Abstract
Background: Neo-adjuvant chemotherapy is an effective strategy for improving treatment of breast cancers. However, the efficacy of this treatment strategy is limited for treatment of triple negative breast cancer (TNBC). Gene therapy may be a more effective strategy for improving the prognosis of TNBC.
Methods: A novel 25 nucleotide sense strand of miRNA was designed to treat TNBC by silencing the Slug gene, and encapsulated into DSPE-PEG2000-tLyp-1 peptide-modified functional liposomes. The efficacy of miRNA liposomes was evaluated on invasive TNBC cells and TNBC cancer-bearing nude mice. Furthermore, functional vinorelbine liposomes were constructed to investigate the anticancer effects of combined treatment.
Results: The functional miRNA liposomes had a round shape and were nanosized (120 nm). Functional miRNA liposomes were effectively captured by TNBC cells in vitro and were target to mitochondria. Treatment with functional liposomes silenced the expression of Slug and Slug protein, inhibited the TGF-β1/Smad pathway, and inhibited invasiveness and growth of TNBC cells. In TNBC cancer-bearing mice, functional miRNA liposomes exerted a stronger anticancer effect than functional vinorelbine liposomes, and combination therapy with these two formulations resulted in nearly complete inhibition of tumor growth. Preliminary safety evaluations indicated that the functional miRNA liposomes did not affect body weight or cause damage to any major organs. Furthermore, the functional liposomes significantly increased the half-life of the drug in the blood of cancer-bearing nude mice, and increased drug accumulation in breast cancer tissues.
Conclusion: In this study, we constructed novel functional miRNA liposomes. These liposomes silenced Slug expression and inhibited the TGF-β1/Smad pathway in TNBC cells, and enhanced anticancer efficacy in mice using combined chemotherapy. Hence, the present study demonstrated a promising strategy for gene therapy of invasive breast cancer.
Keywords: functional miRNA liposomes, Slug, TGF-β1/Smad, gene therapy, TNBC
Introduction
Triple negative breast cancer (TNBC) is an invasive breast cancer that does not express estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor receptor-2 (Her-2).1 TNBC occurs in 10–25% of all patients with breast cancer, and is associated with high rates of metastasis and relapse.2,3 TNBC often requires combination treatment with adjuvant or neo-adjuvant chemotherapy and radiotherapy. TNBC is comprised of heterogeneous subtypes of malignant cells, and is often susceptible to the chemotherapy. However, in some cases treatment response does not correlate with overall survival.4 The lack of good correlation between treatment response and survival necessitates development of new treatment strategies.
In this study, we developed novel functional miRNA liposomes to treat TNBC. A specific miRNA, designed for post-transcriptional silencing of Slug, was encapsulated into cell-penetrating peptide tLyp-1 modified liposomes (referred to as functional liposomes throughout this manuscript) for effective transfection of TNBC cells. In addition, functional vinorelbine liposomes were constructed and used as a positive control or for combination therapy.
MicroRNA (miRNA) are small non-coding RNA molecules typically comprised of approximately 20 nucleotides. miRNA are endogenously produced in the cells of plants, animals, and some viruses, and they are involved in silencing specific RNA and in post-translational regulation of gene expression.5–7 MiRNA act through base-pairing with complementary sequences of intrinsic mRNA, thus interfering transcription and translation of specific genes. Interestingly, extrinsic miRNA, which is often engineered, can be used for silencing specific genes associated with diseases. Recently, a number of miRNA formulations have been studied for use as gene therapies.8,9
Slug encodes the zinc finger protein Slug, and is a subtype of C2H2 type zinc finger transcription factors in the Snail superfamily.10,11 Slug regulates cell migration in concert with other genes (eg, BMPs).12,13 Slug is extensively expressed in human cancers, and correlates with invasiveness and drug resistance.14,15 This study demonstrated that Slug expression is closely associated with TNBC cell invasion and metastasis,16 suggesting that Slug could be a potential target for gene therapy.
TLyp-1 (CGNKRTR) is a cell-penetrating peptide, which is truncated from the LyP-1 (CGNKRTRGC) peptide, and has tumor-targeting properties.17 Further investigations demonstrated that tLyp-1 specifically binds to the transmembrane glycoprotein receptor neuropilin (NRP) in tumors.18,19 Accordingly, tLyp-1 peptide has been used as a targeting molecule for in-drug delivery carriers to increase cellular uptake through NRP-dependent internalization.20–22
The objectives of the present study were to develop functional miRNA liposomes for the treatment of TNBC, to characterize the mechanisms of action of these liposomes, and to verify the efficacy of these liposomes against TNBC. Functional liposomes were prepared by modifying synthetic DSPE-PEG2000-tLyp-1. Moreover, the engineered miRNA was pre-treated with calf thymus DNA and protamine, and then encapsulated in the liposomes to yield functional miRNA liposomes. The physiochemical and biological properties of the liposomes were evaluated in TNBC cells and TNBC cancer-bearing mice using electric microscopy, real-time RT-PCR, Western blotting, and co-immunoprecipitation (co-IP) techniques.
Materials and methods
Materials and reagents
1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethyleneglycol)-2000] (DSPE-PEG2000-MAL) and polyethylene glycol-distearoylphosphatidylethanolamine (DSPE-PEG2000) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). tLyp-1 peptide was purchased from China Peptides (Shanghai, China). Tris-(2-carboxyethyl)-phosphine hydrochloride (1:1) (TCEP hydrochloride, TCEP·HCl) was purchased from BBI Life Sciences Corporation (Shanghai, China). Vinorelbine was purchased from Nanjing Tianzun Zezhong Chemicals (Nanjing, China). Calf thymus DNA and protamine were purchased from Sigma-Aldrich (St. Louis, MO, USA). PCR primers were purchased from Tsingke Biological Technology (Beijing, China).
Phosphate-buffered saline (PBS pH7.4; NaCl 137 mmol/L, KCl 2.7 mmol/L, Na2HPO4 10 mmol/L KH2PO4 2 mmol/L) was purchased from Macgene Biotech (Beijing, China). Anti-Slug antibody (24463) was purchased from SAB (Signalway Antibody; College Park, MD, USA). Anti-Snail (bs-1371R), anti-BMP5 (bs-6614R), anti-Blimp-1 (bs-6466R), and anti-β-Actin (bs-0061R) antibodies were purchased from Bioss Antibodies (Woburn, MA, USA). Anti-IgG-HRP antibody (HS101-01) was purchased from Transgen Biotech (Beijing, China). Anti-Smad2+ Smad3 antibody-ChIP Grade (ab207447) and anti-Smad4 antibody (ab40759) were purchased from Abcam (Shanghai local agent, China). Leibovitz’s L15 medium and DMEM medium were purchased from Macgene Biotech (Beijing, China). Fetal calf serum (FBS) was purchased from PAN-biotech (Beijing local agent, China). Opti-MEM medium was purchased from Gibco (Billings, MT, USA). Transwell chambers were purchased from Invitrogen (Beijing, China). Matrigel matrix was purchased from BD Biosciences (Mountain View, CA, USA). SB431542 (TGF-β pathway inhibitor) was purchased from Dalian Meilun Biotechnology (Dalian, China). BAY 11-7082 (NF-κB pathway inhibitor) was purchased from Beyotime Biotechnology (Beijing, China). All other reagents were of analytical grade, were used without further purification, and obtained from Beijing Chemical Reagents (Beijing, China).
Cells and animals
Human TNBC MDA-MB-231 cells were purchased from Institute of Basic Medical Sciences at Chinese Academy of Medical Sciences (Beijing, China), and grown in Leibovitz’s L15 medium at 37°C without CO2 or grown in DMEM medium supplemented with 10% fetal bovine serum (FBS) at 37°C under 5% CO2 circumstance.
Female BALB/c nude mice (18–20 g, 6–7-week-old) were purchased from Vital River Laboratory Animal Center (Beijing, China) and maintained in a specific pathogen-free (SPF) environment. All animal experiments were performed in accordance with Guide for the Care and Use of Laboratory Animals, and approved by the Committee for Experimental Animal Welfare of Biomedical Ethics of Peking University (LA2017233).
Synthesis of targeting molecule for functional liposomes
DSPE-PEG2000-tLyp-1 was used as a targeting molecule for inclusion in functional liposomes, and synthesized by linking DSPE-PEG2000-MAL with tLyp-1. Briefly, DSPE-PEG2000-MAL (2 μmol), tLyp-1 (10 μmol), and TCEP·HCl (40 μmol) were dissolved in 2 mL of PBS (0.1 M, 10× PBS, pH 7.4). The mixtures were stirred using a magnetic stirrer at room temperature for 24 hrs under argon. The crude product was then transferred to dialysis tubing (3,000 molecular mass cutoff) and dialyzed in deionized water for 48 hrs to remove unreacted tLyp-1, TCEP hydrochloride, and PBS (pH 7.4). The resultant DSPE-PEG2000-tLyp-1 was obtained by freeze-drying, and then stored at −20°C. The identity of the product was confirmed using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) (Bruker Daltonics, Germany). The yield of DSPE-PEG2000-tLyp-1 was determined by calculating the free –SH group concentration using Ellman’s reagent kit (BestBio, Shanghai, China).
Design of miRNA and preparation of functional miRNA liposomes
The region of miRNA used to target Slug was investigated using three computational algorithms including TargetScan,23 miRanda/mirSVR,24 and PicTar,25 and miRNA-203 was identified as a potential regulator of Slug.26–28 Based on the predicted binding sites between miRNA-203 and the target sites of Slug 3′-UTR, the miRNA and negative control miRNA (miRNA-NC) were designed and synthesized using solid-phase synthesis. MiRNA was labeled with the fluorescent probe FAM (carboxyfluorescein-5-succimidyl ester) (Gene Pharma, Shanghai, China).
To prepare miRNA complexes, miRNA and calf thymus DNA (ctDNA) (1:1, mass ratio) were mixed evenly by swirling, and then protamine was gradually dropped into the mixture under swirling. The mass ratio of (miRNA+ctDNA)/protamine was in the range of 0.8–2.1. Afterwards, the mixture was incubated at room temperature for 10 mins to form miRNA complexes.
To prepare functional miRNA liposomes, blank functional liposomes were constructed using a modified film dispersion approach as reported previously.29 The major components of blank functional liposomes were egg phosphatidylcholine (EPC), cholesterol, stearamide, and DSPE-PEG2000-tLyp-1 (70:30:10:5 molar ratio). The blank functional liposomes were further incubated with miRNA complexes (lipids:miRNA =700:1, mol/mol) at room temperature for 30 mins, which yielded functional miRNA liposomes.
FAM-labeled functional miRNA liposomes were prepared in a similar manner (lipids: FAM-miRNA=700:1, mol/mol). As an experimental control, FAM-labeled miRNA liposomes were prepared with DSPE-PEG2000 in place of DSPE-PEG2000-tLyp-1.
