Abstract
Objectives
Tongue squamous cell carcinoma (TSCC) is the most common oral tumours. MicroRNAs play crucial roles in many cell processes including cell viability, development, apoptosis, migration and invasion. The role of miR‐802 in the TSCC is still unknown.
Materials and methods
The miR‐802 expression in TSCC tissues and cell lines was determined by quantitative real‐time polymerase chain reaction. CCK‐8 assay was performed to measure the cell viability, while the cell invasion assay was used to determine the cell invasion. Dual‐luciferase reporter and western blot were used to confirm the potential target gene of miR‐802.
Results
In our study, we demonstrated that miR‐802 expression was downregulated in TSCC tissues and cell lines. Elevated expression of miR‐802 suppressed the TSCC cell viability and invasion. Moreover, enforced expression of miR‐802 increased the expression of E‐cadherin, while suppressed the expression of N‐cadherin, Snail and Vimentin in the TSCC cell. In addition, we identified the mitogen‐activated protein kinase 4 (MAP2K4) as a direct target gene of miR‐802 in the TSCC cell. We also demonstrated that the expression of MAP2K4 was higher in the TSCC tissues than that in the adjacent normal tissues. Furthermore, the expression level of MAP2K4 was inversely associated with the expression of miR‐802 in TSCC tissues. We also demonstrated that the MAP2K4 expression was upregulated in TSCC cell lines. Elevated expression of miR‐802 inhibited TSCC cell viability and invasion through inhibiting MAP2K4 expression.
Conclusions
Our data revealed that miR‐802 played as a tumour suppressor gene and might act as a therapeutic target in TSCC patients.
Keywords: MAP2K4, microRNAs, miR‐802, tongue squamous cell carcinoma
1. Introduction
Squamous cell carcinoma of the oral cavity is the 10th most common solid cancer in the world.1, 2, 3, 4 Tongue squamous cell carcinoma (TSCC) is the most common oral tumours and usually leads to the malfunction of speech, mastication and deglutition.5, 6, 7, 8 Explanation of the molecular mechanism of the metastasis of TSCC is important for the cancer mortality reduction.9, 10 Thus, it is critical to identify new insights into the mechanisms of the carcinogenesis of TSCC.
MicroRNAs (miRNAs) are a group of small, non‐coding endogenous regulatory RNAs that regulate the protein‐coding genes through decreasing the stability or repressing translation of mRNAs.11, 12, 13, 14 Abnormal miRNA expression is found in various tumours such as gastric cancer, renal cell carcinoma, breast cancer, ovarian cancer, osteosarcoma and also the TSCC.15, 16, 17, 18, 19, 20 Increasing studies have demonstrated that miRNAs play a pivotal role in a broad assay of cell processes including cell development, viability, differentiation, migration, invasion and apoptosis.21, 22, 23, 24, 25
Previous study reported that miR‐802 expression was downregulated in the breast cancer cells and tissues.26 Overexpression of miR‐802 suppressed the breast cancer cell viability by inhibiting the Forkhead box protein M1 (FoxM1) expression. Li et al.27 showed that NF‐κB pathway was activated in the cholesteatoma and NF‐κB activation promoted miR‐802 expression in the cholesteatoma. However, the role of miR‐802 in the TSCC is still unknown. In this study, we found that miR‐802 expression was downregulated in TSCC tissues and cell lines. Elevated expression of miR‐802 suppressed the TSCC cell viability and invasion. We also identified mitogen‐activated protein kinase 4 (MAP2K4) as a direct target gene of miR‐802 in TSCC cell.
2. Materials and methods
2.1. Tissues, cell lines and culture and transfection
Tongue squamous cell carcinoma tissues and the adjacent human normal tissues were collected from TSCC cases during surgery from our department. Our study was approved by the Institutional Review Boards of Jinan Stomatological Hospital and all patients provided the written informed consent. Four TSCC cell lines (SCC1, SCC4, Cal27 and UM1) and normal oral keratinocyte cell culture (NHOK) and immortalized NOK16B cell line were bought from ATCC company (American Type Culture Collection) and were maintained in the DMEM/F12 medium with the foetal bovine serum (FBS; Gibco, Grand Island, NY, USA). miR‐802 and the scramble oligonucleotides were purchased from the GenePharma (Shanghai, China) and were transfected into UM1 cell using the Lipofectamine 2000 Transfection kit (Invitrogen Corporation, Carlsbad, CA, USA) following to the manufacturer's recommendations.
