.3, 4 Previous studies have revealed that SOST has anti‐anabolic effects on bone formation.5 Recently, it has been reported that SOST is closely associated with the development of dental tissues.6, 7, 8, 9 For example, SOST inhibition dramatically stimulates alveolar bone regeneration following experimental periodontitis in Sprague‐Dawley (SD) rats.6 Another report has demonstrated that SOST inhibition resulted in significant new deposition of cellular cementum and the formation of well‐organized periodontal ligament fibre in a rat periodontal defect model.7 However, it remains unknown whether SOST plays a role in the development of EMSCs, which are the progenitors of almost all tooth tissues.
Low‐affinity nerve growth factor receptor (LNGFR) is a member of the tumour necrosis factor receptor superfamily and is a receptor for neurotrophins, a family of protein growth factors that stimulate the survival and differentiation of neuronal cells.10, 11 Previous reports have shown that LNGFR is involved in cell death and axonal degeneration in response to injury.12 Increasing numbers of studies have demonstrated that LNGFR can serve as a cell surface marker for mesenchymal stem cells (MSCs) and that LNGFR‐positive MSCs present increased potential for osteogenesis.13, 14, 15, 16, 17, 18 In this study, EMSCs were isolated from embryonic facial processes and sorted to obtain LNGFR+ EMSCs and LNGFR− EMSCs. We further demonstrated that LNGFR+ EMSCs had a higher osteogenic capacity and lower SOST levels compared with LNGFR− EMSCs and that SOST negatively regulated the osteogenic differentiation of EMSCs. Moreover, LNGFR was present upstream of SOST and strengthened the osteogenic differentiation of EMSCs by decreasing SOST. Collectively, this study's results indicate that SOST attenuated the osteogenic differentiation of EMSCs and that LNGFR strengthened the osteogenic differentiation of EMSCs by decreasing SOST, which suggests that the LNGFR/SOST pathway may be a novel target for promoting dental tissue engineering and regeneration.
2. MATERIALS AND METHODS
2.1. EMSC isolation and LNGFR+ and LNGFR− EMSC sorting
EMSCs were isolated from embryonic 12.5‐day (E12.5d) pregnant Sprague‐Dawley (SD) rats from the Third Military Medical University Animal Laboratory. All animal experiments were performed in accordance with procedures approved by the Medical Ethics Committee of the Third Military Medical University. Briefly, the embryonic facial processes were dissected from the E12.5d SD rats. Then, the shredded tissue was digested with a 1% trypsin/1 mM EDTA solution (Sigma‐Aldrich, St Louis, MO, USA) at 37°C for 10 min, followed by the addition of Dulbecco's modified Eagle's medium/Ham's F12 (DMEM/F12, Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco) to stop the digestion. Next, the cell suspension was filtered through a 75 μm mesh filter (BD Biosciences, Oxford, UK) and then centrifuged at 800 rpm for 5 min. Subsequently, the cell pellet was resuspended in DMEM/F12 (with 10% FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin) and cultured at 37°C in 5% CO2 humidified incubator.
LNGFR+ and LNGFR− EMSCs were sorted by fluorescence‐activated cell sorting (FACS) as follows. EMSCs in their third passage (P3) were digested with a 1% trypsin/1 mM EDTA solution, followed by suspension in 1 mL phosphate‐buffered saline (PBS) with 1% BSA. Then, the cells were incubated with anti‐rat LNGFR‐FITC (1:20, Abcam, Cambridge, MA, USA) at 37°C for 60 min. Subsequently, the LNGFR+ and LNGFR− EMSCs were isolated using a FACScalibur flow cytometer (BD Biosciences).
2.2. Identification of LNGFR+ and LNGFR− EMSCs
Flow cytometry analysis was used to detect the cell surface markers (CD29, CD44, CD90, CD146, CD34 and CD45) on LNGFR+ and LNGFR− EMSCs. Briefly, the cells were fixed with 4% paraformaldehyde for 30 min, followed by incubation with anti‐rat CD29, anti‐rat CD44, anti‐rat CD90, anti‐rat CD146, anti‐rat CD34 and anti‐rat CD45 antibodies (1:100; Santa Cruz Biotechnology, Dallas, TX, USA) at 4°C overnight. After incubation with the anti‐mouse IgG‐FITC secondary antibody (1:100), the cells were analysed with a FACScalibur flow cytometer.
2.3. Quantitative real‐time PCR
Total RNA was extracted with the Trizol reagent (Invitrogen, Carlsbad, CA, USA) and first‐strand cDNA was synthesized using 1 μg total RNA in 20 μL 1×PrimeScript™ RT master mix (Takara, Dalian, China). The concentrations of RNA and cDNA were measured by the ultraviolet spectrophotometer NanoDrop Lite (Thermo Scientific, Waltham, MA, USA). Quantitative real‐time PCR (qPCR) was performed using 25 ng (0.5 μL) cDNA/well with the SYBRII qPCR master mix (Takara) according to the manufacturer's instruction using GAPDH as a control. The primer sets are listed in Table S1.
