Abstract
Objectives
Whole‐tooth regeneration for tooth loss has long been a goal of dentistry. There is also an increasing demand to carry out pre‐clinical in vitro and in vivo research methods in large animal model similar to human. The miniature pig has proven to be an alternative as a large mammal model owing to its many similarities to human. However, whole‐tooth regeneration in large animal remains a challenge. Here, we investigated the feasibility of cell re‐association‐based whole‐tooth regeneration in miniature pigs.
Materials and methods
Single cells from the forth deciduous molar germs (p4) of pig were reconstituted to bioengineered tooth bud using different treatment for in vitro culture and in vivo transplantation in mouse subrenal capsules and jawbones.
Results
The bioengineered tooth bud from re‐aggregated epithelial to mesenchymal single cells with and without compartmentalization restored the morphogenesis, interactions or self‐sorting between 2 cells in vitro culture. The pig bioengineered tooth bud transplanted in mouse subrenal capsules and jawbones restored odontogenesis and developed into large size tooth.
Conclusions
We characterized the morphogenesis and interaction of single‐tooth germ cells in vitro, and first addressed efficient long‐term survival and growth through transplantation of pig bioengineered tooth bud under mouse subrenal capsules or in mouse jawbones, where it can develop into large size tooth. Our study extends the feasibility of whole‐tooth regeneration in large animal.
1. INTRODUCTION
The ultimate goal of whole‐tooth regeneration for tooth loss is to provide living, functional and biocompatible tissue that is more in line with the human desire for a third dentition that represents an attractive alternative to classical prosthesis‐based therapies.1, 2 Currently, de novo odontogenesis in humans has been challenging, with many obstacles, although some regenerative attempts have been made using cells from human donors.3, 4, 5, 6, 7 The epithelial and mesenchymal interaction‐based whole‐tooth regenerative approaches in rodents and canine model, hold great promise as a strategy for developing a functional substitute for lost teeth.8, 9, 10, 11, 12, 13 Whether de novo odontogenesisis feasible in humans remains elusive. An essential step is required to move this tooth regeneration strategy from rodents to a large animal model before these regenerative properties are introduced into humans.
Pigs serve as a promising large animal model for studying human diseases and contribute to overcome the shortage of human donor organs.14, 15, 16, 17 The miniature pig has proven to be a valuable animal model for diphyodont development and regeneration owing to its many similarities to human including the morphology, number and size of teeth, particularly its heterodont dentition (incisors, canines, premolars and molars) and diphyodont dentition, which are not available in rodents.18, 19 The morphology and chronology of diphyodont dentition in miniature pigs have been well characterized by our previous studies and other reports.20, 21, 22 Moreover, recent breakthrough in porcine genome engineering aiming to overcome immunological challenges and potential risk of porcine endogenous retrovirus (PERV) transmission make safe clinical xenotransplantation possible.14, 17, 23 However, studying whole‐tooth regeneration using miniature pig as a model remains a significant obstacle. The morphogenesis and interaction of single cells from pig tooth germ in vitro culture remain undefined. Some issues also need to be overcome, such as longer process required for growth and replacement of swine teeth with larger size and dynamic tracking. There is an increasing demand to seek alternative approaches to promote pre‐clinical study.
As in vitro organ culture only provides short term growth and limited functional cytodifferentiation, transplantation of graft under the renal capsule is used for study of development and differentiation of tissue recombinants owing to its high degree of vascularity, suitability for xenografts and convenient examination of organogenesis. However, whether the subrenal capsule microenvironment can bear long‐term growth of pig bioengineered tooth germ remains unknown.
In this study, we traced the morphogenesis, interactions or self‐sorting of cells from pig tooth germs. The pig bioengineered tooth bud achieved long‐term survival and growth, and developed a tooth through transplantation in mouse subrenal capsules and jawbones. Our pilot study for whole‐tooth regeneration in large animal has the potential to be clinically applied and will further promote the use of pig as a diphyodont model similar to humans.
2. MATERIALS AND METHODS
2.1. Animals
Ten pregnant miniature pigs were obtained from the Institute of Animal Science of the Chinese Agriculture University (Beijing, China). The miniature pig embryos (85) were obtained as reported previously.20 Briefly, the pregnant miniature pigs were verified by B‐type ultrasonic inspection, and the staged miniature pig embryos were obtained by caesarean section.
The adult host immunocompromised (SCID) mice (5 week old) were obtained from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences, and maintained in a specific pathogen‐free animal facility with free access to water and food.
All experimental animal procedures were reviewed and approved by the Animal Care and Use Committee of Capital Medical University. (Permit Number: CMU‐2012‐x‐102), and the methods were carried out in accordance with the approved guidelines.
2.2. Isolation of tooth germs from miniature pigs
The forth deciduous molar germs (p4) in mandibles from the same litter of staged miniature pig embryos (embryonic day [E] E40 or E70) were isolated and pooled under stereo microscopy with an attached Olympus DP72 digital camera system (Olympus Corporation, Tokyo, Japan). The morphological stages of the p4 at E40 or E70 corresponded to the cap stage and secretory stage, respectively, and were verified by serial histological sections as previously described.20
2.3. Dissociation of single‐tooth germ cells
Single‐tooth germ cells from miniature pigs were obtained as previously reported.10, 11 Briefly, the epithelium and mesenchyme of isolated lower deciduous molar germs were incubated in PBS containing Dispase II (1.2 U/mL, Sigma‐Aldrich, St. Louis, MO, USA) and DNase I (20 U/mL, Takara Bio, Shiga, Japan) for 15 min at room temperature, then separated under a stereo microscope. The epithelium and mesenchyme were each dissociated into single cells in PBS (−) supplemented with Collagenase type I (3 mg/mL, Sigma, Worthington Biochemical Corp., Lakewood, NJ, USA) and Dispase II (4 mg/mL, Sigma, Roche Diagnostics Corp., Indianapolis, IN, USA) and filtered through a 70‐μm cell strainer (BD Biosciences, San Jose, CA, USA).