2.2. Quantitative real‐time polymerase chain reaction
The total RNA was extracted from tissue or cell using TRIzol reagent (Invitrogen, Kit). The relative expression of miR‐802 and MAP2K4 was detected using quantitative real‐time polymerase chain reaction with SYBR Green on the AB7300 thermo‐system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's recommendations. The relative expression of miRNA and mRNA was calculated using standard DDCT method, where GAPDH and U6 were determined as the control for mRNA and miRNA respectively.
2.3. Cell viability and cell invasion assays
Cell Counting Kit‐8 (CCK8; Dojindo, Tokyo, Japan) was used for cell viability. Cell was cultured on the 96‐well plate and the number of cells was measured by measurement of an absorbance at 450 nm using the microplate reader (Bio‐Rad, Hercules, CA, USA). Transwell matrigel chambers coated with matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were used to perform the invasion assay. Cells were seeded onto the upper matrigel chamber with serum‐free DMEM. The lower chamber contained DMEM with 20% FBS. After 48 hours, the cells that invade to the lower surface were collected. The invasive cells were stained and then counted under a microscope.
2.4. Western blot
Primary antibodies, including MAP2K4 and GAPDH (Abcam, Cambridge, UK), were used in our study. Total protein was extracted from cell or tissue using radioimmunoprecipitation lysis. Equal protein was loaded on 10% SDS‐PAGE and then transferred to the PVDF membrane. After blocking with milk, the PVDF membrane was incubated with primary antibodies (MAP2K4 and GAPDH) overnight. Next, the membrane was incubated with the horseradish peroxidase‐conjugated secondary antibodies and the blot was measured by the ECL (enhanced chemiluminescence solution; GE Healthcare, Kontaktuppgifter, Sweden).
2.5. Dual‐luciferase reporter assay
A fragment from the 3′‐UTR (3′ untranslated region) of MAP2K4 containing the predicted miR‐802‐binding site was cloned into the pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The mutant vector was created through mutating the binding site region of the miR‐802. Using lipofectamine‐2000 (Invitrogen), cell was transfected with wild‐type (WT) MAP2K4 3′‐UTR or mutated (MUT) MAP2K4 3′‐UTR and miR‐802 and the scramble vector. The luciferase activity was then measured by the Dual‐Luciferase Reporter System (Promega).
2.6. Statistical analysis
Result was presented as mean ± SD (standard deviation) and the statistical analysis was performed using the spss 17.0 software (SPSS Inc., Chicago, IL, USA). Student's t‐test and the one‐way analysis of variance were used for detecting the statistical significance of the different groups. P<.05 was indicated the statistically significant.
3. Result
3.1. The expression of miR‐802 was decreased in TSCC tissues
We firstly detected the miR‐802 expression in TSCC tissues. The expression of miR‐802 in 20 pairs of TSCC samples was shown in Figure 1A. The miR‐802 expression level was downregulated in the 12 TSCC cases (12/20; 60%) compared to that in the adjacent normal tissues (Figure 1B). The expression of miR‐802 was lower in the TSCC tissues than that in the adjacent normal tissues (Figure 1C).
Figure 1.
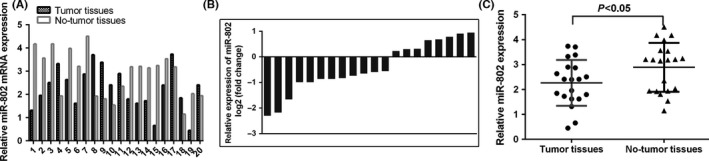
The expression of miR‐802 was decreased in tongue squamous cell carcinoma (TSCC) tissues. A, The expression of miR‐802 in 20 pairs of TSCC samples was shown. miR‐802 expression was detected by quantitative real‐time polymerase chain reaction. B, The miR‐802 expression level was downregulated in the 12 TSCC cases (12/20; 60%) compared to that in the adjacent normal tissues. C, The expression of miR‐802 was lower in the TSCC tissues than that in the adjacent normal tissues
3.2. Overexpression of miR‐802 suppressed the TSCC cell viability
We next demonstrated that the miR‐802 expression was downregulated in four TSCC cell lines (SCC1, SCC4, Cal27 and UM1) compared to that in the normal oral keratinocyte cell culture (NHOK) and immortalized NOK16B cell line (Figure 2A). The expression level of miR‐802 was significantly increased in the SCC1 cell after treated with miR‐802 mimic (Figure 2B). Elevated expression of miR‐802 suppressed SCC1 cell viability (Figure 2C). Furthermore, overexpression of miR‐802 decreased the ki‐67 expression in SCC1 cell.