2.4. Western blot analysis
Total proteins were extracted with RIPA (Beyotime, Shanghai, China) and the protein concentrations were examined by BCA Protein Assay Kit (Beyotime). Then, the proteins (40 μg/lane) were separated via SDS‐PAGE and transferred to PVDF membranes (Pierce, Rockford, IL, USA). After blocking with 5% fat‐free dry milk in PBST (PBS with 0.1% Tween‐20) at 37°C for 1 h, the membranes were separately incubated with primary antibodies against LNGFR (1:1000, Abcam), Runx2 (1:1000, Abcam), ALP (1:1000, Abcam), SOST (1:1000, Santa Cruz Biotechnology) and GAPDH (1:4000, Immunoway, Plano, TX, USA) at 4°C overnight. Next, the membranes were washed with PBST and incubated with the corresponding horseradish peroxidase conjugated secondary antibodies (Proteintech, Wuhan, China) at 37°C for 1 h. Subsequently, the membranes were washed again with PBST, and the signals were visualized with the Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA).
2.5. Osteogenic induction of EMSCs
After being seeded in 6‐well plates for 12 h, P3 LNGFR+ and LNGFR− EMSCs (5 × 104 cells/well) were cultured with osteogenic induction medium, which contains 10% FBS α‐MEM containing 50 mg/mL ascorbic acid, 10 mmol/L b‐glycerol phosphate and 100 nM dexamethasone. The osteogenic induction medium was changed every 3 days.
2.6. Alkaline phosphatase staining
After being seeded in 6‐well plates for 12 h, P3 LNGFR+ and LNGFR− EMSCs (5 × 104 cells/well) were cultured in osteogenic induction medium. At days 0 and 7, the cells were washed twice with PBS, fixed in 4% paraformaldehyde for 30 min and stained with an ALP activity kit (Beyotime). After washing 3 times with distilled water, the cells were observed via phase‐contrast microscopy.
2.7. Alizarin red staining
After being seeded in 6‐well plates for 12 h, P3 LNGFR+ and LNGFR− EMSCs (5 × 104 cells/well) were cultured in osteogenic induction medium. At day 21, the cells were washed twice with PBS, fixed in 4% paraformaldehyde for 30 min and stained with Alizarin red (Sangon, Shanghai, China). After washing 3 times with distilled water, the cells were visualized via phase‐contrast microscopy.
2.8. Transfection assays
After being seeded into 6‐well plates for 12 h, the cells (5 × 104 cells/well) were transfected with siRNAs or expression plasmids (2 μg/well) using Lipofectamine 2000 (Invitrogen) for 24 h. Then, the cells were cultured with or without osteogenic induction medium for 7 or 21 days, followed by the corresponding experiments. The transfection assays were performed every 3 days similar to the osteogenic induction medium replacement. The sequences of the siRNAs are listed in Table S2.
2.9. Construction of LNGFR− EMSCs stably overexpressing LNGFR
The pLJM1‐LNGFR plasmid was co‐transfected with the psPAX2 envelope and CMV VSV‐G packaging plasmids into HEK‐293T cells using Lipofectamine 2000 for 48 h. Then, the virus‐containing supernatants were collected and filtered to remove the cells. Subsequently, P3 LNGFR− EMSCs were infected in the presence of 8 μg/mL polybrene for 24 h, followed by selection with 2 μg/mL puromycin an additional 1 week.
2.10. Statistical analysis
All data were expressed as the mean ± SD. Comparisons between 2 groups were determined with the 2‐tailed unpaired t test. Comparisons among 3 or more groups were made using one‐way analysis of variance (ANOVA). P < .05 was considered to be statistically significant.
3. RESULTS
3.1. LNGFR+ and LNGFR− EMSC characteristics
EMSCs were isolated from embryonic facial processes from an E12.5d pregnant SD rat. Next, the EMSCs were screened using LNGFR as a cell surface marker to obtain LNGFR+ and LNGFR− EMSCs. LNGFR+ and LNGFR− EMSCs separately accounted for 14.54% and 7.02% of the isolated EMSCs (Figure 1A). A series of experiments were performed to identify LNGFR+ and LNGFR− EMSC characteristic. As shown in Figure 1B, LNGFR+ and LNGFR− EMSCs both showed a long spindle morphology, which is morphologically characteristic of MSCs. Moreover, MSC markers (CD29, CD44, CD90 and CD146) were highly expressed in both LNGFR+ and LNGFR− EMSCs (Figure 1C), while hematopoietic markers (CD45 and CD34) were lowly expressed in both LNGFR+ and LNGFR− EMSCs (Figure 1D). The mesenchymal stem markers (CD29, CD44, CD90 and CD146) were also detected in rat hematopoietic stem cells, which were used as a negative control, though they displayed expression (Figure S1A). Moreover, LNGFR mRNA (Figure 1E) and protein (Figures 1F and S1B) levels were higher in LNGFR+ EMSCs compared with LNGFR− EMSCs. These results indicated that LNGFR+ and LNGFR− EMSCs are MSCs.
Figure 1.