2.4. Reconstitution of single‐tooth germ cells and tissue culture in vitro
Single cells of epithelial and mesenchymal origin were then, respectively, pelleted by centrifugation. To characterize the interaction between epithelial and mesenchymal single cells and test whole‐tooth regeneration potential of single‐tooth germ cells, the single‐tooth germ cells were processed using three strategies for bioengineered tooth germs. For re‐association, single cells of epithelial and mesenchymal origin were pelleted separately by centrifugation. Bioengineered tooth germs were constructed by the sequential injection of 0.25 μL mesenchymal and 0.25 μL epithelial single cells pellets into a 50 μL collagen gel drop (Cellmatrix type I‐A, Nitta gelatin, Osaka, Japan) (1:1 ratio, each pellet including approximately 1 × 105 cells) to form compartment contact between the two pellet types.10, 11 For mesenchymal cells alone, 0.5 μL mesenchymal single cells pellet (approximately 2 × 105 cells) were injected into a 50 μL collagen gel drop. For re‐aggregation of tooth germ cells, the tooth germs were directly disassociated into mixed single‐tooth germ cells and were filtered and pelleted, then 0.5 μL cells pellet (approximately 2 × 105 cells) were injected into a 50 μL collagen gel drop without compartmentalization.
The above‐reconstituted pellets embedded into collagen gel drops were placed on a cell culture insert (0.4‐μm pore size, BD, Franklin Lakes, NJ, USA) in 12‐well cell culture plates. The explants were cultured for 3‐8 days in DMEM medium (GIBCO, Grand Island, NY, USA) supplemented with 10% FCS (GIBCO), 100 μg/mL ascorbic acid (Sigma), 2 mM L‐glutamine (Sigma).
2.5. Transplantation of reconstituted explants into SCID mice
The bioengineered tooth germs cultured in vitro for 3 days were transplanted into subrenal capsules or maxillary diastema of adult SCID mice. For subrenal capsule transplantion, a small incision was made with scissors in the skin and body wall over the kidney after SCID mice was anaesthetized. Then the kidney was popped out of the hole in the body wall, an incision was made to create a pocket between the capsule and the underlying renal parenchyma. The bioengineered tooth germ was placed into the pocket. The incision was sutured after return of the kidneys to the peritoneal cavity. For jawbone transplantation, an incision of 2 mm in length was made through the oral mucosa of maxillary diastema and a 0.5‐1.0 mm bony hole was made in alveolar bone surface. The bioengineered tooth germs were then transplanted into the bony hole. The incision was next sutured. The host mice were killed after 8 or 16 weeks, and perfusion fixed with 4% neutral paraformaldehyde.
2.6. Radiographic analysis and three‐dimensional reconstruction
The specimens were examined with cone beam computed tomography (CBCT, NewTom 3G tomography, Summer, Italy). The data analysis and three‐dimensional reconstruction were carried out with Mimics (Materialise, Version 11).
2.7. Histochemical and immunohistochemical analysis
For the histology study, specimens were decalcified, dehydrated, and then embedded in paraffin. Serial sections (5 μm thick) were stained with haematoxylin‐eosin (H&E).
For immunohistochemistry of regenerated tooth, briefly, after the samples were fixed with 4% PFA, decalcified, dehydrated and embedded in paraffin, they were cut into 10‐μm thick sections. Serial sections were permeabilized in 0.4% Triton X‐100 and blocked in PBS containing 5% BSA. Sections were incubated with primary antibodies overnight at 4°C, then washed and incubated for 1 h at 37°C with the respective secondary antibodies followed by haematoxylin or DAPI nuclear stain. Slices were analysed using a microscope (BX43 Olympus) with an attached Olympus DP72 digital camera system.
3. RESULTS
3.1. Isolation of tooth germs from miniature pigs
According to our previous research,20, 24 the staged miniature pig embryos and foetuses at E40 and E70 were obtained by caesarean section. Based on considerations of cellular acquisition, we choose the forth deciduous molar germs (p4) in mandibles with large size. The p4 was at cap stage at E40 (Figure 1A‐E) and reached secretory stage at E70 (Figure S1A‐C). The morphogenetic stages were verified by serial histological sections.
Figure 1.