Figure 2.
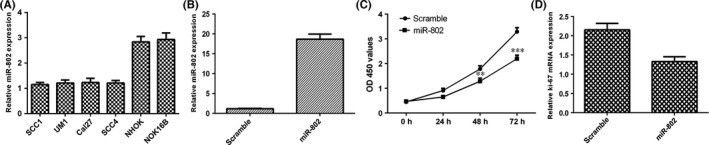
Overexpression of miR‐802 suppressed the tongue squamous cell carcinoma (TSCC) cell viability. A, The miR‐802 expression in four TSCC cell lines (SCC1, SCC4, Cal27 and UM1) and normal oral keratinocyte cell culture (NHOK) and immortalized NOK16B cell line was determined by quantitative real‐time polymerase chain reaction. B, The expression level of miR‐802 was significantly increased in the SCC1 cell after treated with miR‐802 mimic. C, Elevated expression of miR‐802 suppressed SCC1 cell viability. D, Overexpression of miR‐802 decreased the ki‐67 expression in SCC1 cell. P<.01, and ***P<.001
3.3. Elevated expression of miR‐802 inhibited the TSCC cell invasion
Moreover, we demonstrated that overexpression of miR‐802 suppressed SCC1 cell invasion (Figure 3A) and the relative invasive cells were shown in Figure 3B. We showed that overexpression of miR‐802 increased the E‐cadherin expression (Figure 3C), while suppressed the expression of N‐cadherin (Figure 3D), Snail (Figure 3E) and Vimentin (Figure 3F) in the SCC1 cell.
Figure 3.
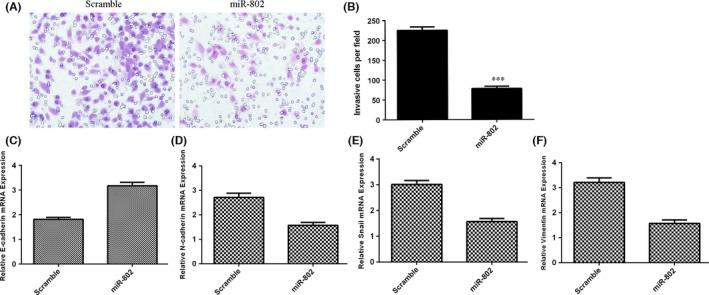
Elevated expression of miR‐802 inhibited the TSCC cell invasion. A, Overexpression of miR‐802 suppressed SCC1 cell invasion. B, The relative invasive cells were shown. C, The expression of E‐cadherin was determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). D, Overexpression of miR‐802 decreased the N‐cadherin expression. E, The expression of Snail was determined by qRT‐PCR. F, Elevated expression of miR‐802 suppressed the Vimentin expression. ***P<.001
3.4. MAP2K4 was a target gene of miR‐802 in TSCC cell and MAP2K4 expression was upregulated in TSCC tissues
The TargetScan database was used to predict the binding sites for miR‐802 within MAP2K4 (Figure 4A). Furthermore, our data showed that the luciferase activity was decreased in SCC1 cell after treated with the miR‐802 mimic and MAP2K4‐WT plasmids, whereas the miR‐802‐mutant plasmids did not change the reporter gene activity (Figure 4B). Overexpression of miR‐802 suppressed the MAP2K4 expression in SCC1 cell (Figure 4C). We next determined the MAP2K4 expression in TSCC tissues. The expression of MAP2K4 in 20 pairs of TSCC samples was shown in Figure 4D. The MAP2K4 expression level was upregulated in 14 TSCC cases (14/20; 70%) compared to that in the adjacent normal tissues (Figure 4E). The expression of MAP2K4 was higher in TSCC tissues than in the adjacent normal tissues (Figure 4F). In addition, the expression level of MAP2K4 was inversely associated with miR‐802 in TSCC tissues (Figure 4G).