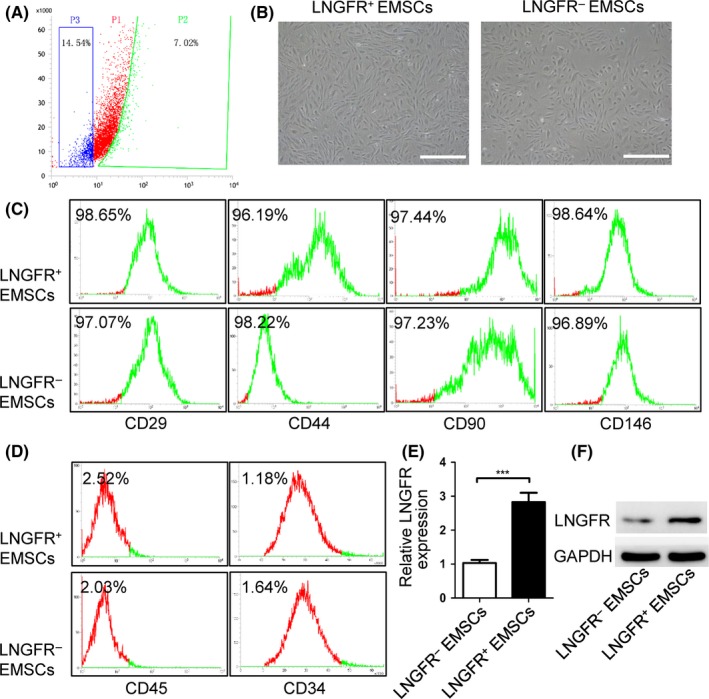
Characteristics of LNGFR + and LNGFR − EMSCs. (A) Flow cytometry sorting of LNGFR + and LNGFR − EMSCs. The cells in right section (P1) represent the LNGFR − EMSCs, while the cells in left section (P3) represent the LNGFR + EMSCs. (B) The morphologies of the LNGFR + and LNGFR − EMSCs were observed by optical microscopy (scale bar = 50 μm). (C, D) The flow cytometry detection of (C) mesenchymal stem cell surface markers (CD29, CD44, CD90 and CD146) and (D) hematopoietic markers (CD45 and CD34) on LNGFR + and LNGFR − EMSCs. (E, F) The (E) qPCR and (F) Western blot analysis of LNGFR mRNA and protein levels, respectively, in LNGFR + and LNGFR − EMSCs using GAPDH as a control. ***P < .001
3.2. LNGFR+ EMSCs have a greater osteogenic capacity than LNGFR− EMSCs
Osteogenic induction experiments were performed to investigate the osteogenic capacity of LNGFR+ and LNGFR− EMSCs. The mRNA (Figure 2A,B) and protein (Figures 2C, S2A and S2B) levels of ALP and RunX2 were elevated in both LNGFR+ and LNGFR− EMSCs after culturing with osteogenic induction medium, though mRNA and protein levels of ALP and RunX2 increased more in LNGFR+ EMSCs compared with LNGFR− EMSCs. Meanwhile, the ALP staining depth was deeper in LNGFR+ EMSCs than that in LNGFR− EMSCs after culturing with osteogenic induction medium (Figure 2D,E). Furthermore, Alizarin red staining showed more mineralized nodules in LNGFR+ EMSCs compared with LNGFR− EMSCs after culturing with osteogenic induction medium (Figure 2F,G). These results revealed that LNGFR+ EMSCs have a greater osteogenic capacity than LNGFR− EMSCs.
Figure 2.
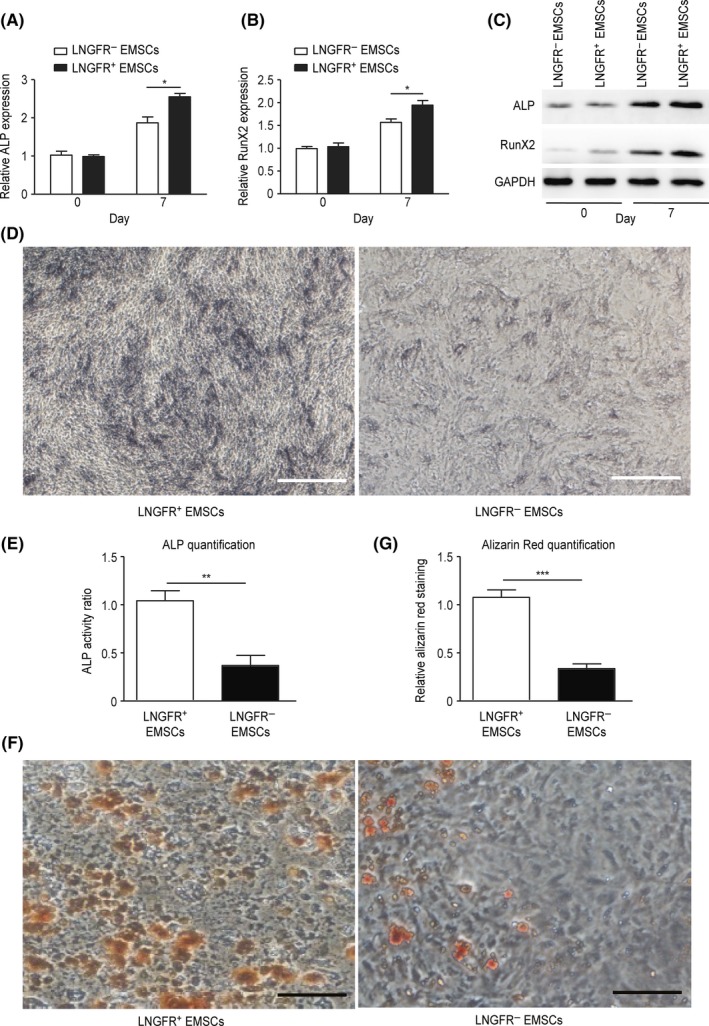
LNGFR + EMSCs have a greater osteogenic capacity than LNGFR − EMSCs. (A‐G) LNGFR + and LNGFR − EMSCs were treated with osteogenic induction medium for (A‐E) 7 days or (F and G) 21 days. On day 0 and day 7, the (A, B) mRNA and (C) protein levels of ALP and RunX2 were detected by qPCR and Western blot, respectively, using GAPDH as a control. (D) On day 7, the ALP staining depth was observed by optical microscopy (Scale bar = 50 μm). (E) The ratio of ALP activity from the triplicate assays were analysed with Imaging‐Q software, and the representative result was shown in (D). (F) On day 21, the mineralized nodules were photographed after Alizarin red staining (ARS staining) (Scale bar = 150 μm). (G) The results of alizarin red staining from the triplicate assays were analysed with Imaging‐Q software, and the representative result was shown in (F). *P < .05
3.3. SOST negatively regulates the osteogenic differentiation of EMSCs