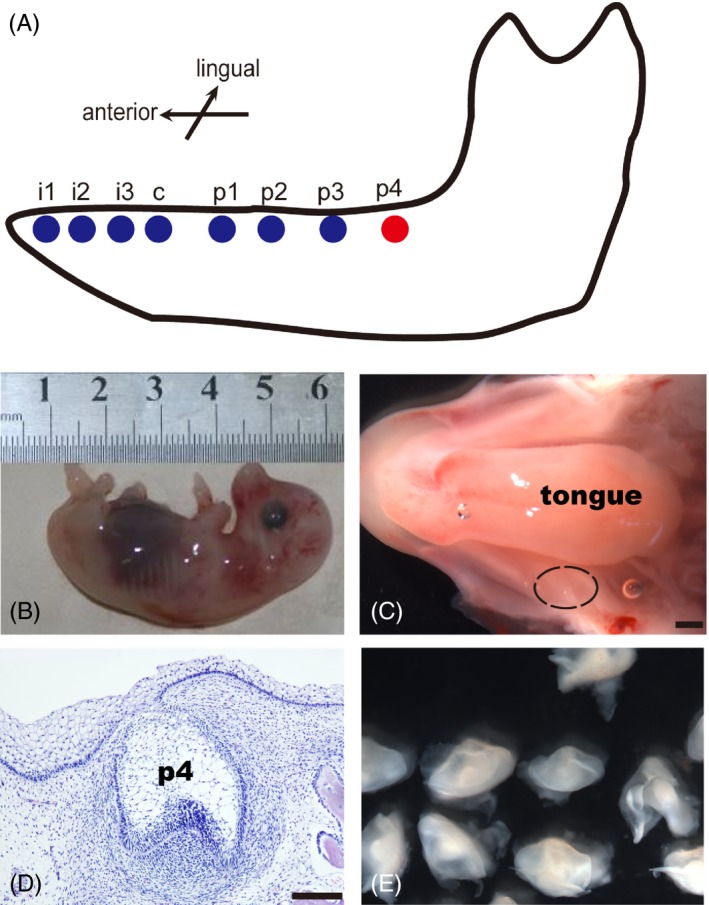
Isolation of deciduous molar germs from miniature pigs at E40. (A), Schematic buccal view of the mandible of miniature pig at E40. (B), The isolated miniature pig embryos at E40. (C), Macro view of mandible from miniature pig at E40. Black dotted line indicated the location of p4, scale bar = 200 μm. (D), Frontal sections showing p4 at cap stage (H&E), scale bar = 200 μm. (E), The isolated p4 at cap stage
3.2. Reconstructed single‐tooth germ cells from cap stage in vitro culture
To characterize the odontogenesis of dissociated single cells from tooth germs of miniature pigs at cap stage, we treated single cells from tooth germs with three methods for in vitro organ culture (Figure 2A). We isolated the p4 at cap stage from mandibles from the same litter of staged miniature pig embryos and separated the epithelium from the mesenchyme. After sequential enzymatic treatment, the epithelial and mesenchymal tissues were dissociated into single cells.
Figure 2.
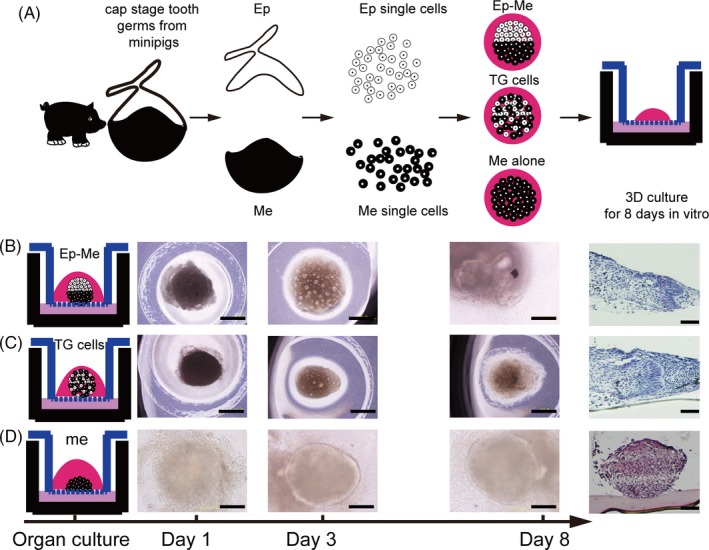
Bioengineered tooth germs from single‐tooth germ cells in vitro culture. (A), Schematic representations of treatment of tooth germ cells for in vitro organ culture. The epithelium and mesenchyme of tooth germs were dissociated to single cells. The single cells were pelleted to reconstruct tooth germs with 3 ways and cultured in vitro for 8 d. (B), Ep‐Me in vitro culture and histological analysis (H&E staining, right panels), bioengineered tooth germs from re‐associated epithelial and mesenchymal single cells with compartment. (C), TG cells in vitro culture and histological analysis (H&E staining, right panels), bioengineered tooth germs from mixed tooth germ single cells. (D), Me in vitro culture and histological analysis (H&E staining, right panels), bioengineered tooth germs from re‐aggregated mesenchymal cells alone. Scale bar = 200 μm
We tracked the in vitro morphogenesis of re‐associated cell pellets with epithelial cell and mesenchymal cell compartment (Ep‐Me) in our organ culture system. Interactions were visible after culture for 3 days, and apparent morphogenesis arose at the 8th day, implying the initiation of odontogenesis identified by histological sections, and polarized epithelial cells were separated from concentrated mesenchymal cells by basement membrane (Figure 2B). Re‐aggregated epithelial and mesenchymal single cells mixed from tooth germs without compartmentalization (TG cells) exhibited a self‐sorting feature; the epithelial cells were able to sort from the mesenchymal cells and organized into a well‐defined dental epithelial structure enveloping a mesenchymal cell mass (Figure 2C). In contrast, the aggregated mesenchymal cells alone (Me alone) exhibited no obvious morphogenesis (Figure 2D).