Figure 4.
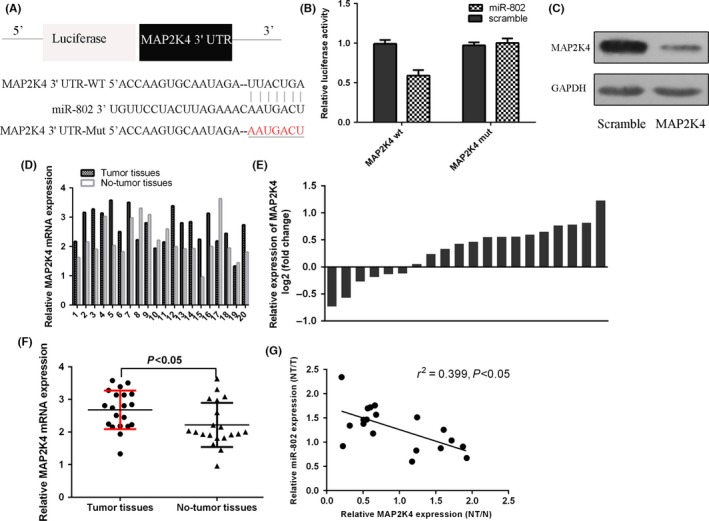
MAP2K4 was a target gene of miR‐802 in tongue squamous cell carcinoma (TSCC) cell and MAP2K4 expression was upregulated in TSCC tissues. A, The binding sites for miR‐802 within MAP2K4 were shown. B, The luciferase activity was decreased in SCC1 cell after treated with the miR‐802 mimic and MAP2K4‐WT plasmids. C, Overexpression of miR‐802 suppressed the MAP2K4 expression in SCC1 cell. D, The expression of MAP2K4 in 20 pairs of TSCC samples was shown. E, The MAP2K4 expression level was upregulated in 14 TSCC cases (14/20; 70%) compared to that in the adjacent normal tissues. F, The expression of MAP2K4 was higher in TSCC tissues than in the adjacent normal tissues. G, The expression level of MAP2K4 was inversely associated with miR‐802 in TSCC tissues
3.5. Elevated expression of miR‐802 inhibited TSCC cell viability and invasion through targeting MAP2K4
We showed that the MAP2K4 expression was upregulated in four TSCC cell lines (SCC1, SCC4, Cal27 and UM1) compared to that in NHOK and NOK16B cell lines (Figure 5A). The expression of MAP2K4 was significantly upregulated in the SCC1 cell treated with MAP2K4 vector (Figure 5B). We further determined whether MAP2K4 was involved in the function of miR‐802 in SCC1 cells and we restored EPHA7 expression by transfecting MAP2K4 expression vectors into the miR‐448‐overexpressing SCC1 cells. Next, we performed the CCK‐8 assay to evaluate the cell viability and the result showed that overexpressed MAP2K4 promoted SCC1 cell viability, reversing miR‐802 inhibition of viability (Figure 5C). Overexpression of MAP2K4 promoted the cell invasion in the miR‐802‐overexpressing SCC1 cell (Figure 5D,E). We also demonstrated that elevated expression of MAP2K4 suppressed the E‐cadherin expression (Figure 5F) and promoted the N‐cadherin (Figure 5G), Snail (Figure 5H) and Vimentin (Figure 5I) in the miR‐802‐overexpressing SCC1 cell.
Figure 5.