Functional vinorelbine liposomes were prepared as a chemotherapy agent using an ammonium phosphate gradient method, as we reported previously.30 The major components of the blank functional liposomes were EPC, cholesterol, stearamide, and DSPE-PEG2000-tLyp-1 (70:30:10:5 molar ratio). Experimental control vinorelbine liposomes were prepared with DSPE-PEG2000 in place of DSPE-PEG2000-tLyp-1.
In addition, several fluorescent probe-labeled liposomes were prepared as indicators including functional coumarin liposomes, regular coumarin liposomes, functional DiR liposomes, and regular DiR liposomes.
Characterization of functional miRNA liposomes
To evaluate the encapsulation efficiency of miRNA, the prepared functional miRNA liposomes were centrifuged (16,000 g), and un-encapsulated free miRNA was collected from the supernatant. The extracted miRNA was reacted with RiboGreen (Invitrogen) and assayed by fluorescence spectrophotometry (500 nm excitation and 525 nm emission). The encapsulation efficiency (EE) of miRNA was calculated using the formula: EE% = (Wi/Wtotal) ×100%, where Wtotal is total miRNA in the un-separated liposomes, and Wi is the measured miRNA in the liposomes.
To measure the encapsulation efficiency of vinorelbine, the functional vinorelbine liposomes were purified using a Sephadex G-50 column equilibrated with PBS (pH 7.4) to remove un-encapsulated vinorelbine. Vinorelbine was measured using high-performance liquid chromatography (HPLC) with UV detection (Agilent Technologies Inc. Cotati, CA, USA). HPLC analysis was performed on an ODS column (Nucleodur 100-5C18 column, 250 mm×4.6 mm, 5.0 μm) at 30°C. The mobile phase consisted of acetonitrile, 0.02 M NaH2PO4 (pH 3.0), and triethylamine (40:60:0.3, v/v). The detection wavelength was set at 268 nm and the flow rate was 1.0 mL/mins. The EE of vinorelbine was calculated by comparing the measured vinorelbine in the liposomes before and after Sephadex G-50 column purification.
Particle sizes, polydispersity index (PDI), and zeta potential were measured using a Nano Series Zen 4003 Zetasizer (Malvern Instruments, Malvern, UK). The miRNA complexes and functional miRNA liposomes were visualized using transmission electron microscope (TEM, JEM1400PLUS, JEOL, Tokyo, Japan). Briefly, the liposomes were diluted with distilled water to form liposome suspensions and then placed on a copper wire mesh with 1% uranyl acetate solution. The samples were air-dried at room temperature and visualized using TEM (120 kV).
Cellular uptake assay by flow cytometry
To determine the cellular uptake of functional miRNA liposomes, FAM-labeled functional miRNA liposomes were used to treat cancer cells. Briefly, MDA-MB-231 cells were seeded into six-well plates at a density of 5×105 cells/well and cultured for 24 hrs. The following solutions were added to the wells: including blank culture medium, FAM-labeled miRNA complexes (100 nM FAM-miRNA), FAM-labeled regular miRNA liposomes (100 nM FAM-miRNA), and FAM-labeled functional miRNA liposomes (100 nM FAM-miRNA). After incubation for 4 hrs, the cells were collected FAM-labeled miRNA fluorescence intensity was measured using a flow cytometer (BD Biosciences). Ten thousand data points were collected.
To determine the cellular uptake of functional vinorelbine liposomes, functional coumarin liposomes were used to treat cancer cells, and the cells were incubated identically to those treated with FAM-miRNA. The treatment and control formulations included blank culture medium, free coumarin (0.5 μM coumarin), regular coumarin liposomes (0.5 μM coumarin), and functional coumarin liposomes (0.5 μM coumarin).
Intracellular localization assay by confocal microscopy
To evaluate cellular localization of functional miRNA liposomes, FAM-labeled functional miRNA liposomes were used to treat cancer cells. Briefly, MDA-MB-231 cells were seeded into confocal cuvettes at 2×105 cells. After incubation for 24 hrs, the cells were treated with blank culture medium, FAM-labeled free miRNA complexes (100 nM FAM-miRNA), FAM-labeled regular miRNA liposomes (100 nM FAM-miRNA), and FAM-labeled functional miRNA liposomes (100 nM FAM-miRNA). After incubation for 2 hrs, the cells were stained with 2 μg/mL Hoechst 33342 (Thermo Scientific, Fremont, CA, USA) for 15 mins, and 200 nM MitoTracker Deep Red FM (Invitrogen) for 30 mins. Localization was visualized using a confocal laser scanning fluorescent microscope (Leica, Heidelberg, Germany). Composite images were created by overlapping the images from individual channels.
To evaluate cellular localization of functional vinorelbine liposomes, functional coumarin liposomes were used to treat cancer cells, which were then incubated identically to the FAM-miRNA liposome-treated cells. The treatment and control formulations included blank culture medium, free coumarin (2 μM coumarin), regular coumarin liposomes (2 μM coumarin), and functional coumarin liposomes (2 μM coumarin).
mRNA expression by qRT-PCR
To evaluate mRNA expression, MDA-MB-231 cells were transfected with functional miRNA-NC liposomes (100 nM miRNA-NC) or functional miRNA liposomes (100 Nm miRNA). The cells were collected after 48 hrs, and rinsed twice with ice-cold PBS (pH 7.4). Total RNA was extracted using Trizol plus RNA purification kit (Invitrogen), and analyzed (260 nm/280 nm ratio) using a spectrophotometer (Nano300, Allsheng, Hangzhou, China). A 500-ng aliquot of total RNA was then reverse-transcribed into cDNA using PrimeScript RT reagent (Takara Bio. Inc., Shiga, Japan), and 5 ng of cDNA was subjected to RT-PCR, using SYBR Premix Ex Taq II (Takara Bio. Inc.) on an Applied Biosystems CFX ConnectTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA).
Results were determined as fold-difference compared to the negative control group. Each experiment was performed in triplicate. Gene expression levels were normalized to β-actin mRNA levels. Relative quantification was calculated using the 2−ΔΔct method.31
Protein expression assay by Western blotting
To evaluate protein expression, MDA-MB-231 cells were transfected with functional miRNA-NC liposomes (100 nM miRNA-NC), functional miRNA liposomes (100 nM miRNA), functional miRNA liposomes (100 Nm miRNA) with SB431542 (TGF-β inhibitor, 10 nM), and functional miRNA liposomes (100 nM) with BAY 11–7082 (NF-κB inhibitor, 5 nM). The cells were collected after 72 hrs, rinsed twice with ice cold PBS (pH7.4), and lysed in 1 mL Pierce RIPA buffer (Applygen, Beijing, China) on ice for 30 mins. Cell lysates were centrifuged at 16,000 g for 20 mins at 4°C to remove the precipitated fragments. Total protein was quantified using the Bradford protein assay. The collected protein was boiled, cooled, and loaded onto the sodium dodecyl sulfate polyacrylamide electrophoresis gels (SDS-PAGE). After the electrophoresis, the gels were transferred to PVDF membranes (Merck-Millipore, Darmstadt, Germany), and the membranes were blocked with 5% skim milk (w/v) at room temperature for 1 hr. The membranes were incubated with primary antibodies against Snail, Slug, BMP5, Blimp-1, and β-actin at 4°C overnight, and then with horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 hr at previously optimized dilution ratios. After washing each membrane with TBST (Applygen, Beijing, China) several times, Immobilon Western Chemiluminescent HRP substrate (Millipore, MA, USA) was added, and the blots were analyzed using a MiniChemi chemiluminescence 610 imaging system (Sage Creation, Beijing, China). Protein expression was normalized to β-actin levels.
Smad protein complex assay by co-IP
To evaluate the Smad2/3/4 complex, MDA-MB-231 cells were transfected with the same reagents as those listed in the section titled “protein expression by Western blotting.” The collected protein samples (crude) were incubated with Smad2/3 antibody-bound immunomagnetic beads for immunoprecipitation (Sino Biological, Beijing, China). The purified protein was boiled, then loaded onto SDS-PAGE gels for Smad4 Western blot analysis.
Cell invasion evaluation by transwell assay
To evaluate the inhibitory effects of functional miRNA liposomes on cell invasion, MDA-MB-231 cells were transfected with functional miRNA liposomes (100 nM miRNA), or functional miRNA liposomes (100 nM miRNA) with TGF-β inhibitor (10 nM SB431542), then cultured for 48 hrs. The upper chambers of 24-well transwell plates were pre-coated with 50 μL of 1 mg/mL Matrigel matrix at 37°C for 1 hr. Then, the transfected cells (1×105) were seeded into the upper chamber with serum-free medium, and culture medium containing 35% serum was added to the lower chamber. After 16 hrs of incubation, the upper chamber was fixed and stained with 0.3% crystal violet (Sigma, Beijing local agent) for 5 mins. Visual fields (n=9) of each chamber were randomly selected and counted at 200× magnification.
To evaluate the inhibitory effects of functional vinorelbine liposomes on cell invasion, MDA-MB-231 cells were treated with blank culture medium or functional vinorelbine liposomes (0.05 μM vinorelbine). Furthermore, MDA-MB-231 cells were transfected with functional miRNA liposomes (100 nM) for 48 hrs in advance, and then treated with functional vinorelbine liposomes (0.05 μM vinorelbine) to evaluate the combined effect.
Cell migration evaluation by transwell assay
To evaluate the inhibitory effects of functional miRNA liposomes on cell migration, MDA-MB-231 cells were treated similar to those in the section titled “Cell invasion evaluation by transwell assay.” For this analysis, the upper chamber was not coated with Matrigel matrix. After 8 hrs of incubation, the upper chamber was fixed and stained with 0.3% crystal violet (Sigma, Beijing local agent) for 5 mins. Visual fields (n=9) of each chamber were randomly selected and counted at 200× magnification.
Cytotoxicity evaluation by SRB assay
To evaluate cytotoxicity, MDA-MB-231 cells were seeded into 96-well culture plates at a density of 5.0×103 cells per well, and then cultured for 24 hrs. The cells were treated with fresh media containing functional vinorelbine liposomes (0–2 μM) with functional miRNA liposomes (0–200 nM miRNA), free miRNA (0–2 μM miRNA), free vinorelbine (0–2 μM vinorelbine), functional miRNA liposomes (0–2 μM miRNA), regular vinorelbine liposomes (0–2 μM vinorelbine), functional vinorelbine liposomes (0–2 μM miRNA), or functional vinorelbine liposomes (0–2 μM vinorelbine) with functional miRNA liposomes (100 nM miRNA). After incubation for 48 hrs, the survival rates of each group were determined using the sulforhodamine B (SRB) staining assay. The absorbance of each well was measured using a microplate reader (Infinite F50, Tecan Group, Beijing, China) at 540 nm. Survival rate = (A540nm for treated cells/A540nm for blank control) ×100%. A540nm was absorbance at 540 nm.