It has been reported that SOST has negative effects on bone formation; thus, we explored whether the difference in the osteogenic capacity of LNGFR+ and LNGFR− EMSCs is associated with SOST. SOST mRNA (Figure 3A) and protein (Figures 3B and S3A) levels were lower in LNGFR+ EMSCs compared with LNGFR− EMSCs. Next, SOST was silenced in LNGFR− EMSCs and overexpressed in LNGFR+ EMSCs during osteogenic differentiation. The mRNA (Figure 3C,D) and protein (Figures 3E, S3B and S3C) levels of ALP and RunX2 were elevated in LNGFR− EMSCs after culturing with osteogenic induction medium, and the increased ALP and RunX2 levels were enhanced by SOST silencing in LNGFR− EMSCs. Meanwhile, the mRNA (Figure 3F,G) and protein (Figures 3H, S3D and S3E) levels of ALP and RunX2 were elevated in LNGFR+ EMSCs after culturing with osteogenic induction medium, and the increased ALP and RunX2 levels were weakened by SOST overexpression in LNGFR+ EMSCs. These results indicated that SOST negatively regulates the osteogenic differentiation of EMSCs.
Figure 3.
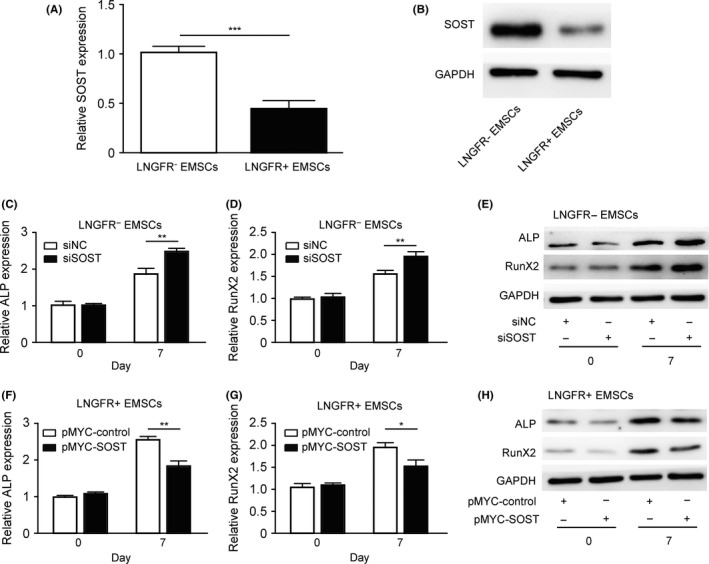
SOST negatively regulates the osteogenic differentiation of EMSCs. (A, B) The (A) qPCR and (B) Western blot analysis of SOST mRNA and protein levels, respectively, in LNGFR + and LNGFR − EMSCs using GAPDH as a control. (C‐E) The LNGFR − EMSCs were transfected with siSOST or siNC for 24 h and then treated with osteogenic induction medium an additional 7 days. On day 0 and day 7, the (C, D) mRNA and (E) protein levels of ALP and RunX2 were detected by qPCR and Western blot, respectively, using GAPDH as a control. (F‐H) The LNGFR + EMSCs were transfected with pMYC‐SOST or pMYC‐NC for 24 h and then treated with osteogenic induction medium for an additional 7 days. On day 0 and day 7, the (F, G) mRNA and (H) protein levels of ALP and RunX2 were detected by qPCR and Western blot, respectively, using GAPDH as a control. siSOST, siRNA for SOST; siNC, negative control siRNA; pMYC‐SOST, SOST expression plasmid; and pMYC‐NC, control plasmid; *P < .05, **P < .01, and ***P < .001
3.4. LNGFR is present upstream of SOST and decreases SOST expression in EMSCs
The above results showed that the levels of LNGFR and SOST were negatively correlated in LNGFR+ and LNGFR− EMSCs; thus, we explored whether a regulatory relationship exists between LNGFR and SOST. The mRNA (Figure 4A) and protein (Figures 4B and S4A) levels of LNGFR were not regulated by SOST silencing in LNGFR− EMSCs or SOST overexpression in LNGFR+ EMSCs. However, the mRNA (Figure 4C) and protein (Figures 4D and S4B) levels of SOST were upregulated by LNGFR silencing in LNGFR+ EMSCs and downregulated by LNGFR overexpression in LNGFR− EMSCs. These results revealed that LNGFR is present upstream of SOST and decreases SOST expression in EMSCs.
Figure 4.
