Abstract
Objectives
KDM6A has been demonstrated critical in the regulation of cell fates. However, whether KDM6A is involved in cartilage formation remains unclear. In this study, we investigated the role of KDM6A in chondrogenic differentiation of PDLSCs, as well as the underlying epigenetic mechanisms.
Methods
KDM6A shRNA was transfected into PDLSCs by lentivirus. The chondrogenic differentiation potential of PDLSCs was assessed by Alcian blue staining. Immunofluorescence was performed to demonstrate H3K27me3 and H3K4me3 levels during chondrogenesis. SOX9, Col2a1, ACAN and miRNAs (miR‐29a, miR‐204, miR‐211) were detected by real‐time RT‐PCR. Western blot was performed to evaluate SOX9, H3K27me3 and H3K4me3.
Results
The production of proteoglycans in PDLSCs was decreased after knockdown of KDM6A. Depletion of KDM6A inhibited the expression of SOX9, Col2a1, ACAN and resulted in increased H3K27me3 and decreased H3K4me3 levels. EZH2 inhibitor rescued the chondrogenic potential of PDLSCs after knockdown of KDM6A by regulating H3K27me3. Additionally, miR‐29a, miR‐204 and miR‐211 were also involved in the process of PDLSCs chondrogenesis.
Conclusions
KDM6A is required in chondrogenic differentiation of PDLSCs by demethylation of H3K27me3, and EZH2 inhibitor could rescue chondrogenesis of PDLSCs after knockdown of KDM6A. It could be inferred that upregulation of KDM6A or application of EZH2 inhibitor might improve mesenchymal stem cell mediated cartilage regeneration in inflammatory tissue destruction such as osteoarthritis.
Keywords: chondrogenesis, KDM6A, periodontal ligment stem cells, SOX9
1. INTRODUCTION
Cartilage defect is frequently caused by inflammation, trauma, tumour, etc. The damaged cartilage cannot be fully regenerated due to its poor intrinsic healing capacity to reconstitute the integrated matrix and regenerate native surface. Till now, many approaches, ranging from symptomatic treatment to structural cartilage regeneration, have obtained limited results. Recently, stem‐cell‐induced tissue regeneration has emerged as a promising strategy for cartilage regeneration or repair.
Mesenchymal stem cells (MSCs) are a reliable resource for tissue regeneration. They are multipotent cells which are capable to differentiate into a variety of cell types, including osteoblasts, chondrocytes, myocytes and adipocytes, etc. Recently, a new population of MSCs has been isolated from dental and craniofacial tissues, including but not limited to periodontal ligament stem cells (PDLSCs), dental pulp stem cells and stem cells from apical papilla. Numerous studies demonstrated that dental stem cells are derived from neural crest cells. Superior proliferation capacity and multipotent differentiation potential were shown in dental stem cells compared with bone marrow MSC.1 Previous study showed that PDLSCs exhibited greater osteo/dentinogenic differentiation potentials compared with Wharton's jelly of umbilical cord stem cells.2, 3 Meanwhile, PDLSCs are convenient to obtain and free of any ethical controversy. Therefore, this population of MSCs might be good resource for cartilage regeneration. However, the molecular mechanisms underlying chondrogenic differentiation of PDLSCs remain unclear, which restrict the potential application of PDLSCs in this field.
The mechanism of MSC differentiation is complicated both in temporal and spatial dimensions, in which covalent histone modifications are thought to play a critical role and be of essential importance in regulating chromatin dynamics and functions.4, 5 Among them, histone methylation occurs on both lysine and arginine residues, participating in diverse biological processes including cell proliferation, differentiation, development, etc. Histone lysine methylation is controlled by histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs). Histone H3 lysine 27 (H3K27) methylation has been mainly linked to transcription repression6 and can be removed by KDM, including KDM6A and KDM6B. H3K27me3 demethylases likely play an important role in driving embryonic stem cell (ESC) differentiation. Lysine‐specific demethylase6A (KDM6A), located on Xp11.3 and also referred as UTX, is one of histone H3K27 specific demethylase. It has been demonstrated to be critical in the regulation of stem cells, including M2 macrophage differentiation, muscle differentiation and neuronal stem cell differentiation.7 Functional studies showed that KDM6A overexpression in MSCs promotes osteogenesis and inhibits adipogenesis, while knockdown of KDM6A inhibits osteogenesis and promotes adipogenesis.8 These findings indicate a positive role of KDM6A in osteogenesis but a negative role in adipogenic differentiation of MSCs. However, the function of KDM6A in chondrogenesis is still poorly understood.
Herein, we hypothesized that KDM6A may play a role in chondrogenic differentiation of MSCs. In this study, PDLSCs were employed to investigate the function of KDM6A and the change of miRNAs in this process. Our results showed that depletion of KDM6A repressed the chondrogenic differentiation potentials in PDLSCs by increasing the histone H3K27me3 level and decreasing the H3K4me3 level. Additionally, miR‐29a, miR‐204 and miR‐211 involved in chondrogenesis of PDLSCs were also investigated by real‐time RT‐PCR.
2. MATERIALS AND METHODS
2.1. Cell cultures
All research involving human stem cells complied with the ISSCR “Guidelines for the Conduct of Human Embryonic Stem Cell Research.” Healthy periodontal ligament (PDL) was obtained from patients (age 16‐20 years) who had no history of periodontal disease and exhibited a relatively healthy periodontium. The remnant or discarded periodontal tissues were obtained from patients following the guidelines set by the Tianjin Medical University School of Stomatology, with informed consent. PDLSCs were isolated, cultured and identified as previously reported.9 Briefly, PDL was gently separated from the surface of the root and then digested in a solution of 3 mg/mL collagenase type I and 4 mg/mL dispase (Sigma‐Aldrich, St. Louis, MO, USA) for 1 hour at 37°C. PDL samples from different individuals were pooled, and single‐cell suspensions were obtained by passing the cells through a 70‐μm strainer (Falcon, BD Labware, Franklin Lakes, NJ, USA), plated on 10‐cm dishes with complete α‐MEM (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA), and cultured at 37°C in 5% carbon dioxide. The culture medium was changed every 3 days. Cells at passages 3‐5 were used in subsequent experiments.
2.2. Virus infection
Virus for shRNA was a kind of gift from Dr. Fan, Capital Medical University School of Stomatology, China. Viral infection was conducted as previously described3, and PDLSCs were plated overnight and then infected with lentiviruses in the presence of polybrene (6 μg/mL; Sigma‐Aldrich) for 6 hours. Cells were then selected with puromycin for 48 hours. Resistant clones were pooled, and knockdown was confirmed via real‐time RT‐PCR. The target sequences for KDM6A shRNA (KDM6Ash) were 5′‐TTTATTCCTTAGTCTATGTGC‐3′.