Evaluation of apoptosis using flow cytometry
To evaluate apoptosis, MDA-MB-231 cells were seeded in 6-well plates at a density of 5×105 cells/well, and cultured for 24 hrs. The cells were treated with regular vinorelbine liposomes, functional vinorelbine liposomes, functional miRNA liposomes, or functional miRNA liposomes with functional vinorelbine liposomes for 24 hrs. The final concentrations of vinorelbine and miRNA were 0.05 μM and 100 nM, respectively. Control experiments were performed by adding culture medium only. After incubation, cells were collected and stained with Annexin V-FITC and PI (BD Biosciences) according to manufacturer’s instructions, and then analyzed using a FACScan Calibur flow cytometer (BD Biosciences). Annexin V-FITC positive/PI negative cells and double-positive cells were considered to be in the early and late stages of apoptosis, respectively. Each assay was performed in triplicate.
Anticancer efficacy in cancer-bearing animals
A cancer-bearing animal model was established by injecting MDA-MB-231 cells (8.0×106) into the right flanks of female BALB/c nude mice. When the tumors reached 80–100 mm3 in volume, the mice were randomly divided into four groups (n=6 each), and treated once at day 21, 23, 25, and 27 with physiological saline, functional vinorelbine liposomes (2 mg vinorelbine/kg), functional miRNA liposomes (20 nmol miRNA/kg), or functional miRNA liposomes (20 nmol miRNA/kg) with functional vinorelbine liposomes (2 mg vinorelbine/kg) via tail vein injection.
Tumors were measured with calipers every two days. Tumor volumes were calculated as follows: length×width2/2.32 Tumor volume ratio was calculated using the formula R = Vi/V0, where Vi is the tumor volume at the ith time-point after drug administration, and V0 represents the tumor volume at the initial time-point before the first drug administration. Tumor volume inhibitory rate was calculated using the formula Refficacy =100%–(Rdrug/Rblank) ×100%, where Rdrug and Rblank are the tumor volume ratios after treatment with drug and physiological saline, respectively.
Histological evaluations by immunostaining
After treatments, the mice were sacrificed by cervical dislocation and tumor tissues and major organ tissues (heart, liver, spleen, lung, and kidney) were harvested. The fresh isolated organ tissues were placed in 10% neutral buffered formalin for hematoxylin and eosin (HE) staining. The tumor tissues were frozen and cut into sections for TUNEL staining and immunohistochemistry (IHC) analysis of Snail, Slug, Blimp-1, and BMP5.
HE staining was used to evaluate the effects of the various treatments on each organ. The organ tissues were immersed in 4% paraformaldehyde for 4 hrs, and then transferred to 70% ethanol. Individual lobes of organ tissues were placed in processing cassettes, dehydrated using a serial alcohol gradient, and embedded in paraffin wax blocks. Prior to staining, 5-μm-thick tissue sections were dewaxed in xylene, rehydrated at decreasing ethanol concentrations, then washed in PBS (pH 7.4). Then, the tissue sections were stained with HE. After staining, sections were dehydrated using increasing concentrations of ethanol and xylene. The sections were photographed using a fluorescence inversion microscope.
Apoptosis was evaluated using TUNEL staining. Tumor tissue sections were fixed with 4% paraformaldehyde for 20 mins and permeabilized with 0.1% sodium citrate containing 0.1% Triton X-100. The sections were then incubated with the TUNEL reaction mixture containing TdT and fluorescein-dUTP. During the incubation period, TdT catalyzed the addition of fluorescein-dUTP to the free 3′-OH groups in single- and double-stranded DNA. After washing, DNA was marked using an anti-fluorescein antibody conjugated with the reporter enzyme peroxidase. After washing to remove unbound enzyme conjugates, the POD retained in the immune complex was visualized using a substrate reaction. The sections were counterstained using Gill-2 hematoxylin (Thermo-Shandon, Pittsburgh, PA, USA). After staining, the sections were dehydrated using increasing concentrations of ethanol and xylene. The sections were photographed using a confocal laser scanning fluorescence microscope.
To evaluate the protein expression of Snail, Slug, Blimp-1, and BMP5 in tumors by IHC, tumor sections were immersed in 10 mmol/L citrate buffer (pH 6.0) and microwaved for 15 mins to unmask the relevant antigens. Then, the tumor sections were immunostained using the avidin-biotinylated enzyme complex method with antibodies against Snail (1 μg/mL), Slug (1 μg/mL), Blimp-1 (1 μg/mL), BMP5 (1 μg/mL), and equivalent concentrations of polyclonal nonimmune IgG controls. After incubation with the appropriate biotin-conjugated secondary antibody and streptavidin solution, a chromogenic reaction was performed using 3,3-diaminobenzidine tetra-hydrochloride (Vector Laboratories, Burlington, CA, USA) as a chromogenic agent. After staining, sections were dehydrated using increasing concentrations of ethanol and xylene. The sections were photographed using a fluorescence inversion microscope.
In vivo imaging analysis in cancer-bearing mice
To observe the real-time distribution and tumor accumulation of functional miRNA liposomes, cancer-bearing mice were evaluated using non-invasive optical imaging systems. The cancer-bearing mice were divided into four groups (n=3 mice). When the volume of the tumors was approximately 400–500 mm3 at day 33, the mice were administered physiological saline, free DiR, regular DiR liposomes, or functional DiR liposomes via tail vein injections, and scanned at 1, 3, 6, 9, 24, and 36 hrs using an IVIS Spectrum Imaging System (PerkinElmer, Waltham, MA, USA). The concentration of DiR in all treatments was 0.04 mg/kg. To determine the distribution of liposomes in tumor masses and major organs, the tumor-bearing mice were sacrificed via cervical dislocation at 36 hrs, the tumor masses, heart, liver, spleen, lung, and kidney were immediately removed. The fluorescence signal intensities in different tissues were visualized using an IVIS Spectrum Imaging System.
Statistical analyses
Data are presented as the mean ± standard deviation (SD). Analysis of variance (ANOVA) was used to determine the significance among three or more groups, post hoc Bonferroni correction was used for multiple comparisons. A value of P<0.05 was considered statistically significant.
Results
Synthesis and characterization of targeting material
To obtain DSPE-PEG2000-tLyp-1 conjugated, tLyp-1-SH was synthesized by reduction reaction of tLyp-1-S-S-tLyp-1 with TCEP·HCl in 10× PBS (pH7.4), followed by coupling of the activated thiol group of tLyp-1-SH to the maleimide groups of DSPE-PEG2000-MAL. The structures or all of the molecules were confirmed using MALDI-TOF-MS (Figure 1A–C). The results indicated that the average mass of DSPE-PEG2000-tLyp-1 was 3860.0 Da (Figure 1C). The difference in mass between DSPE-PEG2000-tLyp-1 (Figure 1C, 3860.0 Da) and DSPE-PEG2000-MAL (Figure 1A, 3041.2 Da) was approximately equal to the mass of tLyp-1 (Figure 1B, 834.7 Da), demonstrating that DSPE-PEG2000-tLyp-1 was successfully synthesized. The yield of DSPE-PEG2000-tLyp-1 was 90.08±8.99%, as determined by calculation of free thiols.
Figure 1.

Preparation and characterization of designed miRNA and functional miRNA liposomes. A. Mass spectrum of DSPE-PEG2000-MAL. (B) Mass spectrum of tLyp-1. (C) Mass spectrum of DSPE-PEG2000-tLyp-1. The results demonstrate that DSPE-PEG2000-tLyp-1 is successfully synthesized. (D) The sense sequences of designed miRNA and meaningless miRNA (miRNA negative control, miRNA-NC). (E) Particle sizes of miRNA complexes in different mass ratios of (miRNA + ctDNA)/protamine in the range of 0.8–2.1. Data are presented as mean ± standard deviation (n=3). (F) Zeta potentials of miRNA complexes in different mass ratios of (miRNA + ctDNA)/protamine in the range of 0.8 −2.1. Data are presented as mean ± standard deviation (n=3). The results indicate that the complex at a mass ratio of 1.8 exhibits a stable status in viewing the suitable particle size and Zeta potential. (G) Particle sizes distributions of different formulations: 1, blank functional liposomes; 2, regular vinorelbine liposomes; 3, miRNA complexes; 4, functional vinorelbine liposomes; 5, functional miRNA liposomes. (H) Images of functional miRNA liposomes by TEM. (I) Images of functional vinorelbine liposomes by TEM.
Preparation and characterization of miRNA and functional miRNA liposomes
To design the objective miRNA, the interactions between the Slug 3′-UTR and miRNA-203 were simulated with a set of softwares (TargetScan, miRanda/mirSVR, and PicTar). The simulation indicated the recognition sites between miRNA-203 and the 3′-UTR of Slug could exist at UUGCUGCCAAAUCAUUUCAA, and all nucleotides in these regions were highly conserved across species (Table S1). According to the predicted recognition region, the miRNA for silencing Slug gene was designed, and the synthesized miRNA was AGUGGUUCUUAACGAUUCAACAGUU. As a negative control, the synthesized negative control miRNA-NC was UUCUCCGAA CGUGUC ACGUTT (Figure 1D).
Table S1.
The design of miRNA through interactions between the binding sites in the Slug 3ʹ-UTR (top) and miR-203 (bottom)
 | ||||
|---|---|---|---|---|
| Slug mRNA 5’ | 5’ | Coding sequence | UUGCUGCCAAAUCAUUUCAA 3’ | 3’ |
| miRNA-203 3’ | 3’ | CACCAGGGAUUUGUAAAGUG 5’ | 5’ | |
| Homo sapiens 5’ | 5’ | Coding sequence | UUGCUGCCAAAUCAUUUCAA 3’ | 3’ |
| Mus musculus 5’ | 5’ | Coding sequence | UUGCUGCCAAAUCAUUUCAA 3’ | 3’ |
Notes: The seed recognition sites are denoted, and all nucleotides in these regions are highly conserved across species. The predictions are performed through the following software and sites: TargetScan: http://www.targetscan.org/vert_72/; miRanda/mirSVR: http://www.microrna.org/microrna/home.do; PicTar: https://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi.
To encapsulate miRNA into the blank functional liposomes, a stable complex of miRNA, ctDNA, and protamine was prepared and characterized. A mass ratio range of 0.8 to 2.1 of (miRNA + ctDNA) to protamine resulted in acceptable stability and suitable particle size and Zeta potential (Figure 1E–G). Hence, the complex at mass ratio 1.8 was chosen as the final stable RNA complex formulation. The encapsulation efficiency of the engineered miRNA was greater than 80% using this complex (Table S2). Furthermore, the particle sizes of functional vinorelbine liposomes and functional miRNA liposomes ranged from 10 to 125 nm with uniform size distributions (0.13–0.28 PDI), and were positively charged (6–10 mV zeta potential). In contrast, regular vinorelbine liposomes were negatively charged (−4 mV zeta potential). In addition, vinorelbine was efficiently encapsulated into the liposomes (>85%). Visualization with TEM showed that the particles of all liposomes were round in shape, and demonstrated a nanoscale structure with sizes ranging from 110 to 130 nm (Figure 1H and I).