We further characterized the reciprocal interaction between the epithelial and mesenchymal cells by re‐associating epithelial cells with mesenchymal cells marked with enhanced green fluorescent protein (eGFP) to identify the morphogenesis of the epithelial compartment (Figure S2A,B).
3.3. Reconstructed single‐tooth germ cells from secretory stage in vitro culture
We also tested the de novo odontogenesis of cells from tooth germ at the secretory stage (E70). Ep‐Me cells pellets from p4 at E70 also restored morphogenesis like Ep‐Me cells pellets from p4 at cap stage (Figure 3A). The morphogenesis and interaction between epithelial cells (marked with anti‐CK14) and mesenchymal cells (marked with anti‐vimentin) was visible after culture for 8 days (Figure 3B).
Figure 3.
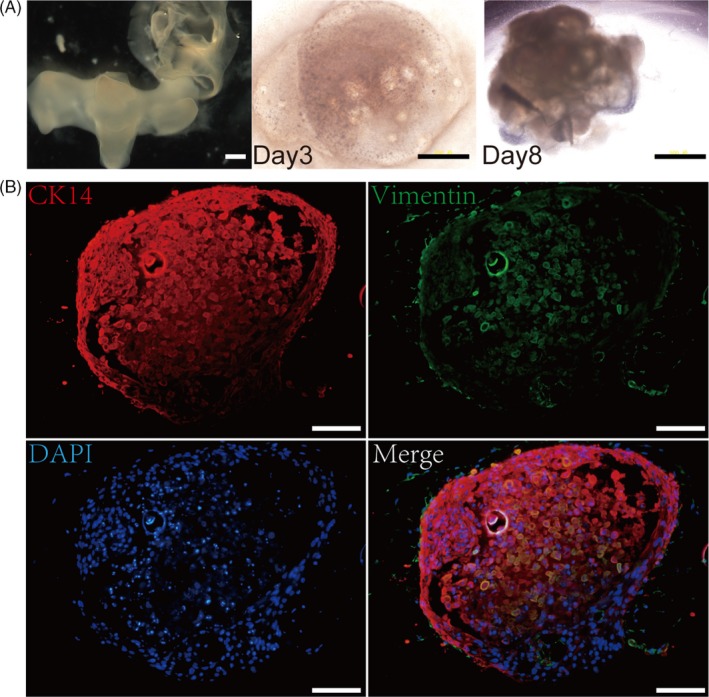
In vitro development of re‐aggregated Ep‐Me cells from p4 at secretory stage. (A), The dissociated epithelium and mesenchyme of p4 (E70, left), Ep‐Me bioengineered tooth germ in vitro culture at 3rd day (middle) and 8th day (right), Scale bar = 500 μm. (B), Morphogenesis and interaction between epithelial cells (red) and mesenchymal cells (green), Scale bar = 100 μm
3.4. Growth and survival of bioengineered tooth germs in subrenal capsules of SCID mice
To investigate the regeneration potential of the constructed tooth germs described above, the bioengineered explants were transplanted into a SCID mouse subrenal capsule after culturing for 3 days in vitro (Figure 4A). The re‐associated explants from cells of tooth germ at cap stage with epithelial and mesenchymal cell compartmentalization in subrenal capsule restored the de novo odontogenesis, as demonstrated by examination of explants at 8 weeks after implantation, the differentiated ameloblasts expressed ameloblastin, and visible Hertwig epithelial root sheaths implied the initiation of root development (5 in 6 grafts, Figure 4B), and developed into tooth‐like structures with crown nearly completed in bony tissues after transplantation for 16 weeks (4 in 6 grafts, Figure 4C). It is noteworthy that no secondary tooth was visible in all bioengineered tooth germ grafts. All 4 re‐aggregation grafts from mixed epithelial and mesenchymal single cells from tooth germ at cap stage without compartmentalization in our experiment were able to form tooth‐like tissues with correctly arranged tooth components, but without normal tooth appearance at 8 weeks after transplantation (Figure 4D). While, re‐aggregation of mesenchymal cells alone from cells of tooth germ at cap stage (Me cells) only formed bone tissue at 8 weeks after transplantation (Figure 4E).
Figure 4.