2.3. Cell viability assay
Wild‐type PDLSCs (PDLSCs‐WT) and PDLSCs‐KDM6Ash were seeded into 96‐well plate (5000/well). Then, the cell counting kit‐8 (CCK‐8) (Dojindo, Kamimashiki‐gun, Kumamoto, Japan) was used to determine cell viability after 1‐ to 8‐day incubation or 3‐day chondrogenic/osteogenic culture. The optical densities (OD) were measured at 450 nm. These experiments were performed 3 times.
2.4. Cell apoptosis assay
Cell apoptosis was determined by staining cells with Annexin V‐fluorescein isothiocyanate (FITC) and counterstaining with propidium iodide (PI), using the eBioscience™ Annexin V‐FITC Apoptosis Detection Kit (Life Technologies, Carlsbad, CA, USA). Briefly, 1 × 105 cells were washed twice with phosphate buffered saline (PBS), resuspended in 200 μL Binding Buffer (1×) and stained with 5 μL Annexin V‐FITC for 10 minutes at room temperature in the dark. Finally, 10 μL PI in 1× binding buffer was added to the cell suspension, and analysis was performed by fluorescence‐activated cell sorting.
2.5. Micromass culture
The micromass culture was based on the methods of Zhu et al.10 The micromass was formed by resuspending 2 × 106 cells in 200 μL of growth medium. The cell suspension was gently pipetted into the middle of a single well of a 6‐cm dish and was left overnight at 37°C, 5% CO2, before adding an additional 1 mL of chondrogenic medium. The medium was changed every 2‐3 days for 21 days.
2.6. Alcian blue stain analysis
Chondrogenic differentiation was induced by using the STEMPRO Chondrogenesis Differentiation Kit (Life Technologies). PDLSCs were grown in the chondrogenic medium for 2 or 3 weeks. For Alcian blue staining, monolayer cultured cells were rinsed with PBS and fixed with 4% formaldehyde solution for 30 minutes. After fixation, cells were rinsed with PBS and stained with 1% Alcian blue solution prepared in 0.1 N HCL for 30 minutes. And then, cells were rinsed with 0.1 N HCl 3 times and distilled water to neutralize the acidity, visualized under light microscope, and images were captured for analysis. Micromasses were rinsed, dehydrated, stained with 1% Alcian blue solution for 30 min, rinsed with PBS for 3 times and fixed on slides. Blue staining indicated synthesis of proteoglycans by chondrocytes. To quantify proteoglycans synthesis, Alcian blue was extracted by 4m guanidine‐HCl overnight at 4°C. Absorbance values were read at 600 nm after temperature equilibration. The final OD value in each group was normalized with the total protein concentrations prepared from a duplicate plate.
2.7. Sirius Red stain
Sirius Red staining was conducted as previously described.11 Briefly, the micromasses were dehydrated, sealed by neutral balsam, stained with 1 g/L picric acid‐Sirius Red at 37°C for 1 hour, and then washed with water. The micromasses were mounted and viewed under a microscope and captured images.
2.8. Real‐time PCR analysis
Total RNA was isolated from PDLSCs with Trizol reagents (Life Technologies). We synthesized cDNA from 2 mg aliquots of RNA, random Hexamers or oligo (dT), and reverse transcriptase, according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). Real‐time RT‐PCR reactions were performed with the QuantiTect SYBR Green PCR kit (Qiagen, Duesseldorf, Germany) and an Icycler iQ Multi‐colour Real‐time PCR detection system. To detect the expression of miRNAs, real‐time PCR was performed using miDETECT A Track™ miRNA qRT‐PCR Starter Kit (RIBOBIO, Guangzhou, China). U6 RNA was used as an internal control for miRNA detection. Table S1 presents the primers for specific genes.
2.9. Western blot analysis
Cells were lysed in RIPA buffer (10 mM Tris‐HCl, 1 Mm EDTA, 1% sodium dodecyl sulphate [SDS], 1% NP‐40, 1:100 proteinase inhibitor cocktail, 50 mM β‐glycerophosphate, 50 mM sodium fluoride). The samples were separated on a 10% SDS polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes with a semidry transfer apparatus (Bio‐Rad, Hercules, CA, USA). The membranes were blotted with 5% dehydrated milk for 2 hours and then incubated with primary antibodies overnight. Primary monoclonal anti‐SOX9, anti‐H3K4me3, anti‐H3K27me3 and the house keeping protein, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) and histone H3 (Abcam, Cambridge, MA, USA) were used. Peroxidase‐conjugated goat anti‐rabbit or goat anti‐mouse were used as the secondary antibody at 1:1000 dilution.
2.10. Immunofluorescence
The monolayer cultured cells were rinsed with PBS buffer, fixed and blocked by BSA solution, and then samples were incubated with primary antibodies (mouse polyclonal antibody against H3K4me3 and H3K27me3 [Abcam], diluted 1:200) overnight at 4°C. After incubation, samples were washed 3 times with PBS. Then, goat anti‐mouse fluorescein isothiocyanate‐conjugated secondary antibody (Abcam) was applied for 1 hour at room temperature. After incubation, samples were washed 3 times with PBS. Then, the samples were incubated by DAPI solution for 5 minutes. Finally, after 3 times wash with PBS, DAPI was used as nuclei‐specific dye. The samples were rinsed with PBS for 3 times and visualized by a fluorescent‐inverted phase‐contrast microscope equipped with a Leica MPS‐30 camera.
2.11. Chromatin immunoprecipitation assays
Briefly, cells were incubated with 1% formaldehyde for 10 minutes at 37°C. Each chromatin immunoprecipitation (ChIP) reaction was performed using 2 × 106 cells. For DNA precipitation, we added 2 mg polyclonal antibodies against trimethyl H3K4 (H3K4me3) and trimethyl H3K27 (H3K27me3, Abcam). The precipitated DNA samples were quantified by real‐time PCR with primers targeting the H3K27me3‐binding region of the SOX9 promoter: forward, 5′‐TGGTGGTCGGTGTAGTCGTA‐3′ and reverse, 5′‐CGAACGCACATCAAGACGGA‐3′. Quantification data are expressed as the percentage of input DNA.
3. RESULTS
3.1. Depletion of KDM6A had no effect on cell viability and apoptosis of PDLSCs
In order to identify the characteristics of the isolated cells, fluorescence‐activated cell sorter analysis was performed and demonstrated that human PDLSCs display specific MSC markers such as CD90, STRO‐1, SSEA‐4 and OCT‐4, while not expressing hematopoietic lineage markers such as CD 45 (Fig. S1C). To investigate the differentiation potential of PDLSCs, cells were induced in osteogenic medium, chondrogenic medium and adipogenic medium, and we found that PDLSCs have the potential of multilineage differentiation (Fig. S1D‐F). These data confirmed the MSC properties of PDLSCs.