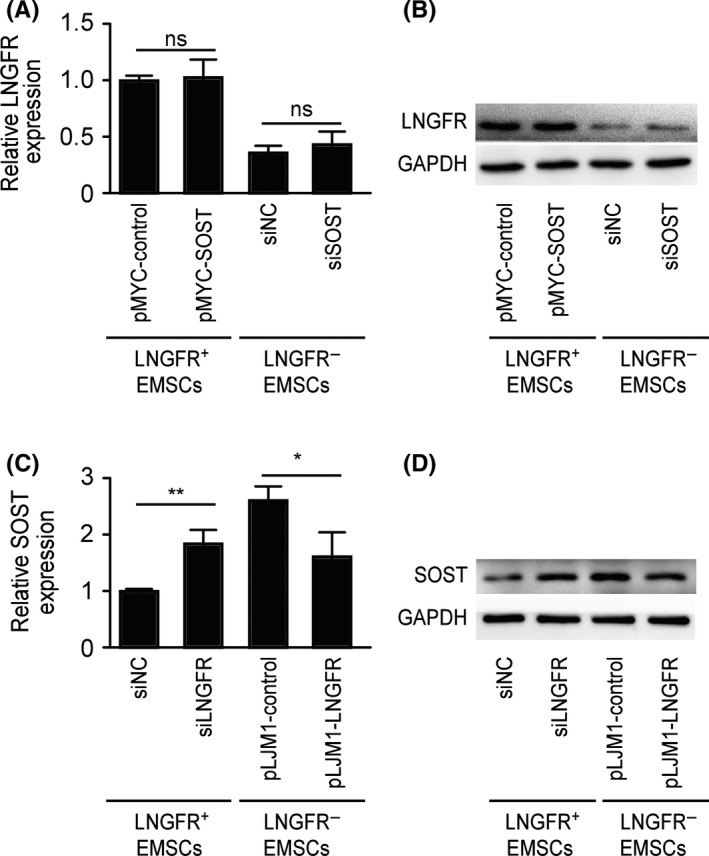
LNGFR is present upstream of SOST and decreases SOST expression in EMSCs. (A, B) The LNGFR + EMSCs were transfected with pMYC‐SOST or pMYC‐NC for 24 h, while LNGFR − EMSCs were transfected with siSOST or siNC for 24 h. Then, LNGFR (A) mRNA and (B) protein levels were detected by qPCR and Western blot, respectively, using GAPDH as a control. (C, D) The LNGFR + EMSCs were transfected with siLNGFR or siNC for 24 h, and LNGFR − EMSCs with or without the stable overexpression of LNGFR were collected. Then, (C) mRNA and (D) protein levels of SOST were detected by qPCR and Western blot, respectively, using GAPDH as a control. siSOST, siRNA for SOST; siNC, negative control siRNA; pMYC‐SOST, SOST expression plasmid; pMYC‐NC, control plasmid; siLNGFR, siRNA for LNGFR; pLJM1‐LNGFR, LNGFR − EMSCs stably overexpressing LNGFR; pLJM1‐NC, LNGFR − EMSCs without the stable overexpression of LNGFR; and ns, no significance. *P < .05 and **P < .01
3.5. LNGFR enhances the osteogenic differentiation of EMSCs by decreasing SOST
Because LNGFR decreased SOST expression in EMSCs and SOST inhibited the osteogenic differentiation of EMSCs, we explored whether LNGFR could regulate the osteogenic differentiation of EMSCs through SOST. LNGFR increased the mRNA (Figure 5A,B) and protein (Figures 5C, S5A and S5B) levels of ALP and RunX2 during the osteogenic differentiation of LNGFR− EMSCs, which was attenuated by SOST overexpression. Similarly, LNGFR augmented ALP staining depth during the osteogenic differentiation of LNGFR− EMSCs, which was alleviated by SOST overexpression in LNGFR− EMSCs (Figure 5D,E). Alizarin red staining showed that LNGFR increased mineralized nodules during the osteogenic differentiation of LNGFR− EMSCs, which was ameliorated by LNGFR overexpression of in LNGFR− EMSCs (Figure 5F,G). These results revealed that LNGFR strengthens the osteogenic differentiation of EMSCs by decreasing SOST expression.
Figure 5.
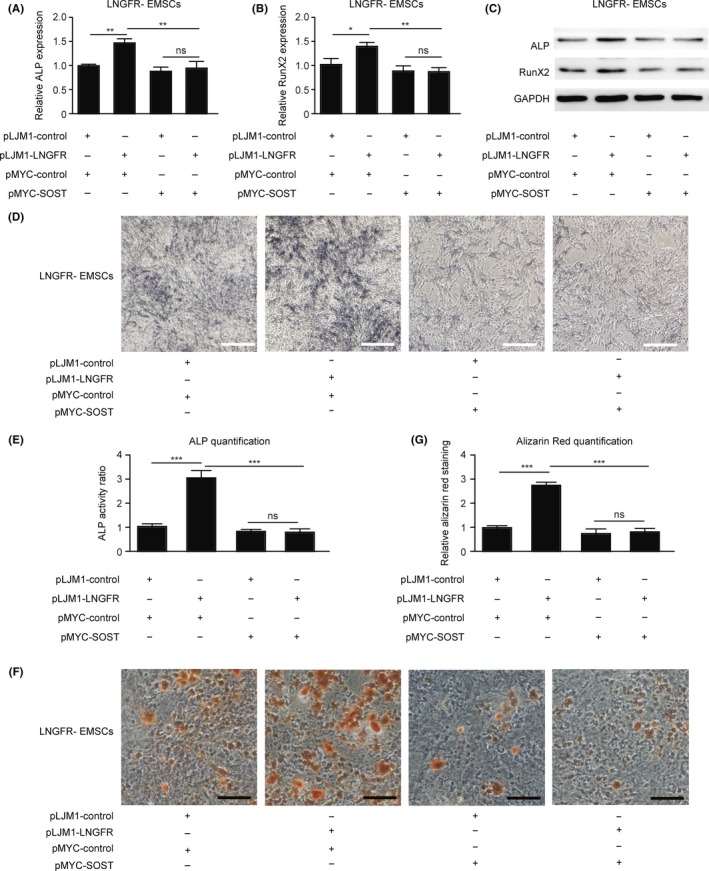
LNGFR enhances the osteogenic differentiation of EMSCs by decreasing SOST. (A‐G) LNGFR − EMSCs with or without the stable overexpression of LNGFR were separately transfected with pMYC‐SOST or pMYC‐NC for 24 h and then treated with osteogenic medium solution for an additional (A‐E) 7 days or (F, G) 21 days. On day 7, the (A, B) mRNA levels of ALP and RunX2 were detected by qPCR, (C) the protein levels of ALP and RunX2 were detected by Western blot, and (D) the ALP staining depth was observed by optical microscopy (scale bar = 50 μm). (E) The ratio of ALP activity from the triplicate assays was analysed with Imaging‐Q software, and the representative result was shown in (D). (F) On day 21, the mineralized nodules were photographed after Alizarin red staining (scale bar = 150 μm). (G) The results of alizarin red staining from the triplicate assays were analysed with Imaging‐Q software, and the representative result was shown in (F). GAPDH was used as the control in qPCR and Western blot. pLJM1‐LNGFR, LNGFR − EMSCs stably overexpressing LNGFR; pLJM1‐NC, LNGFR − EMSCs without the stable overexpression of LNGFR; pMYC‐SOST, SOST expression plasmid; pMYC‐NC, control plasmid; and ns, no significance. *P < .05 and **P < .01
Taken together, we demonstrated that SOST attenuates the osteogenic differentiation of EMSCs and that LNGFR strengthens the osteogenic differentiation of EMSCs. Moreover, LNGFR was present upstream of SOST and strengthens the osteogenic differentiation of EMSCs by decreasing SOST, suggesting that the LNGFR/SOST pathway may be a novel target for dental tissue engineering and regeneration.