Table S2.
Characterization of functional liposomes
| Liposomes | Encapsulation efficiency (%) | Particle size (nm) |
Poly dispersity index | Zeta potential (mV) |
|---|---|---|---|---|
| Blank functional liposomes | – | 113.9±2.1 | 0.284±0.000 | 9.1±0.9 |
| Regular vinorelbine liposomes | 87.2±0.8 | 102.0±1.7 | 0.138±0.020 | −4.1±0.2 |
| Functional vinorelbine liposomes | 99.3±0.1 | 107.4±3.7 | 0.185±0.019 | 8.8±0.4 |
| Functional miRNA liposomes | 84.4±2.2 | 123.7±0.1 | 0.252±0.020 | 6.4±0.4 |
Cellular uptake of functional miRNA liposomes
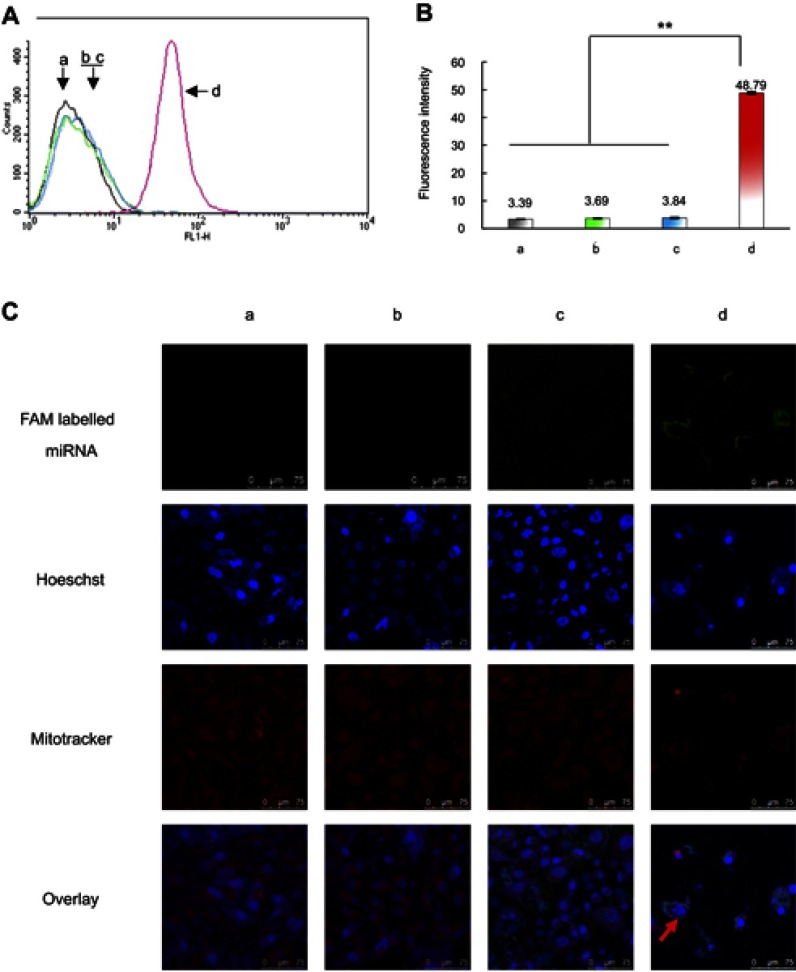
To evaluate the cellular uptake of functional miRNA liposomes, MDA-MB-231 cells were treated with various formulations. The results showed that the fluorescent intensity values after 4 hrs of incubation were 3.39±0.03 for blank medium, 3.69±0.08 for FAM-labeled free miRNA complexes, 3.84±0.19 for FAM-labeled regular miRNA liposomes, and 48.79±0.42 for FAM-labeled functional miRNA liposomes (Figure 2A and B), indicating that the functional miRNA liposomes were internalized to a significantly greater extent than the control formulations.
Figure 2.
Cellular uptake and co-localization of functional miRNA liposomes in TNBC cells. (A). Cellular uptake in MDA-MB-231 cells after treatments. a, blank control; b, FAM labeled free miRNA complexes; c, FAM labeled regular miRNA liposomes; d, FAM labeled functional miRNA liposomes. The results indicate that the FAM labeled functional miRNA liposomes significantly enhance the cellular uptake by MDA-MB-231 cells. (B) Fluorescence intensity values in MDA-MB-231 cells after treatments. a, blank control; b, FAM labeled free miRNA complexes; c, FAM labeled regular miRNA liposomes; d, FAM labeled functional miRNA liposomes. **P<0.01. The results reveal that the FAM labeled functional miRNA liposomes exhibit the strongest cellular uptake by MDA-MB-231 cells. (C) Co-localization in MDA-MB-231 cell mitochondria after treatments. a, blank control; b, FAM labeled free miRNA complexes; c, FAM labeled regular miRNA liposomes; d, FAM labeled functional miRNA liposomes. The results show that only FAM-labelled functional miRNA liposomes were effectively co-localized in cytoplasm and partially in mitochondria.
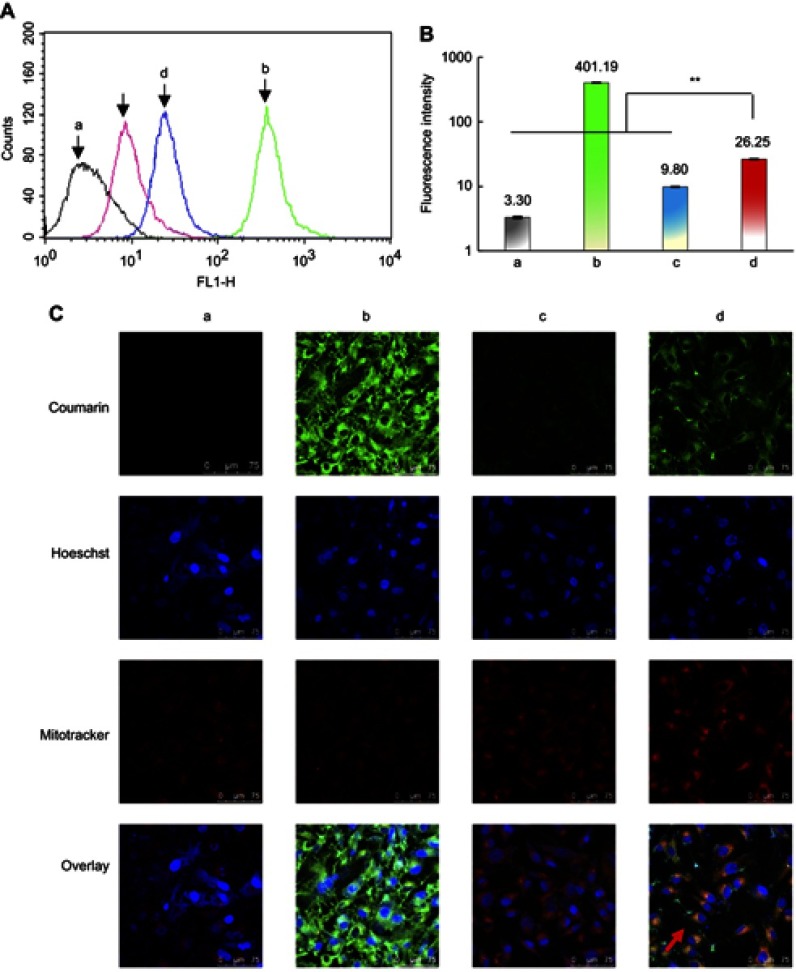
In addition, functional coumarin liposomes were used to evaluate the cellular uptake of functional liposomes. The results showed that the fluorescent intensity values after 4 hrs of incubation were 3.30±0.11 for blank medium, 401.19±2.41 for free coumarin, 9.94±0.10 for regular coumarin liposomes, and 26.25±0.32 for functional coumarin liposomes (Figure S1A and B), demonstrating that the functional coumarin liposomes were internalized to a significantly greater extent than regular coumarin liposomes, but less than free coumarin.
Figure S1.
Cellular uptake and co-localization of functional coumarin liposomes in TNBC cells. (A) Cellular uptake in MDA-MB-231 cells after treatments. a, blank control; b, free coumarin complexes; c, regular coumarin liposomes; d, functional coumarin liposomes. The results indicate that the functional coumarin liposomes are higher internalized than regular coumarin liposomes but less than free coumarin. (B) Fluorescence intensity values in MDA-MB-231 cells after treatments. a, blank control; b, free coumarin complexes; c, regular coumarin liposomes; d, functional coumarin liposomes. **P<0.01. The results show that the functional coumarin liposomes exhibit 2.6-folded cellular uptake than regular coumarin liposomes. (C) Co-localization in MDA-MB-231 cell mitochondria after treatments. a, blank control; b, free coumarin complexes; c, regular coumarin liposomes; d, functional coumarin liposomes. The results show that the functional coumarin liposomes are significantly higher co-localized than regular coumarin liposomes in cytoplasm and in mitochondria.
Intracellular co-localization of functional miRNA liposomes
To further understand intracellular localization, MDA-MB-231 cells were treated with various formulations. Using a confocal microscope, FAM-labeled miRNA, Hoechst dye, and Mitotracker dye were shown in green (indicating miRNA), blue (indicating the nucleus), and red (indicating mitochondria) (Figure 2C), respectively.
The cells were treated with blank medium, FAM-labeled free miRNA complexes, FAM-labeled regular miRNA liposomes, or FAM-labeled functional miRNA liposomes for 2 hrs in OPTI-MEM serum-free culture medium. The results showed that only FAM-labeled functional miRNA liposomes effectively localized in cytoplasm or the mitochondria. Moreover, the mitochondria were partially destroyed by transfection with functional miRNA liposomes, as demonstrated by diffuse structure.
After treatment with blank medium, free coumarin, regular coumarin liposomes, and functional coumarin liposomes for 2 hrs in the DMEM culture medium containing 10% serum, the functional coumarin liposomes were localized to the cytoplasm and mitochondria to a significantly greater extent than regular coumarin liposomes. However, free coumarin exhibited the strongest localization to cytoplasm, but not to mitochondria. Furthermore, the mitochondria were not adversely affected by these treatments (Figure S1C).