After virus infection and antibiotics selection, the knockdown efficiency in PDLSCs‐KDM6Ash was around 94.95% compared with PDLSCs‐WT, which was verified by real‐time RT‐PCR (Figure 1A). Next, we compared the proliferative activity of PDLSCs‐WT with PDLSC‐KDM6Ash by CCK8 assay. We found that during 8 days of culture, the cell viability of PDLSCs‐WT and PDLSCs‐KDM6Ash showed no significant difference (Figure 1B). Then, cells were induced with chondrogenic and osteogenic medium, the cell viability showed no significant difference (Figure 1C). To test the apoptosis of PDLSCs, flow cytometric assay was performed by measuring the percentage of Annexin V positive cells. As observed, there was no significant difference of apoptotic cells percentage between PDLSCs‐WT and PDLSCs‐KDM6Ash (P > .05, Figure 1D).
Figure 1.
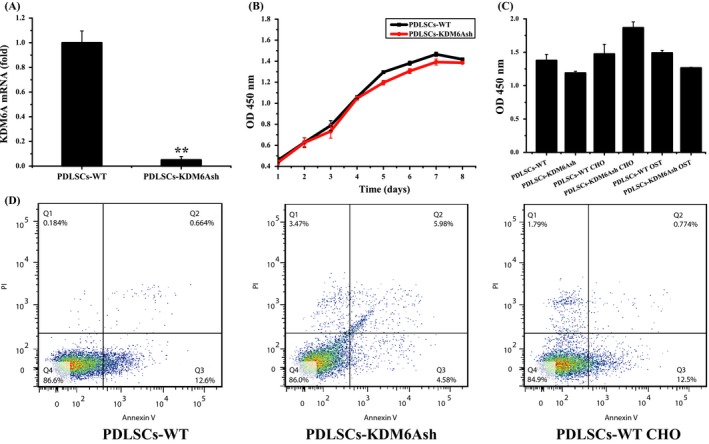
Cell viability and apoptosis detection of PDLSCs after knockdown of KDM6A. (A) Real‐time RT‐PCR confirming depleted expression of KDM6A in PDLSCs‐KDM6Ash compared with PDLSC‐WTs. (B) The cell viability of PDLSCs‐WT and PDLSCs‐KDM6Ash was determined by CCK‐8 assay everyday in 8 days (mean ± SD, n = 3). (C) The cell viability of PDLSCs‐WT and PDLSCs‐KDM6Ash after osteogenic or chondrogenic culture was determined at 3 days. (D) Flow cytometry analysis of PDLSCs‐WT or PDLSCs‐KDM6Ash with or without 3‐day chondrogenic culture. Flow cytometry was performed using a double‐staining method with FITC‐conjugated annexin V and PI
3.2. Depletion of KDM6A repressed chondrogenic differentiation potential of PDLSCs
To assess whether depletion of KDM6A affects the chondrogenic differentiation potential of PDLSCs, we compared the level of chondrogenic‐related protein and mRNAs in PDLSCs‐WT and PDLSCs‐KDM6Ash. Cells of monolayer culture were induced with chondrogenic medium for 2 weeks, and then were stained with Alcian blue. The staining results revealed that proteoglycans production was significantly decreased in PDLSCs‐KDM6Ash compared with PDLSCs‐WT (P < .05, Figure 2A,B). In order to provide cell to cell contact in 3 dimensions, which simulates MSCs condensation to induce chondrogenesis in vivo12, micromass culture was employed to evaluate the influence of KDM6A on chondrogenic differentiation of PDLSCs in vitro. Micromasses were cultured in chondrogenic medium for 3 weeks, and then was stained with Alcian blue and Sirius Red. Similar to the result from monolayer culture, Alcian blue and Sirius Red staining for micromass culture also showed that PDLSCs‐KDM6Ash‐mass exhibited less proteoglycans and collagen formation than PDLSCs‐WT‐mass (Figure 2A,B, P < .05).
Figure 2.
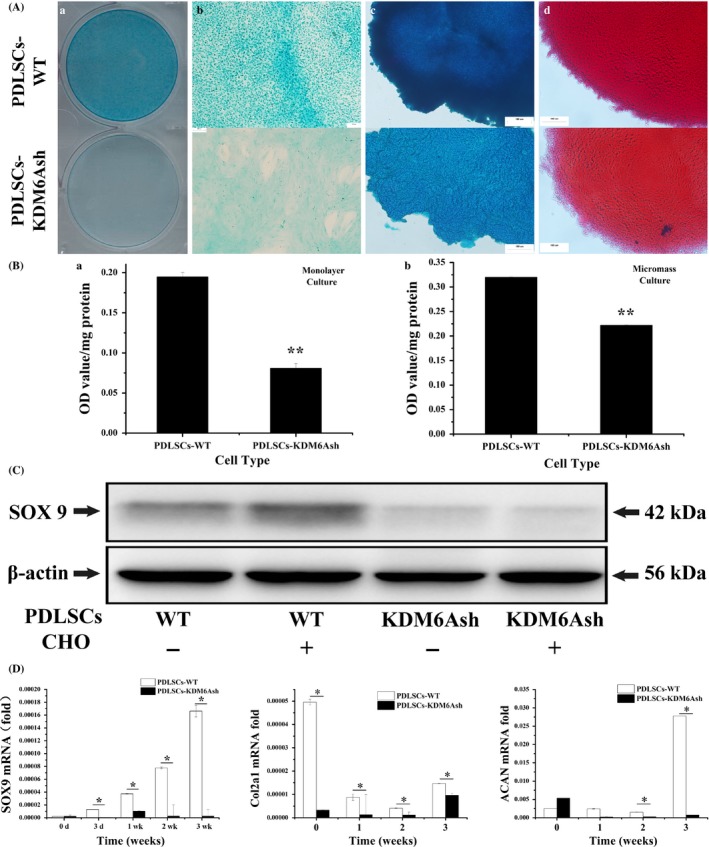
Chondrogeneic differentiation potential of periodontal ligament stem cells (PDLSCs) after knockdown of KDM6A. PDLSCs were cultured with chondrogenic medium for the indicated time periods. (A) Monolayer cultured cells (a, b) were stained with 1% Alcian blue after induced with chondrogenic medium for 2 weeks. Micromasses were stained with alcian blue (c) and sirius Red (d) after 3‐week culture. Scale bar = 100 μm. (B) Quantification of proteoglycans synthesis at 2 weeks. (C) Western blot analysis of PDLSC‐WTs and PDLSCs‐KDM6Ash probed with anti‐SOX9 and anti‐β‐actin showed decreased expression of SOX9 protein. (D) Real‐time RT‐PCR analysis of chondrogenic genes, SOX9, COL2a1 and ACAN normalized to β‐actin in PDLSCs‐WT and PDLSCs‐KDM6Ash induced under chondrogenic differentiation media for 3 weeks. Statistical significance was determined by one‐way ANOVA. All error bars represent SD (n = 3). *P < .05. **P < .01
SOX9 was reported as key transcription factor of chondrogenic differentiation. Therefore, we next investigated the expression of SOX9 in PDLSCs by western blot after 2 weeks of chondrogenic induction. Results showed that protein level of SOX9 was dramatically decreased after knockdown of KDM6A in PDLSCs, both with or without chondrogenic induction (Figure 2C). To further examine the expression of chondrogenic differentiation markers, SOX9, Col2a1 and ACAN mRNA profiles were detected by real‐time RT‐PCR. The results showed that mRNA level of SOX9 was decreased in PDLSCs‐KDM6Ash compared with PDLSCs‐WT after 3 days and 1 week, 2 weeks and 3 weeks of chondrogenic induction. The expression of Col2a1 was decreased in PDLSCs‐KDM6Ash compared with PDLSCs‐WT at time points of 1, 2 and 3 weeks after chondrogenic induction. The expression of ACAN was decreased in PDLSCs‐KDM6Ash compared with PDLSCs‐WT at time points of 2 and 3 weeks (P < .05, Figure 2D).