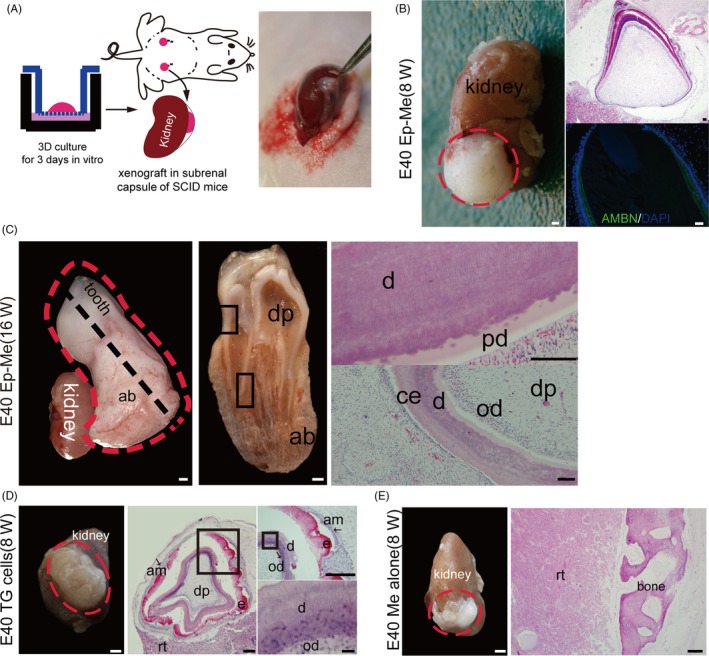
In vivo development of re‐aggregated Ep‐Me cells from tooth germs at cap stage in mouse subrenal capsule. (A), Schematic diagram of the ectopic transplantation of bioengineered tooth germs into a mouse subrenal capsule after culturing for 3 d in vitro. (B), The re‐associated explants with epithelial and mesenchymal cell compartmentalization (E40 Ep‐Me) restore the de novo odontogenesis at 8 wk post‐transplantation. Left panel: macro view of regenerated tissues (red dotted line). Right panels: histological analysis (H&E staining) and immunostaining of the ameloblast‐specific marker ameloblastin (AMBN, green). (C), The re‐associated explants with epithelial and mesenchymal cell compartmentalization (E40 Ep‐Me) developed into a tooth after transplantation for 16 wk in the subrenal capsule, Left panel: macro view. Red dotted line indicates the tooth and supporting tissues. Middle panel: cross view of profile of tooth and supporting tissues at the black dotted cut line in the left panel. Right panels: histological analysis (H&E stain) corresponding to boxed areas in middle panel. (D), Re‐aggregation of mixed tooth epithelial and mesenchymal cells (TG cells) regenerated tooth crown structures by self‐sorting of epithelial and mesenchymal cells at 8 wk post‐transplantation. Left panel: macro view of regenerated tissues (red dotted line). Middle and right panels: histological analysis (H&E stain). (E), Re‐aggregation of mesenchymal cells alone (Me cells) formed bone tissue at 8 wk post‐transplantation. ab, alveolar bone; am, ameloblast; b, bone; d, dentin; dp, dental pulp; e, enamel; od, odontoblast; rt, renal tissue. Scale bars: gross view, 1 mm; histology, 100 μm
Then, we tested the regeneration potential of bioengineered explants from cells of tooth germ at secretory stage. The most of re‐associated explants from cells of tooth germ at E70 with epithelial and mesenchymal cell compartmentalization also restored the de novo odontogenesis in subrenal capsule at 8 weeks after implantation (4/5, Figure 5A) and formed tooth‐like tissues at 16 weeks after implantation (3/5, Figure 5B).
Figure 5.
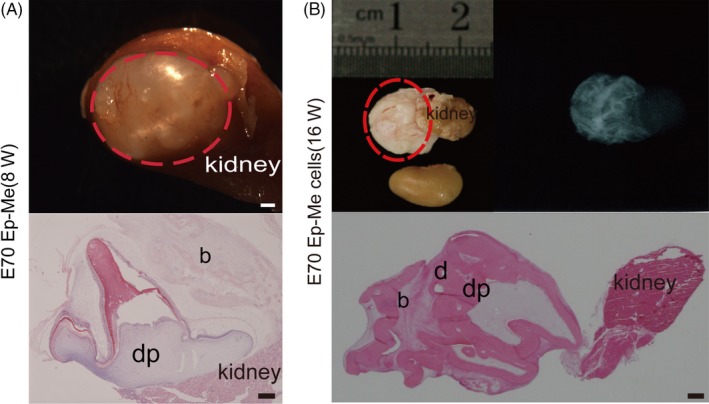
In vivo development of re‐aggregated Ep‐Me cells from p4 at secretory stage in mouse subrenal capsule. (A), The re‐associated explants with epithelial and mesenchymal cell compartmentalization (E70 Ep‐Me) restore the de novo odontogenesis at 8 wk post‐transplantation. Top panel: macro view of regenerated tissues (red dotted line). Bottom panels: histological analysis (H&E staining). (B), The re‐associated explants with epithelial and mesenchymal cell compartmentalization (E70 Ep‐Me) developed into tooth‐like structures (red dotted line) after transplantation for 16 wk in the subrenal capsule, viewed by X‐rays and H&E staining. b, bone; d, dentin; dp, dental pulp. Scale bars: gross view, 1 mm; histology, 100 μm
3.5. Growth of re‐associated explants in jawbone of SCID mouse
A prior study has verified that a bioengineered tooth germ can develop into a fully functioning tooth in mouse jawbone.11 Next, we investigated whether mouse jawbone is also suitable for growing a re‐associated bioengineered tooth germ from miniature pig. We transplanted the re‐associated explants from single‐tooth germ cells at cap stage from miniature pigs into the maxillary diastema in 5 SCID mice (Figure 6A). At 8 weeks post‐transplantation, we found a large bulge in maxillary diastema in 4 mice, which was illustrated with cone beam computed tomography (CBCT) (Figure 6B,C). Histological analysis (haematoxylin/eosin) revealed regenerated tooth tissue with normally arranged structure and tissue elements, including odontoblasts, dentin, dentinal tubules, dental pulp, root analogue, cementum, blood vessels and alveolar bone (Figure 6D,E).
Figure 6.