Silencing slug and inhibition of the TGF-β/Smad pathway
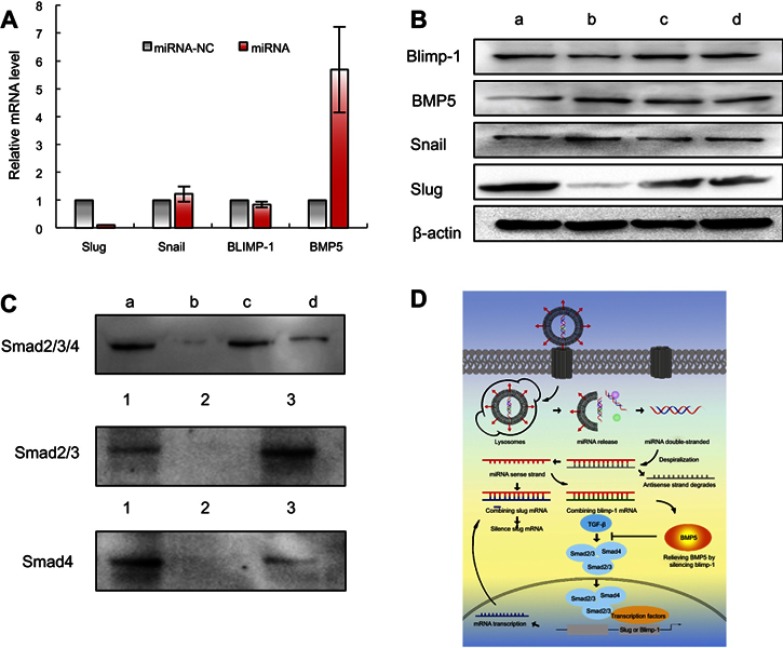
To evaluate mRNA expression of Slug, Snail, Blimp-1, and BMP5, MDA-MB-231 cells were transfected with functional miRNA-NC liposomes and functional miRNA liposomes. Relative mRNA levels were quantified by qRT-PCR (Table S3). The results showed that the functional miRNA liposomes significantly reduced Slug mRNA level but did not affect Snail mRNA. Furthermore, functional miRNA liposomes significantly increased BMP5 mRNA expression, but reduced Blimp-1 mRNA expression (Figure 3A). These results demonstrated that functional miRNA liposomes were able to silence Slug and Blimp-1 mRNA expression through a post-transcriptional mechanism.
Table S3.
Conditions of real-time PCR analysis
| Gene | Primer sequence | Cycle | Tm |
|---|---|---|---|
| Human Blimp1 | Forward 5ʹ- TGGACATGGAGGATGCGGATATG-3ʹ Reverse 5ʹ- AGGTCCTTTCCTTTGGA-GGAGTTG-3’ |
40 | 60℃ |
| Human BMP5 | Forward 5ʹ- CTCTACAATGCCATGACCAATGA-3ʹ Reverse 5ʹ- CCGAGATAACTGTATGCGACGA-3’ |
40 | 60℃ |
| Human Snail | Forward 5ʹ- AATCCAGAGTTTACCTTCCAGC-3ʹ Reverse 5ʹ- AATCCAGAGTTTACCTTCCAGC-3’ |
40 | 60℃ |
| Human Slug | Forward 5ʹ- AGATGCATATTCGGACCCAC-3ʹ Reverse 5ʹ- CCTCATGTTTGTGCAGGAGA-3’ |
40 | 60℃ |
| Human β-actin | Forward 5ʹ-CCTCGCCTTTGCCGATCC-3ʹ Reverse 5ʹ-GGATCTTCATGAGGTAGTCAGTC-3ʹ |
40 | 60℃ |
Note: RNA primers were synthesized by Tsingke Biological Technology (Beijing, China).
Figure 3.
Functional miRNA liposomes enabling to silence Slug gene and suppressing TGF-β1-Smad signaling pathway in TNBC cells. (A) The relative mRNA levels of transcription factor Slug, Snail, Blimp-1, and BMP5 in MDA-MB-231 cells after transfection of functional miRNA-NC liposomes and functional miRNA liposomes. The results indicate that functional miRNA liposomes are able to silence Slug and Blimp-1 and to up-regulate BMP5. (B) The protein expressions of Slug, Snail, Blimp-1, and BMP5 after transfections. a, functional miR-NC liposomes; b, functional miRNA liposomes; c, functional miRNA liposomes with TGF-β1/Smad inhibitor; d, functional miRNA liposomes with NF-κB inhibitor. The results show that the functional miRNA liposomes significantly increase BMP5 protein level while reducing Slug and Blimp-1 proteins. Besides, functional miRNA liposomes do not affect the Snail protein level. In contrast, the silencing effect of functional miRNA liposomes on Slug protein is evidently inhibited by TGF-β inhibitor, and the changed BMP5 and Blimp-1 protein levels are recovered to those of negative control. (C) Smad2/3/4 protein complex in MDA-MB-231 cells. 1, input: positive control, lysates of MDA-MB-231 cells; 2, mouse IgG: negative control; 3, protein samples after immunomagnetic bead sorting by Smad2/3 ChIP grade antibodies. a, functional miR-NC liposomes; b, functional miRNA liposomes; c, functional miRNA liposomes with TGF-β1/Smad inhibitor; d, functional miRNA liposomes with NF-κB inhibitor. The results show that Smad2/3/4 complexes exist in un-treated MDA-MB-231 cells while functional miRNA liposomes inhibit the formation of Smad2/3/4 complex. TGF-β inhibitor and NF-κB inhibitor relieve the inhibition caused by functional miRNA liposomes through a phosphorylation of Smad protein. The results demonstrate that the functional miRNA liposomes are able to silence Slug gene and to inhibit TGF-β1/Smad pathway. (D) Illustration for action mechanism of miRNA.
To evaluate protein expression of Slug, Snail, Blimp-1, and BMP5, MDA-MB-231 cells were transfected with functional miRNA-NC liposomes, functional miRNA liposomes, functional miRNA liposomes with a TGF-β inhibitor, and functional miRNA liposomes with an NF-κB inhibitor. Relative protein levels were evaluated by Western blot analysis. Compared to miRNA-NC liposomes (negative control), the functional miRNA liposomes significantly increased BMP5 protein levels and reduced Slug and Blimp-1 protein levels. The functional miRNA liposomes did not affect Snail protein levels (Figure 3B). In contrast, the silencing effect of functional miRNA liposomes on Slug protein expression was inhibited by a TGF-β inhibitor, and BMP5 and Blimp-1 protein levels were recovered to those of the negative control. Similarly, after transfecting with functional miRNA liposomes with an NF-κB inhibitor, the Slug, BMP5, and Blimp-1 protein levels were returned to those of the negative control by an NF-κB inhibitor. These results indicated that functional miRNA liposomes most likely regulated transcription through the TGF-β signaling pathway.
Co-IP was used to further characterize the mechanisms of action of functional miRNA liposomes. MDA-MB-231 cells were transfected with functional miRNA-NC liposomes, functional miRNA liposomes, functional miRNA liposomes with a TGF-β inhibitor, or functional miRNA liposomes with an NF-κB inhibitor. The results showed that the functional miRNA liposomes significantly inhibited the formation of the Smad2/3/4 complex. In contrast, both the TGF-β inhibitor and the NF-κB inhibitor blocked the inhibition of the Smad2/3/4 complex induced by functional miRNA liposomes (Figure 3C), resulting in formation of the Smad2/3/4 complex through phosphorylation of Smad.33,34 These results indicated that the engineered miRNA was able to silence Slug and to inhibit the TGF-β1/Smad signaling pathway (as illustrated in Figure 3D).
Liposome-induced inhibition of invasion and migration of TNBC cells
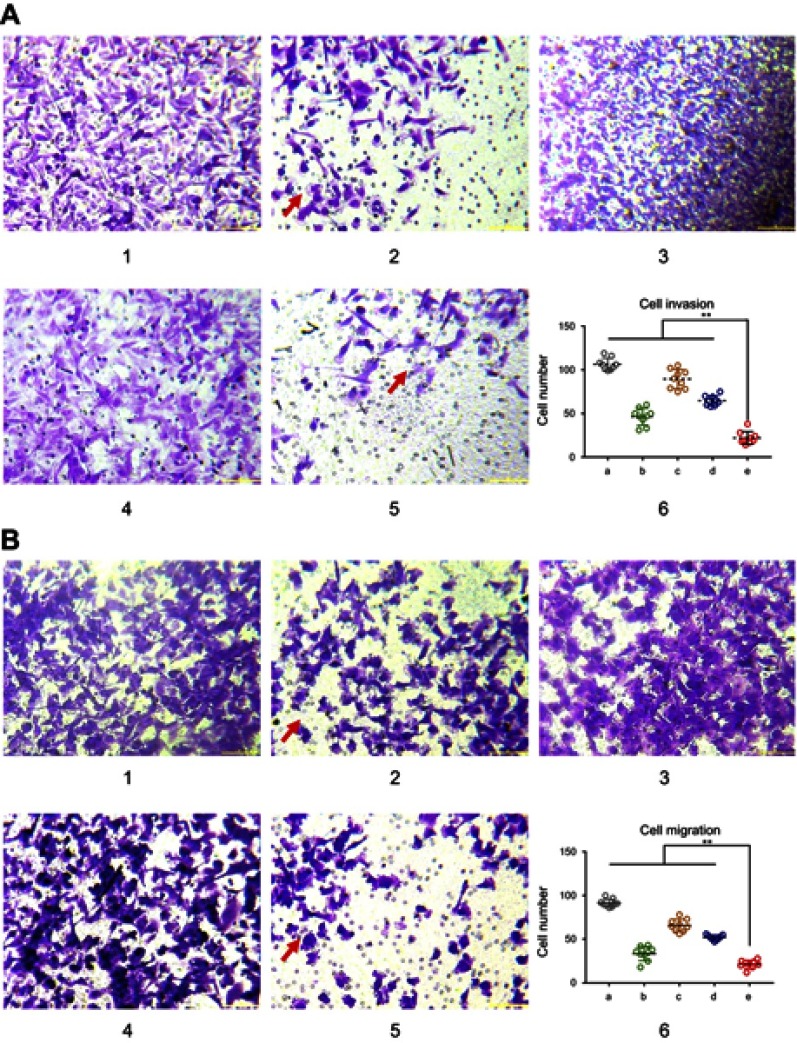
To verify the inhibitory effects of functional miRNA liposomes on cell invasion, various formulations were incubated with MDA-MB-231 cells, including blank medium, functional miRNA liposomes, functional miRNA liposomes with TGF-β inhibitor, functional vinorelbine liposomes, or functional miRNA liposomes with functional vinorelbine liposomes. The results showed that the functional miRNA liposomes were able to significantly inhibit cell invasion. In contrast, the inhibitory effect of functional miRNA liposomes on cell invasion was significantly reduced by the TGF-β inhibitor (Figure 4A). Furthermore, functional vinorelbine liposomes exhibited an inhibitory effect on cell invasion, and combined treatment with functional miRNA liposomes and functional vinorelbine liposomes resulted in a more pronounced inhibitory effect on cell invasion.
Figure 4.