3.3. Depletion of KDM6A resulted in increased H3K27me3 and decreased H3K4me3 in PDLSCs
To investigate the mechanism of KDM6A promoting chondrogenic differentiation potential of PDLSCs, we examined the H3K27me3 and H3K4me3 level during the chondrogenic differentiation process of PDLSCs by immunofluorescence and western blot. Results showed that H3K27me3 was significantly upregulated in the PDLSCs‐KDM6Ash compared with PDLSCs‐WT (Figure 3A,B,E). However, the level of H3K4me3 was unequivocally downregulated in PDLSCs‐KDM6Ash compared with PDLSCs‐WT (Figure 3C‐E). The level of H3K27me3 remains higher and H3K4me3 remains lower in the PDLSCs‐KDM6Ash after 3 days of chondrogenic induction (Fig. S2).
Figure 3.
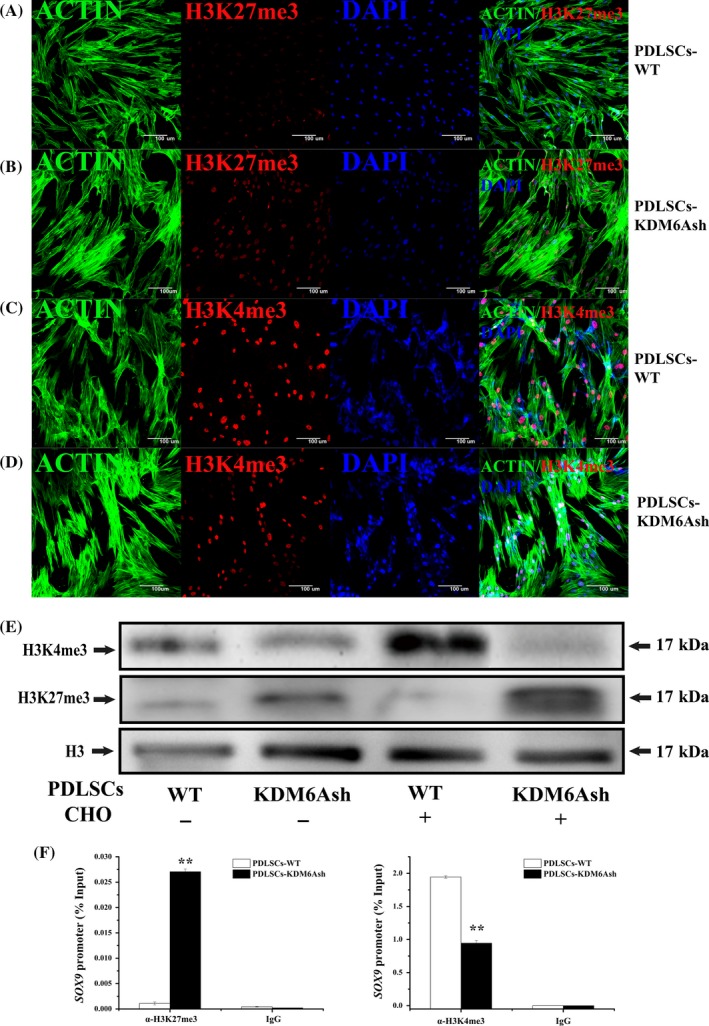
H3K27me3 and H3K4me3 level of PDLSCs after knockdown of KDM6A. (A, B) Cytoskeleton/actin staining (green), H3K27me3 staining (red) and DAPI staining (blue) in PDLSCs‐WT (A) and PDLSCs‐KDM6Ash (B). (C, D) Cytoskeleton/actin staining (green), H3K4me3 staining (red) and DAPI staining (blue) in PDLSCs‐WT (C) and PDLSCs‐KDM6Ash (D). (E) Western blot analysis probed with anti‐H3K27me3, anti‐H3K4me3 and anti‐H3 showed the expression of H3K27me3 and H3K4me3. (F) ChIP assay showed the change of H3K27me3 and H3K4me3 after knockdown of KDM6A in PDLSCs on the promoter of SOX9. Student's t test was performed to determine statistical significance. Error bars represent SD (n = 3). *P < .05. **P < .01
3.4. Depletion of KDM6A resulted in increased H3K27me3 and decreased H3K4me3 in SOX9 promoter
Aim to prove that KDM6A promotes chondrogenic differentiation of PDLSCs by demethylation of SOX9, we next performed a ChIP assay. The results indicated that KDM6A silencing increased the H3K27me3 level in the SOX9 promoter (Figure 3F), suggesting that KDM6A promoted SOX9 transcription by decreasing H3K27 trimethylation. Interestingly, we also found that depletion of KDM6A in PDLSCs resulted in decreased level of H3K4me3 in the SOX9 promoter.
3.5. EZH2 inhibitor rescued the chondrogenic potential of PDLSCs by decreased H3K27me3 after depletion of KDM6A
EPZ‐6438 is a potent, selective and orally bioavailable small‐molecule inhibitor of H3K27me3 methyltransferases (EZH2), and it could inhibit the activity of human PRC2‐containing wild‐type EZH2. Herein, we hypothesis that EPZ‐6438 could rescue the repression of chondrogenic differentiation potential of PDLSCs after knockdown of KDM6A. Thus, we treated PDLSCs with series EPZ‐6438 solution, of which concentration ranged from 0 mM to 1.5 mM. During 8 days of culture, PDLSCs‐WT with 0.5 mM and 1.0 mM EPZ‐6438 showed similar proliferation activity with PDLSCs‐WT. However, cell proliferation activity of PDLSCs was significantly lower when treated with 1.5 mM EPZ‐6438 (Fig. S3). Therefore, 1.0 mM solution of EPZ‐6438 was employed in the following procedures.