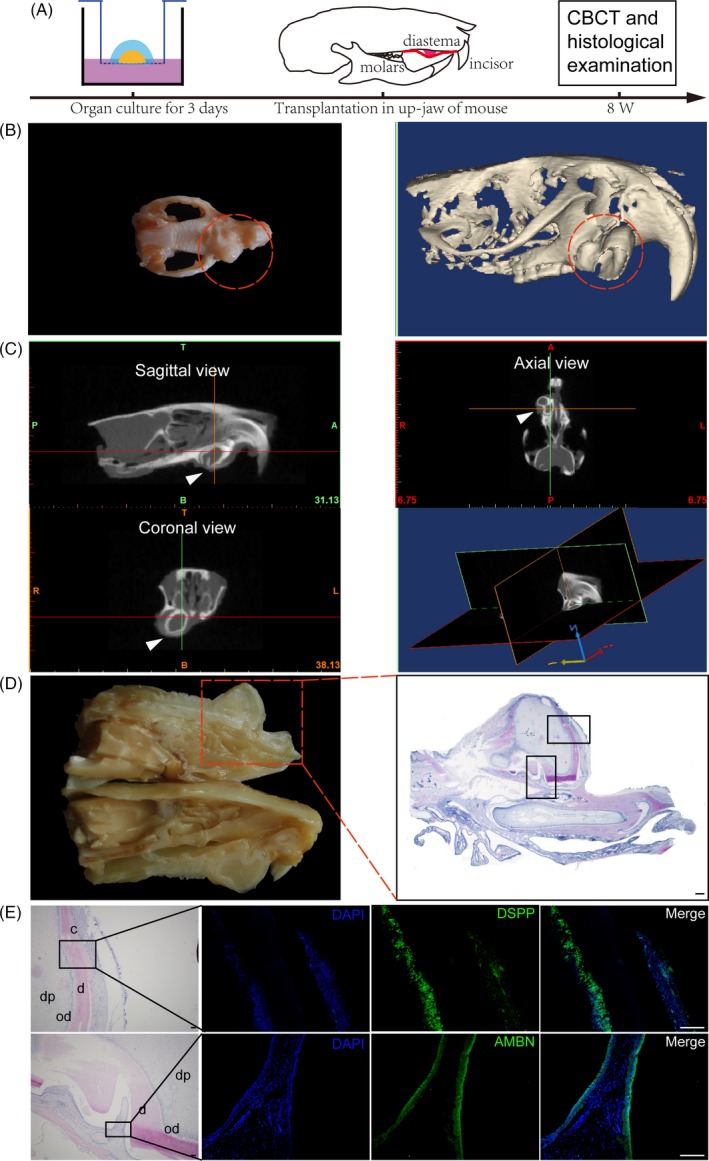
Orthotopic regeneration of re‐associated explants in the maxilla of SCID mice. (A), Schematic diagram of experimental design. (B), Macro view and 3D reconstruction of a regenerated tooth (red dotted line) in murine maxilla at 8 wk post‐transplantation. (C), Coronal, sagittal and axial views of regenerated tooth with CBCT. A and P, anterior and posterior; T and B, top and bottom; R and L, right and left. (D), Cross view of profile in (B) and histological sections. (E), Histological sections of the boxed areas in (D) and immunostaining of the amnioblast‐specific marker ameloblastin (AMBN, green), the odontoblast‐specific marker dentin sialophosphoprotein (DSPP, green) and DAPI (blue), showing the regenerated tooth‐like structure. c, cementum; d, dentin; dp, dental pulp; Od, odontoblast. Scale bars: 100 μm
4. DISCUSSION
The restoration of lost teeth through whole‐tooth regeneration in humans is the ultimate goal.3, 11, 12, 25, 26, 27, 28, 29 The successes of de novo odontogenesis in mice provide an attractive alternative to the future manipulation of cell‐based whole‐tooth regeneration in humans for the recovery of tooth function.10, 11, 30, 31 Despite some significant advances, the cell‐based recombination approach to clinical whole‐tooth regeneration remains a challenge, including larger tooth size, longer developmental time and so on.32 It is essential for pre‐clinical study in large animal model similar to human for tooth regeneration. Compared with mice, Pigs (Sus scrofa) serve as a promising large animal model for studying human diseases and pre‐clinical therapies owing to its comparability with humans in many respects.17, 33 Previous studies have proven that miniature pigs can serve as an attractive alternative to rodents for understanding tooth development and replacement.18, 20, 21, 34 Studying tooth regeneration in miniature pigs can contribute to understanding processes of human tooth development that are applicable to regenerative medicine.
The miniature pig's deciduous molars are similar to humans’; both are larger and require a longer developmental period than those of mice. Our previous results showed that approximately 3 months were required to grow p4 from the cap stage at E40 to eruption with nearly completed roots at postnatal day 20 (P20).20 It is critical to develop an environment using alternative method where can contribute to long‐term observation for large size tooth growth. Currently, ectopic tooth regeneration is predominant. The omentum, anterior eye chamber, subcutaneous tissues, jawbone and kidney capsule of rodents have traditionally been used for the long‐term culture of tooth germ grafts.35, 36, 37 The subrenal capsule graft is suitable for tissue recombinants which permits dynamic examination of development from morphogenesis to functional differentiation and overcomes the inaccessible restrictions in pig embryos. Our results suggested that the subrenal capsules of mice are sufficient to sustain the growth and survival of bioengineered tooth germs.