Functional miRNA liposomes inhibiting invasion and migration of TNBC cells. (A) The cell invasion of MDA-MB-231 cells after treatments. 1, blank control; 2, functional miRNA liposomes; 3, functional miRNA liposomes with TGF-β1/Smad inhibitor; 4, functional vinorelbine liposomes; 5, functional miRNA liposomes with functional vinorelbine; 6, counted invasion cell numbers of 9 visual fields, **P<0.01. The red arrows indicate the invading cells transfected with functional miRNA liposomes are significantly reduced compared with blank control group. The results demonstrate that both functional miRNA liposomes and functional vinorelbine liposomes enable to inhibit cell invasion, and the combination treatment using functional miRNA liposomes with functional vinorelbine liposomes exhibit the strongest inhibitory effect. (B) The cell migration of MDA-MB-231 cells after treatments. 1, blank control; 2, functional miRNA liposomes; 3, functional miRNA liposomes with TGF-β1/Smad inhibitor; 4, functional vinorelbine liposomes; 5, functional miRNA liposomes with functional vinorelbine; 6, counted migration cell numbers of 9 visual fields, **P<0.01. The red arrows indicate the migrating cells transfected with functional miRNA liposomes are significantly reduced compared with blank control group. The results demonstrate that both functional miRNA liposomes and functional vinorelbine liposomes enable to inhibit cell migration, and the combination treatment using functional miRNA liposomes with functional vinorelbine liposomes exhibit the strongest inhibitory effect.
To further verify the inhibitory effects of functional miRNA liposomes on cell migration, MDA-MB-231 cells were treated with the same formulations used to evaluate cell invasion. The results showed that functional miRNA liposomes were able to significantly inhibit cell migration. In contrast, the inhibitory effect of functional miRNA liposomes on cell migration was significantly reduced by the TGF-β inhibitor (Figure 4B). Similarly, functional vinorelbine liposomes also inhibited cell migration, and combined treatment with functional miRNA liposomes and functional vinorelbine liposomes resulted in a more pronounced inhibitory effect on cell migration.
Growth inhibition of TNBC cells by functional mirna liposomes
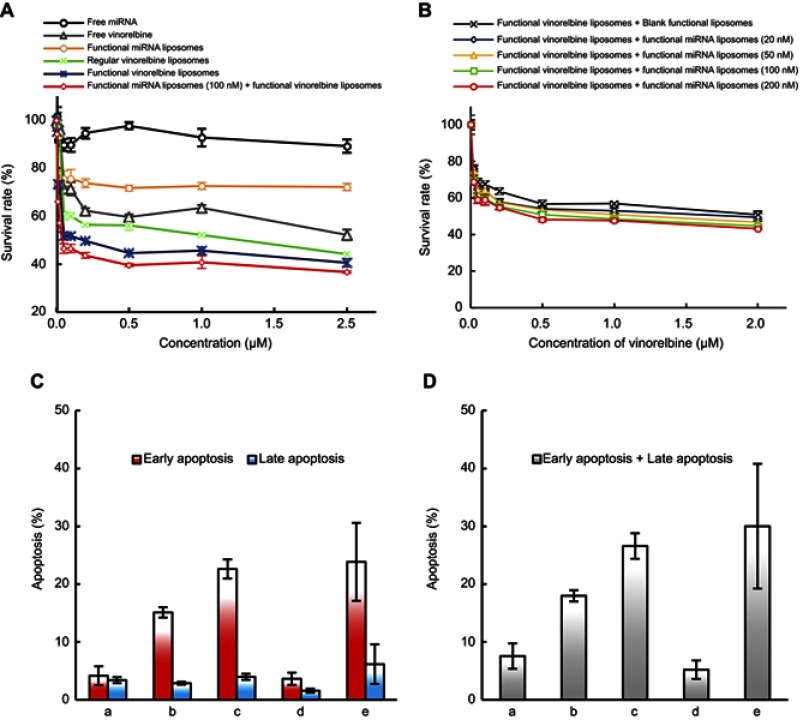
To confirm cytotoxicity of functional miRNA liposomes on TNBC cells, MDA-MB-231 cells were treated with free miRNA, free vinorelbine, functional miRNA liposomes, regular vinorelbine liposomes, functional vinorelbine liposomes, or functional miRNA liposomes with functional vinorelbine liposomes. The results showed that the functional miRNA liposomes inhibited cell growth, and the combination of functional miRNA liposomes with functional vinorelbine liposomes demonstrated the strongest inhibitory effect on the cell growth. In contrast, free miRNA did not affect cell growth. The inhibitory effect of functional vinorelbine liposomes on cell growth was more pronounced than that of regular vinorelbine liposomes and free vinorelbine (Figure 5A).
Figure 5.
Cytotoxicity and apoptosis in TNBC cells after treatment with functional miRNA liposomes and functional vinorelbine liposomes. (A) Cytotoxicity in MDA-MB-231 cells in vitro after treatments. The results reveal that the combination treatment of functional miRNA liposomes and functional vinorelbine liposomes has the strongest efficacy in killing MDA-MB-231 cells. (B) Cytotoxicity in MDA-MB-231 cells in vitro after transfections. The results demonstrate that the concentration of 100 nM miRNA is the optimal transfection concentration, with the highest transfection efficiency and lower cytotoxicity. (C) Early apoptosis and late apoptosis ratios in MDA-MB-231 cells after treatments at 12 hrs. Data are the mean ± standard deviation (n=3). a, blank control; b, regular vinorelbine liposomes; c, functional vinorelbine liposomes; d, functional miRNA liposomes; e, functional miRNA liposomes with functional vinorelbine liposomes. (D) Total apoptosis ratios in MDA-MB-231 cells after treatments at 12 hrs. Data are the mean ± standard deviation (n=3). a, blank control; b, regular vinorelbine liposomes; c, functional vinorelbine liposomes; d, functional miRNA liposomes; e, functional vinorelbine liposomes with functional miRNA liposomes. The results indicate that the combination treatment of functional miRNA liposomes and functional vinorelbine liposomes result in the strongest apoptosis in MDA-MB-231 cells.
To investigate the effect of miRNA concentration on cell growth, MDA-MB-231 cells were treated with functional vinorelbine liposomes in blank medium or functional vinorelbine liposomes with functional miRNA liposomes (20, 50, 100, and 200 nM miRNA). The results showed that the inhibitory effect increased in a functional miRNA liposomes concentration-dependent manner across the range of 20–100 nM miRNA. AAs such as 100 nM miRNA were used for further experiments (Figure 5B).
Induction of apoptosis by functional mirna liposomes
MDA-MB-231 cells were treated with blank medium, functional miRNA liposomes, regular vinorelbine liposomes, functional vinorelbine liposomes, or functional miRNA liposomes with functional vinorelbine liposomes. The results showed that functional miRNA liposomes slightly induced the apoptosis of TNBC cells. In contrast, functional vinorelbine liposomes strongly induced apoptosis in TNBC cells (Figure 5C and D).
Anticancer efficacy and silencing slug gene in cancer-bearing animals
MDA-MB-231 cancer-bearing mice were treated with various formulations of functional miRNA liposomes. The results showed that the inhibitory rates at day 33 were 41.24±12.5% for functional vinorelbine liposomes, 64.33±8.18% for functional miRNA liposomes, and 79.71±5.98% for the combination of functional miRNA liposomes and functional vinorelbine liposomes (Figure 6A).
Figure 6.
Anticancer efficacy and silencing Slug gene of functional miRNA liposomes in TNBC cancer-bearing mice. (A) Anticancer efficacy in cancer-bearing nude mice. P<0.05; a, vs physiological saline; b, vs functional miRNA liposomes; c, vs functional vinorelbine liposomes. Data are presented as mean ± standard deviation (n=6). (B) Body weight changes in cancer-bearing nude mice, Data are presented as mean ± standard deviation (n=6). (C) Apoptosis in the tumor tissues of breast cancer-bearing nude mice. a, physiological saline; b, functional miRNA liposomes; c, functional vinorelbine liposomes; d, functional miRNA liposomes with functional vinorelbine liposomes. The number of red arrows indicate the level of cell apoptosis rate. (D) IHC staining of Slug, Snail, Blimp-1, and BMP5 in tumor tissue. a, physiological saline; b, functional miRNA liposomes; c, functional vinorelbine liposomes; d, functional miRNA liposomes with functional vinorelbine liposomes. Light blue indicates no protein expression, light yellow indicates lower protein expression, brown-yellow color indicates middle expression, and dark brown indicates higher protein expression. The red up-arrow indicates the increase of protein expression while the number of red down-arrow indicates the decrease of the protein expression. The results demonstrate that the functional miRNA liposomes have a stronger anticancer effect than functional vinorelbine liposomes, and the combination treatment of functional miRNA liposomes with functional vinorelbine liposomes result in nearly a full inhibition to the growth of tumor. Furthermore, functional miRNA liposomes significantly silence the Slug and Blimp-1 proteins while increase BMP5 protein in tumor tissue.
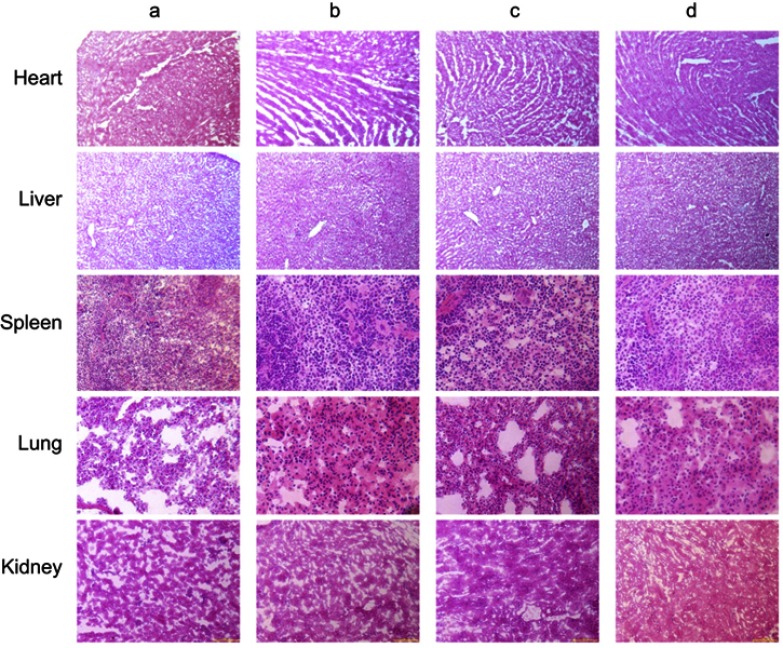
The body weights of cancer-bearing mice were monitored during drug administrations to estimate the safety of functional miRNA liposomes. The results showed that the functional miRNA liposomes did not affect the body weights of the animals, and none of the treatments resulted in significant loss of body weight (Figure 6B). To further evaluate the safety of functional miRNA liposomes, the major organs (heart, liver, spleen, lung, and kidney) of cancer-bearing mice were isolated for pathological observation. The results showed that none of the drug formulations caused obvious pathological abnormalities or lesion in the major organs compared with physiological saline treatment (Figure S2).