Next, we investigated the effect of EPZ‐6438 on chondrogenic differentiation potential of PDLSCs‐WT and PDLSCs‐KDM6Ash. After induction with chondrogenic medium for 2 weeks, the Alcian blue staining and Sirius Red staining for monolayer cultures and micromasses results revealed that proteoglycans and collagen production in PDLSCs‐KDM6Ash was significantly increased when treated with EPZ‐6438; however, EPZ‐6438 has shown no effect on chondrogenic differentiation of PDLSCs‐WT (P < .01, Figure 4A,B). During 3 weeks of chondrogenic differentiation, real‐time RT‐PCR results showed that mRNA levels of SOX9, Col2A1 and ACAN were significantly increased in PDLSCs‐WT and PDLSCs‐KDM6Ash when treated with EPZ‐6438 (P < .05, Figure 4C). After 2 weeks of chondrogenic differentiation, SOX9 protein showed similar results with proteoglycans and collagen (Figure 4D). Next, we examined histone methylation during the chondrogenic differentiation process by immunofluorescence and western blot. Results showed that the H3K27me3 level was obviously decreased in PDLSCs‐KDM6Ash when treated with EPZ‐6438, while H3K4me3 level was increased in PDLSCs‐KDM6Ash when treated with EPZ‐6438 (Figure 5A‐E). Together, these results support the hypothesis that EZH2 inhibitor could rescue impaired chondrogenic potential of KDM6A‐depleted PDLSCs by decreasing H3K27me3.
Figure 4.
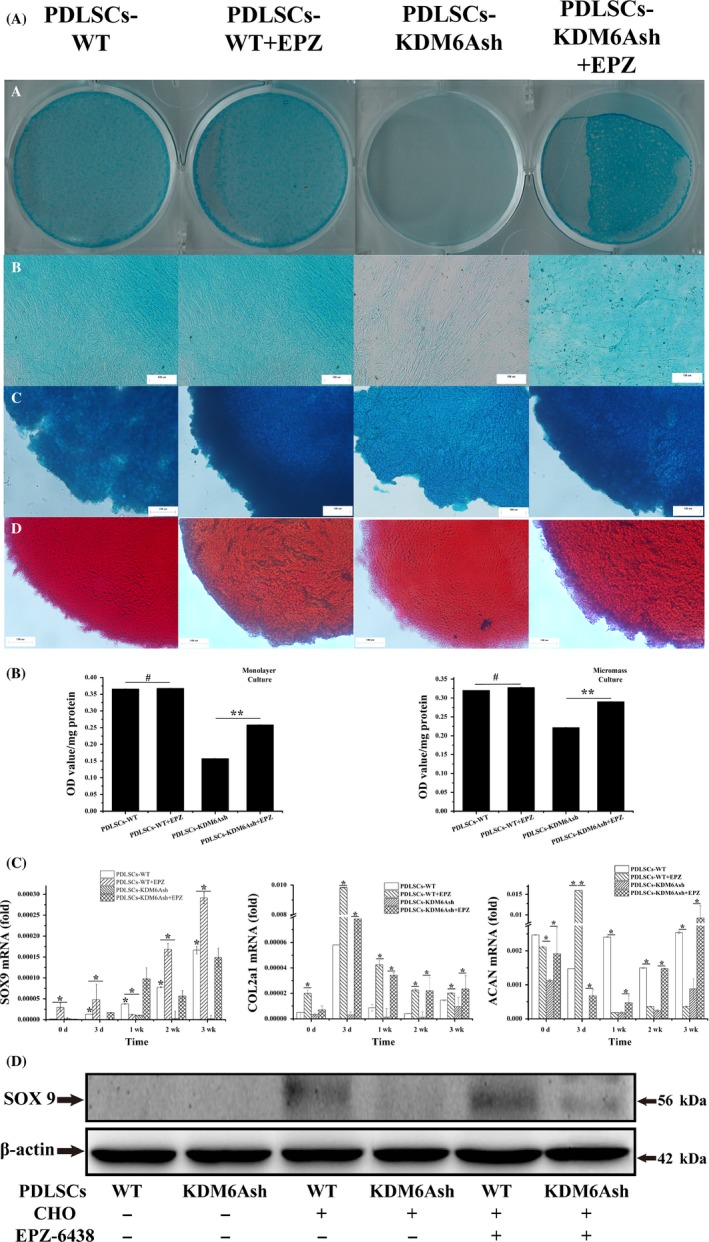
Chondrogenic differentiation potential of periodontal ligament stem cells (PDLSCs) after using EZH2 inhibitor EPZ‐6438. (A) PDLSCs cultured in monolayer culture (a, b) and micromass culture (c) were stained with 1% Alcian blue, and micromasses were stained with Sirius Red (d) at 2 weeks after induced with chondrogenic medium with/without 1 mm EPZ‐6438. (B) Quantification of proteoglycans synthesis at 2 weeks. (C) Real‐time RT‐PCR analysis of chondrogenic genes, SOX9 (a), COL2a1 (b), ACAN (c) normalized to β‐actin in PDLSC‐WTs and PDLSCs‐KDM6Ash induced under chondrogenic differentiation media for 3 weeks. (D) Western blot analysis of PDLSC‐WTs and PDLSCs‐KDM6Ash probed with anti‐SOX9 and anti‐β‐actin showed decreased expression of SOX9 protein. β‐actin was used as an internal control. Statistical significance was determined by one‐way ANOVA. Error bars represent SD (n = 3). *P < .05. **P < .01
Figure 5.

The expression of H3K4me3 and H3K27me3 after using EZH2 inhibitor EPZ‐6438. (A, B) Cytoskeleton/actin staining (green), H3K27me3 staining (red) and DAPI staining (blue) in PDLSCs‐KDM6Ash (A) and PDLSCs‐KDM6Ash with EPZ‐6438 (B). (C, D) Cytoskeleton/actin staining (green), H3K4me3 staining (red) and DAPI staining (blue) in PDLSCs‐KDM6Ash (C) and PDLSCs‐KDM6Ash with EPZ‐6438 (D). (E) Western blot analysis probed with anti‐H3K27me3, anti‐H3K4me3 and anti‐H3 showed reversed expression of H3K27me3 and H3K4me3 after using EPZ‐6438, and histone H3 was employed as internal control
3.6. miRNAs were involved in chondrogenesis of PDLSCs
Runx2 is the target gene of miR‐204 and miR‐211 and is the key transcription factor regulating osteogenesis and chondrogenesis. Real‐time RT‐PCR showed that the mRNA level of Runx2 was increased in PDLSCs at time point of 3 d of chondrogenic induction, and was decreased after 1, 2 and 3 weeks of induction (P < .05, Figure 6A). During 3 weeks of chondrogenic differentiation, real‐time PCR showed that the levels of miR‐204 and miR‐211 were decreased after knockdown of KDM6A, while inducing PDLSCs‐KDM6Ash with EPZ‐6438, the expressions of miR‐204 and miR‐211 were rescued, without significant difference with PDLSCs‐WT at the time point of 3 weeks (P < .05, Figure 6C,D). And it was also revealed that while inducing PDLSCs with chondrogenic medium for 3 weeks, the expression of miR‐29a was decreased. After knockdown of KDM6A, the expression of miR‐29a was increased, while inducing PDLSCs‐KDM6Ash with EPZ‐6438, the expression of miR‐29a was decreased, showed no significant difference with PDLSCs‐WT (P < .05, Figure 6B).
Figure 6.