4. DISCUSSION
As a neurotrophin receptor, LNGFR has been reported to be associated with the osteogenic differentiation in several types of cells.19, 20, 21 Alexander et al have reported that LNGFR is a differentiation marker for distinguishing mineralizing jaw periosteum‐derived cells (JPCs) and non‐mineralizing JPCs during the first phase of osteogenesis, and can be considered an early surface marker of in vitro osteogenic capacity.19 Another study has shown that LNGFR overexpression promotes the proliferation and osteogenic differentiation of the MC3T3‐E1 pre‐osteoblast cell line through the tyrosine kinase pathway.20 Recent evidence has demonstrated that LNGFR overexpression induces ALP activity and increases the mRNA levels of osteoblast‐related genes, resulting in the enhancement of osteoblast differentiation in the human MG63 osteoblast cell line.21 In this study, we found that LNGFR+ EMSCs had higher osteogenic capacity compared with LNGFR− EMSCs. Moreover, LNGFR can promote the osteogenic differentiation of EMSCs; this was attenuated by SOST overexpression, suggesting that the LNGFR/SOST pathway may be a potent target for advancing dental tissue engineering.
SOST is a secreted glycoprotein that is synthesized and secreted by a series of steps.22 The synthesis involves the transcription of deoxyribonucleic acid (DNA) to messenger ribonucleic acid (mRNA) in the nucleus and the conversion of the mRNA base sequence into the amino acid sequence in the protein or polypeptide chain in the ribosome.23 After processing in the endoplasmic reticulum (ER), the protein is enclosed in vesicles, bulged by the ER and then transferred to the Golgi apparatus (GA) for further processing.24 Subsequently, small vesicles enclosing the protein form on the edge of the GA, transport the protein to the cell membrane, fuse with the cell membrane and release the protein outside of the cell, thereby completing the protein secretion.25 Recent studies have shown that SOST is regulated at the transcriptional level.26, 27, 28, 29, 30 Two reports have revealed that Mef2 transcription factors bind a distal enhancer (ECR5) in the SOST locus, leading to increased SOST expression.26, 27 Another study has reported that TGF‐β also regulates SOST expression via the ECR5 enhancer.28 Moreover, Cohen‐Kfir et al have demonstrated that Sirt1 negatively regulates SOST expression by deacetylating histone 3 at lysine 9 in the SOST promoter.29 Yang and his colleagues have shown that Osterix activates the SOST promoter by directly binding to the GC‐rich sequence.30 Aside from transcription factors, parathyroid hormone (PTH) has been reported to be a potent negative regulator of SOST.31, 32, 33, 34 For example, PTH infusion in healthy men induced a reduction in circulating SOST and mouse osteocytes lacking the PTH receptor display increased SOST expression.33, 34 Moreover, prostaglandin E2, oncostatin M, cardiotrophin‐1 and leukaemia inhibitory factor have also been shown to be negative regulators of SOST.35, 36, 37 In this study, we revealed that SOST mRNA and protein levels were negatively regulated by LNGFR in EMSCs, suggesting that LNGFR may regulate SOST expression at the transcriptional level. More studies are warranted regarding the molecular mechanism by which LNGFR regulates SOST expression in EMSCs.