Previous study has suggested that a fully functioning tooth can be achieved by transplantation of a bioengineered tooth germ into the alveolar bone in an adult mouse.11 Our results also indicate that the orthotopic transplantation of bioengineered tooth bud from large animals into the jawbone in SCID mouse is also feasible. However, we did not obtain functioning tooth at 16 weeks post‐transplantation in jawbone, due to the large size and long growth stage of regenerated tissue which increase the potential for damage in the mice.
Previous study in mice suggested that the odontogenesis potential of single cells from tooth germ after cap stage reduced and develops into teeth at a lower frequency.12 Our results suggested that cells from pig late‐stage tooth germ seemed to still retain better odontogenesis potential than it in mouse, more work needs to be done for studying the differences in cell properties from both animal tooth germs. In 3D cell culture, the pellet from mixed epithelial and mesenchymal cells can form highly ordered structure by selective aggregation of cells (homing) or by rearrangement of the relative positions of cells within the structure (self‐sorting), suggesting the self‐sorting and homing ability of two cell types.38, 39, 40
We used deciduous tooth germ cells from the miniature pig for ectopic and orthotopic whole‐tooth regeneration, suggesting the feasibility of whole‐tooth regeneration in a large animal model. Our pilot study to manipulate swine tooth germ cells in vitro and in vivo offer an alternative model and a reference for whole‐tooth regeneration with large size and long growth stage in large animals.
CONFLICT OF INTEREST
The authors declare no competing financial interests or conflicts in relation to the work described.
AUTHOR CONTRIBUTIONS
FW and ZW performed studies and interpreted data. ZF and TW participated in in vivo experiments and performed analysis. JW and CZ assisted with in vitro experiments. SW designed study, analysed and interpreted data, obtained funding. SW and FW drafted manuscript. All authors reviewed and approved manuscript.
Supporting information
ACKNOWLEDGEMENTS
We thank J. Xiao and J. He for their helpful suggestions. This study was supported by grants from Beijing Municipality Government grants (Beijing Scholar Program PXM2016_014226_000034, PXM2016_014226_000006, PXM2015_014226_000116, PXM2015_014226_000055, PXM2015_014226_000052, PXM2014_014226_000048, PXM2014_014226_000013, PXM2014_014226_000053, Z121100005212004, PXM2013_014226_000055, PXM2013_014226_000021, PXM2013_014226_07_000080 and TJSHG201310025005); the National Natural Science Foundation of China (91649124 to S. W.); the National Natural Science Foundation of China (81771032 to F. W.).
Wang F, Wu Z, Fan Z, et al. The cell re‐association‐based whole‐tooth regeneration strategies in large animal, Sus scrofa . Cell Prolif. 2018;51:e12479 10.1111/cpr.12479
Fu Wang and Zhifang Wu contributed equally to this work.
REFERENCES
- 1. Mitsiadis TA, Harada H. Regenerated teeth: the future of tooth replacement. An update. Regen Med. 2015;10:5‐8. [DOI] [PubMed] [Google Scholar]
- 2. Smith EE, Yelick PC. Progress in bioengineered whole tooth research: from bench to dental patient chair. Curr Oral Health Rep. 2016;3:302‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angelova Volponi A, Kawasaki M, Sharpe PT. Adult human gingival epithelial cells as a source for whole‐tooth bioengineering. J Dent Res. 2013;92:329‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang W, Ahluwalia IP, Yelick PC. Three dimensional dental epithelial‐mesenchymal constructs of predetermined size and shape for tooth regeneration. Biomaterials. 2010;31:7995‐8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang B, Li L, Du S, et al. Induction of human keratinocytes into enamel‐secreting ameloblasts. Dev Biol. 2010;344:795‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snead ML. Whole‐tooth regeneration: it takes a village of scientists, clinicians, and patients. J Dent Educ. 2008;72:903‐911. [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Chen Y. Bioengineering of a human whole tooth: progress and challenge. Cell Regen (Lond). 2014;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu B, Nadiri A, Kuchler‐Bopp S, Perrin‐Schmitt F, Peters H, Lesot H. Tissue engineering of tooth crown, root, and periodontium. Tissue Eng. 2006;12:2069‐2075. [DOI] [PubMed] [Google Scholar]
- 9. Hu B, Nadiri A, Bopp‐Kuchler S, Perrin‐Schmitt F, Lesot H. Dental epithelial histomorphogenesis in vitro. J Dent Res. 2005;84:521‐525. [DOI] [PubMed] [Google Scholar]
- 10. Hu B, Nadiri A, Bopp‐Kuchler S, Perrin‐Schmitt F, Wang S, Lesot H. Dental epithelial histo‐morphogenesis in the mouse: positional information versus cell history. Arch Oral Biol. 2005;50:131‐136. [DOI] [PubMed] [Google Scholar]
- 11. Ikeda E, Morita R, Nakao K, et al. Fully functional bioengineered tooth replacement as an organ replacement therapy. Proc Natl Acad Sci USA. 2009;106:13475‐13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakao K, Morita R, Saji Y, et al. The development of a bioengineered organ germ method. Nat Methods. 2007;4:227‐230. [DOI] [PubMed] [Google Scholar]