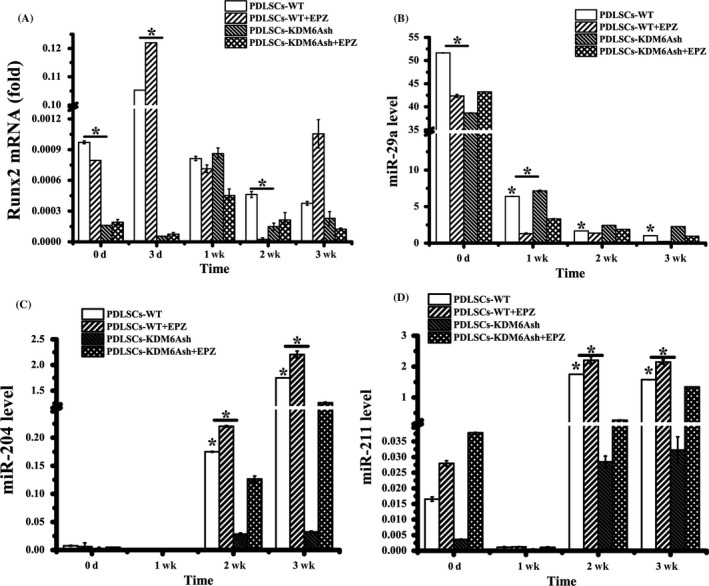
The expression of miRNA in chondrogenic differentiation of PDLSCs. (A) Real‐time RT‐PCR analysis of osteogenic gene Runx2 normalized to β‐actin in PDLSCs‐WT and PDLSCs‐KDM6Ash induced under chondrogenic differentiation media for 3 weeks. qRT‐PCR analysis of miR‐29a (B), microRNA‐204 (C) and microRNA‐211 (D) normalized to U6 in PDLSCs‐WT and PDLSCs‐KDM6Ash with or without EPZ‐6438 induced with chondrogenic medium for 3 weeks. Statistical significance was determined by one‐way ANOVA. Error bars represent SD (n = 3). *P < .05. **P < .01
4. DISCUSSION
Recently, dental stem cell‐induced tissue regeneration attracts much attention, and dental stem cells have been employed to successfully regenerate certain tissues in vitro or in large animal models.13 PDLSCs are one kind of the dental stem cells and can be harvested easily from oral cavity and also can be obtained from discarded biological samples in dental clinics.14 Moshaverinia et al have reported that PDLSCs showed superior chondrogenic differentiation potential than human bone marrow mesenchymal stem cells (hBMMSCs) in vitro and in vivo.15 Our study also verified PDLSCs have chondrogenic differentiation potential, suggesting this population of dental stem cells could be a promising candidate for cartilage regeneration.
Epigenetic regulation of gene expression plays a pivotal role in multiple differentiation of stem cells. Histone methylation is one of the epigenetic regulations in diverse biological processes including transcription.16 H3K27me3 is associated with inactivation of gene promoters, leading to a polycomb‐mediated repression. H3K27me3 demethylases, mainly include KDM6A, KDM6B and KDM7A, specifically target trimethyl groups and overcome the repression of master transcription factors during development and tissue specification. KDM6A has been functionally linked to stem cell differentiation including osteogenic differentiation, cardiac differentiation, M2 macrophage differentiation, muscle differentiation and neuronal stem cell differentiation, etc.17, 18, 19 However, the effect of KDM6A on stem cell chondrogenic differentiation still remains unclear. Therefore, we sought to investigate if KDM6A has an impact on MSCs chondrogenesis in vitro. In this study, shRNA was performed to deplete KDM6A and chemical inhibitor of EZH2 was employed to reverse the effect of KDM6A depletion in PDLSCs. We found that depletion of KDM6A resulted in increasing H3K27me3 and decreasing expression of chondrogenic markers including SOX9, Col2a1 and ACAN. Meanwhile, EZH2 inhibitor led to decreasing H3K27me3 in PDLSCs‐KDM6Ash and rescued the decreased expression of chondrogenic markers, which suggested that KDM6A is required in chondrogenic differentiation by removal of H3K27me3 in target gene promoters.
The trithorax (trx) group proteins are primarily involved in gene activation, although some can also have repressive effects. Mixed‐lineage leukaemia (MLL) is the human homologue of trx. Several trx proteins are able to covalently modify histones, and these proteins are frequently found in complexes which are able to perform more than one type of modification reaction. KDM6A is a member of trx proteins family and is contained in trx‐like group complex in drosophila. Agger et al reported that a histone demethylase for H3K27 in a complex with a H3K4 methyltransferase can lead to the activation of a target gene.20 And it is reported that KDM6A could function as the histone demethylase for H3K27 in MLL3/4 complex which may employ 2 distinct histone‐modifying activities to synergistically activate target gene expression.21 Previous studies showed that during cellular differentiation in NT2/D1, the occupancy of UTX at HOX gene promoters was increased, paralleled with a decrease in trimethyl H3K27 and an increase in trimethylation of H3K4, leading to the activation of transcription.22 Li et al reported that knockdown of KDM6A decreased H3K4me2 and MLL4 (KMT2D) enrichments,23 and Kim had demonstrated that knockdown of KDM6A resulted in decreased recruitment of MLL4 and decreased H3K4me3 levels on the promoters of MMP‐9 et al.24 Our study showed that knockdown of KDM6A led to decreased levels of H3K4me3 and MLL4 in PDLSCs, which is consistent with previous report that KDM6A, interacting with MLL4, also influence H3K4 methylation.
Trx group complex containing KDM6A acts antagonistically to polycomb‐repressive complex 2 (PRC2) containing EZH2 in maintaining the dynamics of the activation and repression of gene expression through H3K27 methylation. EZH2 is the only histone methyltransferase for H3K27, which plays a central role in inflammation, autoimmunity and a wide range of malignancies, and manifests upregulated level during osteoarthritis (OA).25, 26, 27 Afterward, EPZ‐6438, an efficient and selective EZH2 inhibitor, controls important inflammatory gene targets and protects neonatal microglial activation, and EPZ‐6438 (Tazemetostat) is the first EZH2 inhibitor to enter human clinical trials as a single therapeutic agent in subjects with advanced NHL (https://clinicaltrials.gov/ct2/show/NCT02601950). Studies have demonstrated that EPZ‐6438 could be a useful therapeutic approach for the treatment of neuro‐inflammatory diseases associated with microglial activation.28 It was reported that EPZ‐6438 delayed tumour growth in a colorectal cancer xenograft model by inhibiting proliferation and clonogenicity of stem‐like cells.29 Consequently, our aim was to investigate whether inhibition of EZH2 with EPZ‐6438 could protect chondrogenic differentiation potential against knockdown of KDM6A in PDLSCs. The results suggested that EZH2 inhibitor could reversed the decreased level of H3K4me3 and increased level of H3K27me3 in PDLSCs after knockdown of KDM6A, and could rescue the decreased expression of chondrogenic differentiation markers, such as SOX9, Col2a1, ACAN and proteoglycans. In addition, we observed that SOX9, Col2a1 and aggrecan proteins in EPZ‐6438‐treated PDLSCs‐WT were neither decreased nor increased compared with PDLSCs‐WT. The same phenomenon was also reported by Arifuzzaman. In Arifuzzaman's study, EPZ‐6438 had no effect on normal primary microglial cells but could significantly downregulate the key inflammatory mediators released by LPS‐stimulated microglial cells.28 Based on the above observations, it is possible that H3K27me3 maintains a poised status in PDLSCs‐WT rather than the high expression status after knockdown of KDM6A; therefore, EPZ‐6438 had no impact on PDLSCs‐WT.