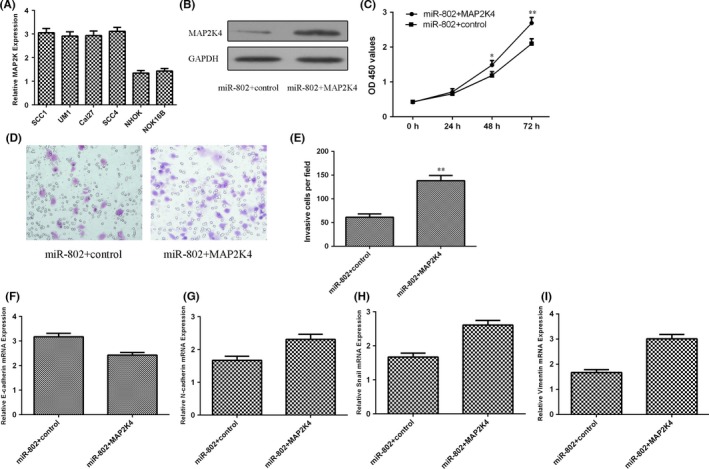
Elevated expression of miR‐802 inhibited tongue squamous cell carcinoma (TSCC) cell viability and invasion through targeting MAP2K4. A, The MAP2K4 expression in four TSCC cell lines (SCC1, SCC4, Cal27 and UM1) and NHOK and NOK16B cell lines was determined by quantitative real‐time polymerase chain reaction (qRT‐PCR). B, The protein expression of MAP2K4 was detected by western blot. C, Overexpressed MAP2K4 promoted miR‐802‐transfected SCC1 cell viability. D, Overexpression of miR‐802 suppressed the TSCC cell invasion. E, The relative invasive cells were shown. F, The expression of E‐cadherin was determined by qRT‐PCR. G, Overexpression of MAP2K4 increased the N‐cadherin expression in the miR‐802‐transfected SCC1 cell. H, The expression of Snail was determined by qRT‐PCR. I, Elevated expression of MAP2K4 promoted the Vimentin expression in the miR‐802‐transfected SCC1 cell. *P<.05 and **P<.01
4. Discussion
In the present study, we demonstrated that miR‐802 expression was downregulated in TSCC tissues and cell lines. Elevated expression of miR‐802 suppressed TSCC cell viability and invasion. Moreover, enforced expression of miR‐802 increased the E‐cadherin expression and suppressed the expression of N‐cadherin, Snail and Vimentin in TSCC cells. In addition, we identified the MAP2K4 as a direct target gene of miR‐802 in TSCC cell. We also demonstrated that the expression of MAP2K4 was higher in the TSCC tissues than that in the adjacent normal tissues. Furthermore, the expression level of MAP2K4 was inversely associated with the expression of miR‐802 in TSCC tissues. We also demonstrated that the MAP2K4 expression was upregulated in TSCC cell lines. Elevated expression of miR‐802 inhibited TSCC cell viability and invasion through suppressing the MAP2K4 expression. Our data revealed that miR‐802 played as a tumour suppressor gene and might act as a therapeutic target in TSCC patients.
Previous study reported that miR‐802 expression level was downregulated in the breast cancer cells and tissues.26 Overexpression of miR‐802 suppressed the breast cancer cell viability by inhibiting the FoxM1 expression. Li et al.27 demonstrated that NF‐κB pathway was activated in the cholesteatoma and NF‐κB activation promoted miR‐802 expression in the cholesteatoma. Overexpression of miR‐802 increased keratinocyte cell cycle progression and cell viability through directly repressing the PTEN expression in cholesteatoma. However, Cao et al.28 found that miR‐802 expression was upregulated in osteosarcoma tissues. Elevated expression of miR‐802 promoted osteosarcoma cell viability through directly targeting p27. However, the expression and biological function of miR‐802 in the TSCC remain unclear. Here, miR‐802 expression was downregulated in TSCC cells and tissues. Furthermore, elevated expression of miR‐802 suppressed TSCC cell viability and invasion. Moreover, overexpression of miR‐802 increased the E‐cadherin expression and inhibited the expression of N‐cadherin, Snail and Vimentin in the TSCC cell. These results suggest that miR‐802 acts as tumour suppressor in TSCC.