SOST is a vital negative regulator of bone formation. It has recently been reported that SOST inhibition increases bone formation in several disorders.38, 39, 40, 41, 42 Delgado‐Calle et al have shown that SOST inhibition augmented osteoblast numbers, promoted new bone formation and decreased osteoclast number in multiple myeloma‐colonized bone.38 McDonald and his colleague have revealed that the SOST antibody prevented myeloma‐induced bone loss, decreased osteolytic bone lesions and enhanced fracture resistance.39 Another study has shown that osteoclast‐derived leukaemia inhibitory factor suppresses SOST expression to regulate bone remodelling.40 Moreover, SOST antibodies increase osteoclast numbers and stimulate the bone formation rate in a mouse model of recessive osteogenesis imperfect (OI) and in adults with moderate OI from a randomized phase 2a trial.41, 42 In this study, we demonstrated that SOST negatively regulated osteogenesis in EMSCs, which are the progenitors of all tooth tissues, suggesting that targeting SOST may have great prospects in dental tissue regeneration and engineering.
In summary, we revealed that LNGFR+ EMSCs had a higher osteogenic capacity and lower SOST levels compared with LNGFR− EMSCs and that SOST negatively regulated the osteogenic differentiation of EMSCs. We have further shown that LNGFR is present upstream of SOST and enhanced the osteogenic differentiation of EMSCs by decreasing SOST expression. Altogether, these findings illustrated that the LNGFR/SOST pathway may be a novel target for dental tissue regeneration and engineering.
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
Supporting information
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (81470032). All experiments were performed in the Laboratories of Oral Diseases and Biomedicine at Chongqing Medical University.
Li G, Liu J, Zhao M, et al. SOST, an LNGFR target, inhibits the osteogenic differentiation of rat ectomesenchymal stem cells. Cell Prolif. 2018;51:e12412 10.1111/cpr.12412
Contributor Information
Xiujie Wen, Email: wenxiujie@tom.com.
Luchuan Liu, Email: liuluchuan1957@126.com.
REFERENCES
- 1. Kaukua N, Shahidi MK, Konstantinidou C, et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature. 2014;513:551‐554. [DOI] [PubMed] [Google Scholar]
- 2. Egbuniwe O, Idowu BD, Funes JM, Grant AD, Renton T, Di Silvio L. P16/p53 expression and telomerase activity in immortalized human dental pulp cells. Cell Cycle. 2011;10:3912‐3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drake MT, Farr JN. Inhibitors of sclerostin: emerging concepts. Curr Opin Rheumatol. 2014;26:447‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winkler DG, Sutherland MK, Geoghegan JC, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267‐6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hay E, Bouaziz W, Funck‐Brentano T, Cohen‐Solal M. Sclerostin and bone aging: a mini‐review. Gerontology. 2016;62:618‐623. [DOI] [PubMed] [Google Scholar]
- 6. Taut AD, Jin Q, Chung JH, et al. Sclerostin antibody stimulates bone regeneration after experimental periodontitis. J Bone Miner Res. 2013;28:2347‐2356. [DOI] [PubMed] [Google Scholar]
- 7. Han P, Ivanovski S, Crawford R, Xiao Y. Activation of the Canonical Wnt signaling pathway induces Cementum regeneration. J Bone Miner Res. 2015;30:1160‐1174. [DOI] [PubMed] [Google Scholar]
- 8. Ahn Y, Sims C, Murray MJ, et al. Multiple modes of Lrp4 function in modulation of Wnt/beta‐catenin signaling during tooth development. Development. 2017;144:2824‐2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao N, Nociti FH Jr., Duan P, et al. Isolation and functional analysis of an immortalized Murine Cementocyte Cell Line, IDG‐CM6. J Bone Miner Res. 2016;31:430‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nikoletopoulou V, Plachta N, Allen ND, Pinto L, Gotz M, Barde YA. Neurotrophin receptor‐mediated death of misspecified neurons generated from embryonic stem cells lacking Pax6. Cell Stem Cell. 2007;1:529‐540. [DOI] [PubMed] [Google Scholar]
- 11. Park KJ, Grosso CA, Aubert I, Kaplan DR, Miller FD. p75NTR‐dependent, myelin‐mediated axonal degeneration regulates neural connectivity in the adult brain. Nat Neurosci. 2010;13:559‐566. [DOI] [PubMed] [Google Scholar]
- 12. Ibanez CF, Simi A. p75 neurotrophin receptor signaling in nervous system injury and degeneration: paradox and opportunity. Trends Neurosci. 2012;35:431‐440. [DOI] [PubMed] [Google Scholar]
- 13. Vaculik C, Schuster C, Bauer W, et al. Human dermis harbors distinct mesenchymal stromal cell subsets. J Invest Dermatol. 2012;132:563‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Churchman SM, Ponchel F, Boxall SA, et al. Transcriptional profile of native CD271+ multipotential stromal cells: evidence for multiple fates, with prominent osteogenic and Wnt pathway signaling activity. Arthritis Rheum. 2012;64:2632‐2643. [DOI] [PubMed] [Google Scholar]
- 15. Churchman SM, Kouroupis D, Boxall SA, et al. Yield optimisation and molecular characterisation of uncultured CD271+ mesenchymal stem cells in the reamer irrigator aspirator waste bag. Eur Cell Mater. 2013;26:252‐262. [DOI] [PubMed] [Google Scholar]