SOX9 was reported to be an activator in cartilage tissue‐specific gene expression during chondrogenic differentiation.30 For ESCs, to maintain self‐renewal, the promoter of various key developmental genes is marked by H3K27me3 catalysed by EZH2,31, 32 and these promoters often carry the activating H3K4 mark as well and are thus called “bivalent.”33, 34 At the end of ESC lineage commitment, these bivalent domains have been resolved to carry either activating (H3K4me3) or inactivating (H3K27me3) marks in a lineage‐specific manner.33 The expression of SOX9 in limb bud mesenchymal cells cultured in chondrogenic medium is high and its promoter is accordingly marked by increased levels of H3K4me3 and decreased levels of H3K27me3.35 Our results also showed that knockdown of KDM6A in PDLSCs led to increase of H3K27 methylation and decrease of H3K4 methylation in SOX9 promoter, and finally led to the reduced expression of chondrogenic differentiation markers after chondrogenic induction. Although we do not have the direct evidence to state the relationship between KDM6A and SOX9, Yoo has reported that a putative enhancer of the SOX9 gene is bound by KDM6A.36 KDM6A may have both direct and indirect effects on SOX9 transcription. H3K27me3 is present within the SOX9 promoter, and SOX9 and Col2a1 have previously been shown to contain H3K27me3 sites that undergo demethylation during chondrogenesis of human primary bone marrow‐derived MSCs.37 Based on our results, KDM6A may also bind to SOX9 promoter in PDLSCs, implementing H3K4 methylation and H3K27me3 demethylation, and finally results in SOX9 transcription. Besides, demethylating H3K27me3 at promoter sites of accessory factors such as the orphan nuclear receptor N2RF1, gene can also lead to increased SOX9 transcription via N2RF1 binding to the SOX9 promoter.38
In cartilage development, extracellular matrix protein Col2a1 and cartilage‐specific proteoglycan core protein ACAN are specific for cartilaginous tissues and necessary for the normal embryonic development of the skeleton and chondrocyte differentiation. The expression of Col2a1 has been shown to be dependent on SOX9. Accumulating evidences showed that combination of SOX9/5/6 could enhance Col2a1 transcription through binding to its enhancer region in intron 1 and intron 6.39 MicroRNAs (miRs), as small molecular regulators of gene expression, exert critical influence in post‐transcriptional regulation in stem cell function.40, 41 The miR‐29a was reported to be an important regulator of collagen expression, and it was revealed to directly target 3′ UTR of Col2a1 encoding type II collagen in mouse MSCs, finally promoting the mRNA expression of Col2a1 and ACAN.42, 43, 44, 45 MiR‐204 and miR‐211 are close homologues and have 2 nucleotides difference in human. Huang et al reported that SOX9 can promote the expression of miR‐204 and miR‐211, which act as attenuators of Runx2.46 Runx2 is identified as the key transcription factor regulating osteogenesis and chondrogenesis.47, 48, 49 These 2 miRNAs act as endogenous negative regulators, which inhibit the expression of Runx2 protein to restrain BMSCs from differentiating into osteoblasts.50 In this study, we investigated the expression of miR‐204 and miR‐211 to prove the involvement of miR‐204 and miR‐211 in PDLSCs during chondrogenic differentiation. During 3 weeks of chondrogenic differentiation, SOX9 kept increasing, while the transcription of Runx2 was increased at 3 days and subsequently decreased gradually. Relatively, miR‐29a was significantly decreased after chondrogenic culture, the miR‐204 and miR‐211 decreased at 1 week after chondrogenic differentiation culture, and increased at 2 and 3 weeks. These results were consistent with Huang's report. Our results also showed that miR‐204 and miR‐211 were decreased after knockdown of KDM6A; however, EPZ‐6438 could rescue the decrease.
Together, KDM6A was required in chondrogenic differentiation of PDLSCs by demethylation of H3K27me3, and EZH2 inhibitor could rescue chondrogenesis of PDLSCs after knockdown of KDM6A, which suggests that upregulation of KDM6A or utilize of EZH2 inhibitor might improve MSC‐mediated cartilage regeneration in chondrocytic deficient diseases such as OA. Future study should focus on the define mechanism by which KDM6A and its protein complexes regulate SOX9 transcription, thus promote chondrogenic differentiation of MSCs.
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
AUTHORS' CONTRIBUTIONS
Pingting Wang and Yanjing Li: collection and/or assembly of data and manuscript writing; Tingting Meng, Junjiang Zhang, Yuanyuan Wei and Zhaosong Meng: collection and/or assembly of data; Yunfeng Lin: data analysis and interpretation and manuscript writing; Dayong Liu and Lei Sui: conception and design, data analysis and interpretation, financial support and final approval of manuscript.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by grants from the Science Foundation of Tianjin Medical University (grant no. 2015KYZM11 to P.T.W), grant from The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (grant no. 2016YD19 to P.T.W), grant from the National Natural Science Foundation of China (81371109 and 81670953 to D.Y.L, 81700927 to Z.S.M), grant from Tianjin Research Program of Application Foundation and Advanced Technology (grant no. 15JCYBJC50200 to D.Y.L), grant from Beijing Key Laboratory of Tooth Regeneration and Function Reconstruction Open Project (grant no. 2014QYZS02 to D.Y.L), and grant from Tianjin Natural Science Foundation (grant no. 16JCZDJC32800 to L.S).
Wang P, Li Y, Meng T, et al. KDM6A promotes chondrogenic differentiation of periodontal ligament stem cells by demethylation of SOX9. Cell Prolif. 2018;51:e12413 10.1111/cpr.12413
Pingting Wang and Yanjing Li contribute equally to this work.
Contributor Information
Dayong Liu, Email: dyliuperio@tmu.edu.cn.
Lei Sui, Email: suilei@tmu.edu.cn.