Figure S2.
HE staining of organs tissues (heart, liver, spleen, lung, and kidney) in TNBC cancer-bearing mice. (a) Physiological saline; (b) Functional miRNA liposomes; (c) Functional vinorelbine liposomes; (d) Functional vinorelbine liposomes plus functional miRNA liposomes. The results reveal that, after treatments, heart, liver, spleen, lung, and kidney tissues in the cancer-bearing rats have no obvious histopathological abnormalities or lesions as compared with those in the healthy rats.
Evaluation of apoptosis showed that both functional miRNA liposomes and functional vinorelbine liposomes significantly induced apoptosis in the tumor masses. Furthermore, the combination of functional miRNA liposomes and functional vinorelbine liposomes resulted in induced apoptosis of tumor masses to a greater extent than either treatment alone (Figure 6C).
Analysis of protein expression showed that the functional miRNA liposomes reduce the expression of Slug and Blimp-1 proteins, and increased the expression of BMP5 protein. Combination treatment with functional miRNA liposomes and functional vinorelbine liposomes induced a stronger effect than that of functional miRNA liposomes alone. In contrast, functional vinorelbine liposomes had a less-pronounced effect on the expression of Slug, Blimp-1, and BMP5 protein (Figure 6D).
In vivo and ex vivo imaging analysis
To evaluate the distribution of functional miRNA liposomes in vivo, DiR was used as a fluorescent probe to determine localization of the functional liposomes in MDA-MB-231 cancer-bearing mice. After intravenous injection of functional DiR liposomes, a strong fluorescent signal was observed in the blood and in the tumor location at the earliest time-point (3 hrs). Moreover, the signal was maintained for up to 36 hrs at the tumor site. In contrast, after administration of free DiR the fluorescent signal gradually weakened and disappeared within 24 hrs (Figure 7A).
Figure 7.
In vivo real-time imaging the distribution of fluorescent probe-labeled functional liposomes in TNBC cancer-bearing mice. (A) In vivo real-time imaging the cancer-bearing mice after intravenous injections. a, physiological saline; b, free DiR; c, regular DiR liposomes; d, functional DiR liposomes. (B) Ex vivo imaging tumor masses and organs after intravenous injections. a, physiological saline; b, free DiR; c, regular DiR liposomes; d, functional DiR liposomes. The results reveal that the functional liposomes have long circulatory circulation in blood system and a higher accumulation in tumor site.
To observe the distribution of functional miRNA liposomes in ex vivo tumor masses and major organs, the cancer-bearing mice were sacrificed at 36 hrs after drug administration. The results showed that the fluorescence signals of functional DiR liposomes were pronounced in the tumor masses and in the liver, spleen, lung, and kidney. In contrast, the fluorescent signal of free DiR was only weakly visible in tumor masses (Figure 7B).
Discussion
Neo-adjuvant chemotherapy has allowed for substantial advances in treatment of breast cancers. However, the efficacy of neo-adjuvant chemotherapy is limited in treatment of patients with TNBC.35,36 Gene therapy may provide an effective strategy for improving the prognosis of TNBC. Therefore, we developed a novel functional miRNA liposome which may effectively treat TNBC through post-transcriptional silencing of Slug and inhibition of the TGF-β1/Smad pathway. Furthermore, this novel formulation may be effective when used in combination with chemotherapy.
To produce an effective gene therapy agent, a highly efficient transfection vector and a specific mRNA are required. Therefore, a cell-penetrating peptide (tLyp-1 peptide) was synthesized and incorporated into a cationic lipid-based liposome to enhance gene transfection efficiency (Figure 1A–C) because this peptide has been shown to specifically bind to NPR receptors on tumors.37 Slug was identified as the target gene because Slug protein has been associated with invasion and metastasis of TNBC cells.38 Furthermore, loss of miRNA-203 in cells correlates with development of TNBC. In addition, recent studies have demonstrated that miRNA-203 plays an important role in inhibiting cell proliferation, inducing cell apoptosis, and decreasing aggressiveness of human breast cancer.39–41 A number of mRNAs were predicted using algorithms (TargetScan, miRanda/mirSVR, and PicTar) and public data libraries. Therefore, a novel 25 nucleotide sense strand of miRNA (Table S1 and Figure 1D) was designed based on the sequence of miRNA-203, and this sequence was capable of silencing Slug in vitro and in vivo.
For gene therapy, the transfection vector determines the transfection efficiency and treatment effects. In this study, a safe and highly efficient non-viral functional liposome was designed to encapsulate miRNA. To enhance encapsulation, calf thymus DNA was used to compress the structure of miRNA with the aid of protamine. By fine-tuning the ratio of (miRNA + ctDNA) to protamine (Figure 1E and F), the miRNA was effectively encapsulated into the blank functional liposomes (Table S2), resulting in a uniform, positively charged, and nanosized miRNA delivery vector (Figure 1G and H). The functional vinorelbine liposomal formulation was constructed identically to the functional miRNA liposomal formulation for efficient delivery of drug to the tumor site (Figure 1I) to improve duration of circulation in the blood, and to enhance the permeability and retention (EPR) effect.42–44
Moreover, TNBC is a refractory breast cancer and it exhibits tolerance to paclitaxel treatment. Accordingly, the first-line chemotherapy drug usually includes vinorelbine or gemcitabine combined with carboplatin. As one of the most effective drugs for treatment of metastatic and recurrent breast cancer, vinorelbine was included in the present study as a positive control and a combinational chemotherapeutic agent.
To validate the transfection efficiency of functional miRNA liposomes in TNBC cells, cellular uptake and intracellular localization are evaluated. The results confirmed that the functional miRNA liposomes were effectively internalized by the cells, and localized to the cytoplasm, and to a lesser extent the mitochondria (Figure 2A–C; Figure S1A–C), which may have resulted from electrostatic interactions between the positively charged liposomes and negatively charged mitochondria.45
The results from the silencing experiments and evaluations of the mechanisms of action of the functional liposomes demonstrated that the functional miRNA liposomes were capable of effectively silencing Slug through a post-transcriptional process (Figure 3A) and inhibition of the TGF-β1/Smad pathway (Figure 3B and C).
The likely mechanisms of action of the functional miRNA liposomes (shown in Figure 3D) are as follows: (i) functional miRNA liposomes were transfected into the cancer cells through specific interaction of tLyp-1 peptide with NRP receptors; (ii) the functional miRNA liposomes were internalized by endosomes, captured by lysosomes, and escaped from the lysosomes by the proton-sponge effect;46 (iii) miRNA was released from ruptured or intact liposomes; (iv) the sense miRNA strand bound to Slug and Blimp-1 mRNA, and their antisense miRNA strands were degraded in the cytoplasm; (v) the newly combined Slug mRNA was degraded, thereby silencing Slug by post-transcriptional regulation; (vi) the newly combined mRNA relieves BMP5 by silencing Blimp-1,47 and (vii) the relieved BMP5 blocked the TGF-β1/Smad signaling pathway. As a result, the Slug and Blimp-1 transcription factors were also inhibited.
Functional miRNA liposomes were evaluated for their effects on the following cellular properties: invasiveness (invasion and migration); growth; and apoptosis of TNBC cells. The results indicated that the functional miRNA liposomes were able to effectively inhibit the invasiveness and growth of TNBC cells, but were less effective at inducing apoptosis in vitro. In contrast, the functional miRNA liposomes demonstrate a substantial ability to induce apoptosis in vivo in cancer-bearing mice. Moreover, combination treatment with functional miRNA liposomes and functional vinorelbine liposomes significantly enhanced inhibition of invasiveness, and increased apoptosis of TNBC cells (Figure 4A and B; Figure 5A–D).
The anticancer efficacy and Slug silencing effects of functional miRNA liposomes were evaluated in cancer-bearing mice. The results demonstrated that the functional mRNA liposomes exerted a stronger anticancer effect than functional vinorelbine liposomes in TNBC cancer-bearing mice, and that combined therapy with functional miRNA liposomes and functional vinorelbine liposomes resulted in nearly complete inhibition of tumor growth, indicating a remarkable anticancer effect (Figure 6A). In addition, the results from the preliminary safety evaluations indicated that the functional miRNA liposomes did not affect body weight (Figure 6A) or major organs (Figure S2) of the mice.
Furthermore, functional miRNA liposomes strongly induced apoptosis in tumors in vivo (Figure 6C). The observed differences between the in vitro and in vivo results were most likely due to differences in timing. In vivo treatment with functional miRNA liposomes resulted in significant apoptosis of the cancer cells because of a relatively long duration of action (eg, 48 hrs). In contrast, in vitro dosing with functional miRNA liposomes induced less apoptosis of cancer cells due to a 24 hrs duration of treatment, suggesting that the miRNA requires an extended time period (such as 48 hrs) to induce post-transcriptional effects. Furthermore, apoptosis experiments in vitro require viable cells for flow cytometry, so they often need to be evaluated within a relatively short time (24 hrs); otherwise, there may not be enough living cells to perform the analysis.
After administration of functional miRNA liposomes, the silencing effect on Slug and Blimp-1 protein and up-regulation of BMP5 protein were confirmed in TNBC cancer-bearing mice (Figure 6D). The results indicated the same trend as that seen in vitro in TNBC cells. Clinical investigations demonstrated that overexpression of Slug protein in patients with TNBC was associated with higher clinical malignant grade, lymph node metastasis, post-operative relapse, and shorter patient survival.48,49 In addition, BMP5 has been shown to be downregulated in patients with breast cancer,50 and the down-regulation of BMP5 correlated with the overall survival of patients with TNBC.51 Consequently, Slug and BMP5 could be used as potential biomarkers to determine the effectiveness of functional miRNA liposomes in gene therapy.
The results from in vivo imaging demonstrated that the functional miRNA liposomes had long duration of circulation after intravenous injection, and accumulate significantly at the tumor site (Figure 7A and B). Furthermore, the results of ex vivo imaging further confirmed that the functional liposomes accumulated in tumors. In addition, the functional liposomes were enriched in the liver, suggesting that the functional miRNA liposomes may be eliminated by the liver-bile excretion route.
The results from in vitro and in vivo evaluations suggest that the formulation of functional miRNA liposomes deserves further development and is promising for future clinical trial. To reach clinical application, the quality specification and the clinical dosage regimen of functional miRNA liposomes await to be developed. On the one hand, the quality specification could be established by performing the pre-clinical evaluations on long-term stability, pharmacology, toxicology, etc. On the other hand, the clinical dosage regimen could be developed by a converted dose calculation. Namely, a dose in nude mice is equivalent to 0.11-fold dose in human body, depending on the individual body surface area of patients, and the dose escalation in the Phase I clinical study.