- 16. Calabrese G, Giuffrida R, Lo Furno D, et al. Potential effect of CD271 on human mesenchymal stromal cell proliferation and differentiation. Int J Mol Sci. 2015;16:15609‐15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko KI, Coimbra LS, Tian C, et al. Diabetes reduces mesenchymal stem cells in fracture healing through a TNFalpha‐mediated mechanism. Diabetologia. 2015;58:633‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuoka Y, Nakatsuka R, Sumide K, et al. Prospectively isolated human bone marrow cell‐derived MSCs support primitive human CD34‐negative hematopoietic stem cells. Stem Cells. 2015;33:1554‐1565. [DOI] [PubMed] [Google Scholar]
- 19. Alexander D, Schafer F, Munz A, et al. LNGFR induction during osteogenesis of human jaw periosteum‐derived cells. Cell Physiol Biochem. 2009;24:283‐290. [DOI] [PubMed] [Google Scholar]
- 20. Mikami Y, Suzuki S, Ishii Y, et al. The p75 neurotrophin receptor regulates MC3T3‐E1 osteoblastic differentiation. Differentiation. 2012;84:392‐399. [DOI] [PubMed] [Google Scholar]
- 21. Akiyama Y, Mikami Y, Watanabe E, et al. The P75 neurotrophin receptor regulates proliferation of the human MG63 osteoblast cell line. Differentiation. 2014;87:111‐118. [DOI] [PubMed] [Google Scholar]
- 22. Paccou J, Mentaverri R, Renard C, et al. The relationships between serum sclerostin, bone mineral density, and vascular calcification in rheumatoid arthritis. J Clin Endocrinol Metab. 2014;99:4740‐4748. [DOI] [PubMed] [Google Scholar]
- 23. Lewis CJ, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA‐protein interactions. Nat Rev Mol Cell Biol. 2017;18:202‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brandizzi F, Barlowe C. Organization of the ER‐Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Campelo F, Malhotra V. Membrane fission: the biogenesis of transport carriers. Annu Rev Biochem. 2012;81:407‐427. [DOI] [PubMed] [Google Scholar]
- 26. Fulzele K, Krause DS, Panaroni C, et al. Myelopoiesis is regulated by osteocytes through Gsalpha‐dependent signaling. Blood. 2013;121:930‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu L, van der Valk M, Cao J, et al. Sclerostin expression is induced by BMPs in human Saos‐2 osteosarcoma cells but not via direct effects on the sclerostin gene promoter or ECR5 element. Bone. 2011a;49:1131‐1140. [DOI] [PubMed] [Google Scholar]
- 28. Loots GG, Keller H, Leupin O, Murugesh D, Collette NM, Genetos DC. TGF‐beta regulates sclerostin expression via the ECR5 enhancer. Bone. 2012;50:663‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cohen‐Kfir E, Artsi H, Levin A, et al. Sirt1 is a regulator of bone mass and a repressor of Sost encoding for sclerostin, a bone formation inhibitor. Endocrinology. 2011;152:4514‐4524. [DOI] [PubMed] [Google Scholar]
- 30. Yang F, Tang W, So S, de Crombrugghe B, Zhang C. Sclerostin is a direct target of osteoblast‐specific transcription factor osterix. Biochem Biophys Res Comm. 2010;400:684‐688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kramer I, Loots GG, Studer A, Keller H, Kneissel M. Parathyroid hormone (PTH)‐induced bone gain is blunted in SOST overexpressing and deficient mice. J Bone Miner Res. 2010;25:178‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Costa AG, Cremers S, Rubin MR, et al. Circulating sclerostin in disorders of parathyroid gland function. J Clin Endocrinol Metab. 2011;96:3804‐3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu EW, Kumbhani R, Siwila‐Sackman E, Leder BZ. Acute decline in serum sclerostin in response to PTH infusion in healthy men. J Clin Endocrinol Metab. 2011b;96:E1848‐1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Powell WF Jr., Barry KJ, Tulum I, et al. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J Endocrinol. 2011;209:21‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galea GL, Sunters A, Meakin LB, et al. Sost down‐regulation by mechanical strain in human osteoblastic cells involves PGE2 signaling via EP4. FEBS Lett. 2011;585:2450‐2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Genetos DC, Yellowley CE, Loots GG. Prostaglandin E2 signals through PTGER2 to regulate sclerostin expression. PLoS ONE. 2011;6:e17772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walker EC, McGregor NE, Poulton IJ, et al. Oncostatin M promotes bone formation independently of resorption when signaling through leukemia inhibitory factor receptor in mice. J Clin Investig. 2010;120:582‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delgado‐Calle J, Anderson J, Cregor MD, et al. Genetic deletion of Sost or pharmacological inhibition of sclerostin prevent multiple myeloma‐induced bone disease without affecting tumor growth. Leukemia. 2017. 10.1038/leu.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald MM, Reagan MR, Youlten SE, et al. Inhibiting the osteocyte‐specific protein sclerostin increases bone mass and fracture resistance in multiple myeloma. Blood. 2017;129:3452‐3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koide M, Kobayashi Y, Yamashita T, et al. Bone formation is coupled to resorption via suppression of sclerostin expression by osteoclasts. J Bone Miner Res. 2017;32:2074‐2086. [DOI] [PubMed] [Google Scholar]
- 41. Grafe I, Alexander S, Yang T, et al. Sclerostin antibody treatment improves the bone phenotype of Crtap(−/−) Mice, a model of recessive osteogenesis imperfecta. J Bone Miner Res. 2016;31:1030‐1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glorieux FH, Devogelaer JP, Durigova M, et al. BPS804 anti‐sclerostin antibody in adults with moderate osteogenesis imperfecta: results of a randomized phase 2a trial. J Bone Miner Res. 2017;32:1496‐1504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