REFERENCES
- 1. Mantesso ASB. Dental stem cells for tooth regeneration and repair. Expert Opin Biol Ther. 2009;9:1143‐1154. [DOI] [PubMed] [Google Scholar]
- 2. Yu S, Long J, Yu J, et al. Analysis of differentiation potentials and gene expression profiles of mesenchymal stem cells derived from periodontal ligament and Wharton's jelly of the umbilical cord. Cells Tissues Organs. 2013;197:209. [DOI] [PubMed] [Google Scholar]
- 3. Liu D, Wang Y, Jia Z, et al. Demethylation of IGFBP5 by histone demethylase KDM6B promotes mesenchymal stem cell‐mediated periodontal tissue regeneration by enhancing osteogenic differentiation and anti‐inflammation potentials. Stem Cells. 2015;33:2523. [DOI] [PubMed] [Google Scholar]
- 4. Fan Z, Yamaza T, Lee JS, et al. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol. 2009;11:1002‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ye L, Fan Z, Yu B, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao R, Zhang Y. The functions of E(Z)/EZH2‐mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155‐164. [DOI] [PubMed] [Google Scholar]
- 7. Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hemming S, Cakouros D, Isenmann S, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32:802‐815. [DOI] [PubMed] [Google Scholar]
- 9. Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149. [DOI] [PubMed] [Google Scholar]
- 10. Zhu S, Chen P, Yan W, et al. Programmed application of transforming growth factor β3 and Rac1 inhibitor NSC23766 committed hyaline cartilage differentiation of adipose‐derived stem cells for osteochondral defect repair. Stem Cells Transl Med. 2014;3:167‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Sun L, Wang W, Ma X. Quantitative analysis of fibrosis formation on the microcapsule surface with the use of Picro‐Sirius Red staining, polarized light microscopy, and digital image analysis. J Biomed Mater Res, Part A. 2006;76:120‐125. [DOI] [PubMed] [Google Scholar]
- 12. Lengner CJ, Lepper C, Wijnen AJV, Stein JL, Stein GS, Lian JB. Primary mouse embryonic fibroblasts: a model of mesenchymal cartilage formation. J Cell Physiol. 2004;200:327. [DOI] [PubMed] [Google Scholar]
- 13. Ding G, Liu Y, Wang W, et al. Allogeneic periodontal ligament stem cell therapy for periodontitis in swine. Stem Cells. 2010;28:1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu D, Xu J, Liu O, et al. Mesenchymal stem cells derived from inflamed periodontal ligaments exhibit impaired immunomodulation. J Clin Periodontol. 2012;39:1174. [DOI] [PubMed] [Google Scholar]
- 15. Moshaverinia A, Xu X, Chen C, Akiyama K, Snead ML, Shi S. Dental mesenchymal stem cells encapsulated in alginate hydrogel co‐delivery microencapsulation system for cartilage regeneration. Acta Biomater. 2013;9:9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838. [DOI] [PubMed] [Google Scholar]
- 17. Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3‐Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936. [DOI] [PubMed] [Google Scholar]
- 18. Seenundun S, Rampalli S, Liu QC, et al. UTX mediates demethylation of H3K27me3 at muscle‐specific genes during myogenesis. EMBO J. 2010;29:1401‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S, Lee JW, Lee SK. UTX, a histone H3‐lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731‐734. [DOI] [PubMed] [Google Scholar]
- 21. Cho YW, Hong S, Ge K. Affinity purification of MLL3/MLL4 histone H3K4 methyltransferase complex. Methods Mol Biol. 2012;809:465‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447. [DOI] [PubMed] [Google Scholar]
- 23. Lee JE, Wang C, Xu S, et al. H3K4 mono‐ and di‐methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705‐1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trenkmann M, Brock M, Gay RE, et al. Expression and function of EZH2 in synovial fibroblasts: epigenetic repression of the Wnt inhibitor SFRP1 in rheumatoid arthritis. Ann Rheum Dis. 2011;70:1482. [DOI] [PubMed] [Google Scholar]
- 26. Chen L, Wu Y, Wu Y, Wang Y, Sun L, Li F. The inhibition of EZH2 ameliorates osteoarthritis development through the Wnt/β‐catenin pathway. Sci Rep. 2016;6:29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aury‐Landas J, Bazille C, Allas L, et al. Anti‐inflammatory and chondroprotective effects of the S‐adenosylhomocysteine hydrolase inhibitor 3‐Deazaneplanocin A, in human articular chondrocytes. Sci Rep. 2017;7:6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arifuzzaman S, Das A, Kim SH, et al. Selective inhibition of EZH2 by a small molecule inhibitor regulates microglial gene expression essential for inflammation. Biochem Pharmacol. 2017;137:61‐80. [DOI] [PubMed] [Google Scholar]
- 29. Chen JF, Luo X, Xiang LS, et al. EZH2 promotes colorectal cancer stem‐like cell expansion by activating p21cip1‐Wnt/β‐catenin signaling. Oncotarget. 2016;7:41540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co‐activators CREB‐binding protein and p300 regulate chondrocyte‐specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224‐27229. [DOI] [PubMed] [Google Scholar]
- 31. Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349‐353. [DOI] [PubMed] [Google Scholar]
- 32. Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome‐wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123‐1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315‐326. [DOI] [PubMed] [Google Scholar]
- 34. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome‐wide maps of chromatin state in pluripotent and lineage‐committed cells. Nature. 2007;448:553‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar D, Lassar AB. Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Rep. 2014;8:1419‐1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoo KH, Oh S, Kang K, et al. Histone demethylase KDM6A controls the mammary luminal lineage through enzyme‐independent mechanisms. Mol Cell Biol. 2016;36:2108‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herlofsen SR, Bryne JC, Høiby T, et al. Genome‐wide map of quantified epigenetic changes during in vitro chondrogenic differentiation of primary human mesenchymal stem cells. BMC Genom. 2013;14:1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sosa MS, Parikh F, Maia AG, et al. NR2F1 controls tumour cell dormancy via SOX9‐ and RARbeta‐driven quiescence programmes. Nat Commun. 2015;6:6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yasuda H, Oh CD, Chen D, De CB, Kim JH. A novel regulatory mechanism of type II collagen expression via a SOX9‐DEPENDENT ENHANCER in Intron 6. J Biol Chem. 2016;292:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clark EA, Kalomoiris S, Nolta JA, Fierro FA. Concise review: microRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32:1074‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maurer B, Stanczyk J, Jüngel A, et al. MicroRNA‐29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733‐1743. [DOI] [PubMed] [Google Scholar]
- 43. Yan C, Wang Y, Shen XY, et al. MicroRNA regulation associated chondrogenesis of mouse MSCs grown on polyhydroxyalkanoates. Biomaterials. 2011;32:6435‐6444. [DOI] [PubMed] [Google Scholar]
- 44. Le LTT, Swingler TE, Natalie C, et al. The microRNA‐29 family in cartilage homeostasis and osteoarthritis. J Mol Med. 2016;94:583‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Zhen Z, Tang G, Zheng C, Yang G. MiR‐29a and MiR‐140 protect chondrocytes against the anti‐proliferation and cell matrix signaling changes by IL‐1β. Mol Cell. 2016;39:103‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang J, Zhao L. MicroRNA‐204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755‐764. [DOI] [PubMed] [Google Scholar]
- 48. Yoshida CA, Yamamoto H, Fujita T, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952‐963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jonason JH, Xiao G, Zhang M, Xing L, Chen D. Post‐translational regulation of Runx2 in bone and cartilage. J Dent Res. 2009;88:693‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA. 2011;108:9863‐9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