- 13. Ono M, Oshima M, Ogawa M, et al. Practical whole‐tooth restoration utilizing autologous bioengineered tooth germ transplantation in a postnatal canine model. Sci Rep. 2017;7:44522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu J, Platero‐Luengo A, Sakurai M, et al. Interspecies chimerism with mammalian pluripotent stem cells. Cell. 2017;168(473–486):e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kemter E, Wolf E. Pigs pave a way to de novo formation of functional human kidneys. Proc Natl Acad Sci USA. 2015;112:12905‐12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan S, Tu Z, Liu Z, et al. A Huntingtin knockin pig model recapitulates features of selective neurodegeneration in huntington's disease. Cell. 2018. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ekser B, Ezzelarab M, Hara H, et al. Clinical xenotransplantation: the next medical revolution? Lancet. 2012;379:672‐683. [DOI] [PubMed] [Google Scholar]
- 18. Wang S, Liu Y, Fang D, Shi S. The miniature pig: a useful large animal model for dental and orofacial research. Oral Dis. 2007;13:530‐537. [DOI] [PubMed] [Google Scholar]
- 19. Stembirek J, Kyllar M, Putnova I, Stehlik L, Buchtova M. The pig as an experimental model for clinical craniofacial research. Lab Anim. 2012;46:269‐279. [DOI] [PubMed] [Google Scholar]
- 20. Wang F, Xiao J, Cong W, et al. Morphology and chronology of diphyodont dentition in miniature pigs, Sus scrofa . Oral Dis. 2014;20:367‐379. [DOI] [PubMed] [Google Scholar]
- 21. Stembirek J, Buchtova M, Kral T, Matalova E, Lozanoff S, Misek I. Early morphogenesis of heterodont dentition in minipigs. Eur J Oral Sci. 2010;118:547‐558. [DOI] [PubMed] [Google Scholar]
- 22. Putnova I, Odehnalova S, Horak V, et al. Comparative morphology of normal and cleft minipigs demonstrates dual origin of incisors. Arch Oral Biol. 2011;56:1624‐1634. [DOI] [PubMed] [Google Scholar]
- 23. Niu D, Wei HJ, Lin L, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR‐Cas9. Science. 2017;357:1303‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang F, Xiao J, Cong W, et al. Stage‐specific differential gene expression profiling and functional network analysis during morphogenesis of diphyodont dentition in miniature pigs, Sus scrofa . BMC Genom. 2014;15:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakahara T, Idei Y. Tooth regeneration: implications for the use of bioengineered organs in first‐wave organ replacement. Hum Cell. 2007;20:63‐70. [DOI] [PubMed] [Google Scholar]
- 26. Kuo TF, Lin HC, Yang KC, et al. Bone marrow combined with dental bud cells promotes tooth regeneration in miniature pig model. Artif Organs. 2011;35:113‐121. [DOI] [PubMed] [Google Scholar]
- 27. Arakaki M, Ishikawa M, Nakamura T, et al. Role of epithelial‐stem cell interactions during dental cell differentiation. J Biol Chem. 2012;287:10590‐10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dimmeler S, Ding S, Rando TA, Trounson A. Translational strategies and challenges in regenerative medicine. Nat Med. 2014;20:814‐821. [DOI] [PubMed] [Google Scholar]
- 29. Park YJ, Cha S, Park YS. Regenerative applications using tooth derived stem cells in other than tooth regeneration: a literature review. Stem Cells Int. 2016;2016:9305986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oshima M, Mizuno M, Imamura A, et al. Functional tooth regeneration using a bioengineered tooth unit as a mature organ replacement regenerative therapy. PLoS ONE. 2011;6:e21531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang YD, Chen Z, Song YQ, Liu C, Chen YP. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 2005;15:301‐316. [DOI] [PubMed] [Google Scholar]
- 32. Tucker A, Sharpe P. The cutting‐edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499‐508. [DOI] [PubMed] [Google Scholar]
- 33. Groenen MA, Archibald AL, Uenishi H, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buchtova M, Stembirek J, Glocova K, Matalova E, Tucker AS. Early regression of the dental lamina underlies the development of diphyodont dentitions. J Dent Res. 2012;91:491‐498. [DOI] [PubMed] [Google Scholar]
- 35. Young CS, Terada S, Vacanti JP, Honda M, Bartlett JD, Yelick PC. Tissue engineering of complex tooth structures on biodegradable polymer scaffolds. J Dent Res. 2002;81:695‐700. [DOI] [PubMed] [Google Scholar]
- 36. Duailibi MT, Duailibi SE, Young CS, Bartlett JD, Vacanti JP, Yelick PC. Bioengineered teeth from cultured rat tooth bud cells. J Dent Res. 2004;83:523‐528. [DOI] [PubMed] [Google Scholar]
- 37. Cunha GR, Baskin L. Use of sub‐renal capsule transplantation in developmental biology. Differentiation. 2016;91:4‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitsiadis TA, Graf D. Cell fate determination during tooth development and regeneration. Birth Defects Res C Embryo Today. 2009;87:199‐211. [DOI] [PubMed] [Google Scholar]
- 39. Balic A, Thesleff I. Tissue interactions regulating tooth development and renewal. Curr Top Dev Biol. 2015;115:157‐186. [DOI] [PubMed] [Google Scholar]
- 40. Song Y, Zhang Z, Yu X, et al. Application of lentivirus‐mediated RNAi in studying gene function in mammalian tooth development. Dev Dyn. 2006;235:1334‐1344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