Conclusion
In summary, we developed a novel functional miRNA liposome, in which a new 25 nucleotide sense strand of miRNA was designed and encapsulated into tLyp-1 peptide-modified functional liposomes. The functional miRNA liposomes had a round shape and were nanosized (120 nm). In TNBC cells, the functional liposomes silenced Slug and inhibited the TGF-β1/Smad pathway, resulting in inhibition of invasiveness and growth of TNBC cells. In TNBC cancer-bearing mice, functional miRNA liposomes exhibited a stronger anticancer effect than functional vinorelbine liposomes, and combination treatment with functional miRNA liposomes and functional vinorelbine liposomes resulted in nearly complete inhibition of tumor growth. Preliminary safety evaluations indicated that the functional miRNA liposomes did not affect body weight or major organs of the mice. In conclusion, functional miRNA liposomes effectively treated TNBC cells by post-transcriptional silencing of Slug and inhibition of the TGF-β1/Smad pathway. Furthermore, these functional liposomes had enhanced anticancer efficacy when used in combination with another chemotherapy. Hence, the present study described a promising new gene therapy strategy for treatment of TNBC.
Acknowledgments
This study was funded by Beijing Natural Science Foundation (Key Grant No.7181004) and National Natural Science Foundation of China (Grant 81673367, and 81874303).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary Materials
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. New Eng J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI57873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granados-Principal S, Liu Y, Guevara ML, et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17:25. doi: 10.1186/s13058-015-0527-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albergaria A, Ricardo S, Milanezi F, et al. Nottingham Prognostic Index in triple-negative breast cancer: a reliable prognostic tool? BMC Cancer. 2011;11:299. doi: 10.1186/1471-2407-11-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 9.Mathiyalagan P, Sahoo S. Exosomes-based gene therapy for microRNA delivery. Methods Mol Biol. 2017;1521:139–152. doi: 10.1007/978-1-4939-6588-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhim H, Savagner P, Thibaudeau G, Thiery JP, Pavan WJ. Localization of a neural crest transcription factor, Slug, to mouse chromosome 16 and human chromosome 8. Mamm Genome. 1997;8(11):872–873. [DOI] [PubMed] [Google Scholar]
- 11.Cohen ME, Yin M, Paznekas WA, Schertzer M, Wood S, Jabs EW. Human SLUG gene organization, expression, and chromosome map location on 8q. Genomics. 1998;51(3):468–471. doi: 10.1006/geno.1998.5367 [DOI] [PubMed] [Google Scholar]
- 12.Richter A, Valdimarsdottir L, Hrafnkelsdottir HE, et al. BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells. 2014;32(3):636–648. doi: 10.1002/stem.1592 [DOI] [PubMed] [Google Scholar]
- 13.Bai L, Chang HM, Zhu YM, Leung PCK. Bone morphogenetic protein 2 increases lysyl oxidase activity via up-regulation of snail in human granulosa-lutein cells. Cell Signal. 2019;53:201–211. doi: 10.1016/j.cellsig.2018.10.009 [DOI] [PubMed] [Google Scholar]
- 14.Chang TH, Tsai MF, Su KY, et al. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med. 2011;183(8):1071–1079. doi: 10.1164/rccm.201009-1440OC [DOI] [PubMed] [Google Scholar]
- 15.Findlay VJ, Wang C, Nogueira LM, et al. SNAI2 modulates colorectal cancer 5-fluorouracil sensitivity through miR145 repression. Mol Cancer Ther. 2014;13(11):2713–2726. doi: 10.1158/1535-7163.MCT-14-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari-Amorotti G, Chiodoni C, Shen F, et al. Suppression of invasion and metastasis of triple-negative breast cancer lines by pharmacological or genetic inhibition of slug activity. Neoplasia. 2014;16(12):1047–1058. doi: 10.1016/j.neo.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat Med. 2002;8(7):751–755. doi: 10.1038/nm720 [DOI] [PubMed] [Google Scholar]
- 18.Laakkonen P, Akerman ME, Biliran H, et al. Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells. Proc Natl Acad Sci U S A. 2004;101(25):9381–9386. doi: 10.1073/pnas.0403317101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth L, Agemy L, Kotamraju VR, et al. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene. 2012;31(33):3754–3763. doi: 10.1038/onc.2011.537 [DOI] [PubMed] [Google Scholar]
- 20.Dong P, Cai H, Chen L, et al. Biodistribution and evaluation of 131 I-labeled neuropilin-binding peptide for targeted tumor imaging. Contrast Media Mol Imaging. 2016;11(6):467–474. doi: 10.1002/cmmi.1708 [DOI] [PubMed] [Google Scholar]
- 21.Liang DS, Su HT, Liu YJ, Wang AT, Qi XR. Tumor-specific penetrating peptides-functionalized hyaluronic acid-d-α-tocopheryl succinate based nanoparticles for multi-task delivery to invasive cancers. Biomaterials. 2015;71:11–23. doi: 10.1016/j.biomaterials.2015.08.035 [DOI] [PubMed] [Google Scholar]
- 22.Choi KM, Kwon IC, Ahn HJ. Selfassembled amphiphilic DNAcholesterol/DNApeptide hybrid duplexes with liposomelike structure for doxorubicin delivery. Biomaterials. 2013;34(16):4183–4190. doi: 10.1016/j.biomaterials.2013.02.044 [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microrna targets. Cell. 2003;115(7):787–798. [DOI] [PubMed] [Google Scholar]
- 24.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 26.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9(4):293–302. doi: 10.1038/nrc2619 [DOI] [PubMed] [Google Scholar]
- 27.Shiraishi T, Matsuyama S, Kitano H. Large-scale analysis of network bistability for human cancers. PLoS Comput Biol. 2010;6(7):e1000851. doi: 10.1371/journal.pcbi.1000851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Béchec A, Portales-Casamar E, Vetter G, et al. MIR@NT@N: a framework integrating transcription factors, microRNAs and their targets to identify sub-network motifs in a meta-regulation network model. BMC Bioinformatics. 2011;12:67. doi: 10.1186/1471-2105-12-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Sen J, Bathula SR, Yang Q, Fittipaldi R, Huang L. Novel cationic lipid that delivers siRNA and enhances therapeutic effect in lung cancer cells. Mol Pharm. 2009;6(3):696–705. doi: 10.1021/mp800136v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi JF, Sun MG, Li XY, et al. A combination of targeted sunitinib liposomes and targeted vinorelbine liposomes for treating invasive breast cancer. J Biomed Nanotechnol. 2015;11(9):1568–1582. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Ahn RW, Chen F, Chen H, et al. A novel nanoparticulate formulation of arsenic trioxide with enhanced therapeutic efficacy in a murine model of breast cancer. Clin Cancer Res. 2010;16(14):3607–3617. doi: 10.1158/1078-0432.CCR-10-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watt KI, Jaspers RT, Atherton P, et al. SB431542 treatment promotes the hypertrophy of skeletal muscle fibers but decreases specific force. Muscle Nerve. 2010;41(5):624–629. doi: 10.1002/mus.21573 [DOI] [PubMed] [Google Scholar]
- 34.Huang RL, Yuan Y, Zou GM, Liu G, Tu J, Li Q. LPS-stimulated inflammatory environment inhibits BMP-2-induced osteoblastic differentiation through crosstalk between TLR4/MyD88/NF-κB and BMP/Smad signaling. Stem Cells Dev. 2014;23(3):277–289. doi: 10.1089/scd.2013.0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- 36.Podo F, Buydens LMC, Degani H, et al; FEMME Consortium. Triple-negative breast cancer: present challenges and new perspectives. Mol Oncol. 2010;4(3):209–229. doi: 10.1016/j.molonc.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, von Maltzahn G, Xu MJet al,. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc Natl Acad Sci U S A. 2010;107(3):981–986. doi: 10.1073/pnas.0909565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao S, Zhao X, Zhang X, et al. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer. 2015;14:28. doi: 10.1186/s12943-014-0278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res. 2012;31:58. doi: 10.1186/1756-9966-31-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S, Han J, Zheng L, Yang Z, Zhao L, Lv Y. MicroRNA-203 regulates growth and metastasis of breast cancer. Cell Physiol Biochem. 2015;37(1):35–42. doi: 10.1159/000430331 [DOI] [PubMed] [Google Scholar]
- 41.Taipaleenmäki H, Browne G, Akech J, et al. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75(7):1433–1444. doi: 10.1158/0008-5472.CAN-14-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koning GA, Eggermont AMM, Lindner LH, ten Hagen TL. Hyperthermia and thermosensitive liposomes for improved delivery of chemotherapeutic drugs to solid tumors. Pharm Res. 2010;27(8):1750–1754. doi: 10.1007/s11095-010-0154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang J, Nakamura H, Maeda H. The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Deliv Rev. 2011;63(3):136–151. doi: 10.1016/j.addr.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 44.Danquah MK, Zhang XA, Mahato RI. Extravasation of polymeric nanomedicines across tumor vasculature. Adv Drug Deliv Rev. 2011;63(8):623–639. doi: 10.1016/j.addr.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 45.Modica-Napolitano JS, Aprille JR. Delocalized lipophilic cations selectively target the mitochondria of carcinoma cells. Adv Drug Deliv Rev. 2001;49(1–2):63–70. [DOI] [PubMed] [Google Scholar]
- 46.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The possible “proton sponge “ effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol Ther. 2013;21(1):149–157. doi: 10.1038/mt.2012.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bramlage CP, Müller GA, Tampe B, et al. The role of bone morphogenetic protein-5 (BMP-5) in human nephrosclerosis. J Nephrol. 2011;24(5):647–655. doi: 10.5301/JN.2011.6330 [DOI] [PubMed] [Google Scholar]
- 48.Zhou S, Sun X, Yu L, et al. Differential expression and clinical significance of epithelial-mesenchymal transition markers among different histological types of triple-negative breast cancer. J Cancer. 2018;9(3):604–613. doi: 10.7150/jca.19190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai JW, Chen MN, Wei XL, et al. The zinc-finger transcriptional factor Slug transcriptionally downregulates ERα by recruiting lysine-specific demethylase 1 in human breast cancer. Oncogenesis. 2017;6(5):e330. doi: 10.1038/oncsis.2017.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travers MT, Barrett-Lee PJ, Berger U, et al. Growth factor expression in normal, benign, and malignant breast tissue. Br Med J (Clin Res Ed). 1988;296(6637):1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romagnoli M, Belguise K, Yu Z, et al. Epithelial-to-mesenchymal transition induced by TGF-β1 is mediated by Blimp-1-dependent repression of BMP-5. Cancer Res. 2012;72(23):6268–6278. doi: 10.1158/0008-5472.CAN-12-2270 [DOI] [PMC free article] [PubMed] [Google Scholar]