MAP2K4 is one member of MAPK signalling pathway family and is located on the chromosome 17, which encodes a 399‐amino‐acids protein.29, 30, 31 MAP2K4 is a kind of protein kinase with tyrosine residues, threonine and phosphorylates serine.32, 33, 34 MAP2K4 was an important biology signal and played a critical role in various cellular processes including cell development, viability, invasion, migration and differentiation.35, 36, 37, 38 Previous studies showed that MAP2K4 acted as an important role in a lot of tumours such as prostate cancer, breast cancer, osteosarcoma and lung cancer.29, 30, 35, 39 In our study, we identified MAP2K4 as a direct target gene of miR‐802 in TSCC cell by using dual‐luciferase reporter assay and western blot. Moreover, we demonstrated that the MAP2K4 expression was upregulated in TSCC tissues compared to that in the adjacent normal tissues. In addition, the expression level of MAP2K4 was inversely associated with the expression of miR‐802 in TSCC tissues. Furthermore, we performed the rescue experiment to study the role MAP2K4 in TSCC cell. Our result demonstrated that elevated expression of miR‐802 inhibited TSCC cell viability and invasion through inhibiting MAP2K4. These data suggested that miR‐802 suppressed cell viability and invasion in TSCC cells partially by inhibiting MAP2K4 expression.
In conclusion, our result showed that miR‐802 expression was downregulated in TSCC tissues and cell lines. Elevated expression of miR‐802 suppressed TSCC cell viability and invasion by inhibiting MAP2K4. These data revealed that miR‐802 acted as a tumour suppressor gene and might act as a therapeutic target in TSCC patients.
Acknowledgements
This study was supported by “Health Science and Technology Development Program Project of Jinan” (no. 2011‐14) and “Science and Technology Development Program Project of Jinan” (no. 201401082).
Wu X, Gong Z, Sun L, Ma L, Wang Q. MicroRNA‐802 plays a tumour suppressive role in tongue squamous cell carcinoma through directly targeting MAP2K4. Cell Prolif. 2017;50:e12336 10.1111/cpr.12336
Xiaozhen Wu and Zuo‐de Gong are co‐first authors.
References
- 1. Ratovitski EA. Phospho‐DeltaNp63alpha‐dependent microRNAs modulate chemoresistance of squamous cell carcinoma cells to cisplatin: at the crossroads of cell life and death. FEBS Lett. 2013;587:2536–2541. doi: 10.1016/j.febslet.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 2. Jia LF, Wei SB, Gan YH, et al. Expression, regulation and roles of miR‐26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer. 2014;135:2282–2293. doi: 10.1002/ijc.28667. [DOI] [PubMed] [Google Scholar]
- 3. Jia LF, Wei SB, Gong K, Gan YH, Yu GY. Prognostic implications of micoRNA miR‐195 expression in human tongue squamous cell carcinoma. PLoS ONE. 2013;8:e56634. doi: 10.1371/journal.pone.0056634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu X, Wang A, Heidbreder CE, et al. MicroRNA‐24 targeting RNA‐binding protein DND1 in tongue squamous cell carcinoma. FEBS Lett. 2010;584:4115–4120. doi: 10.1016/j.febslet.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang L, Liu X, Chen Z, et al. MicroRNA‐7 targets IGF1R (insulin‐like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432:199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu JY, Yang LL, Ma C, Huang YL, Zhu GX, Chen QL. MiR‐25‐3p attenuates the proliferation of tongue squamous cell carcinoma cell line Tca8113. Asian Pac J Trop Med. 2013;6:743–747. doi: 10.1016/S1995-7645(13)60130-3. [DOI] [PubMed] [Google Scholar]
- 7. Kai Y, Peng W, Ling W, Jiebing H, Zhuan B. Reciprocal effects between microRNA‐140‐5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem Biophys Res Commun. 2014;448:308–314. doi: 10.1016/j.bbrc.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 8. Yan B, Fu Q, Lai L, et al. Downregulation of microRNA 99a in oral squamous cell carcinomas contributes to the growth and survival of oral cancer cells. Mol Med Rep. 2012;6:675–681. doi: 10.3892/mmr.2012.971. [DOI] [PubMed] [Google Scholar]
- 9. Jiang L, Dai Y, Liu X, et al. Identification and experimental validation of G protein alpha inhibiting activity polypeptide 2 (GNAI2) as a microRNA‐138 target in tongue squamous cell carcinoma. Hum Genet. 2011;129:189–197. doi: 10.1007/s00439-010-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tseng SH, Yang CC, Yu EH, et al. K14‐EGFP‐miR‐31 transgenic mice have high susceptibility to chemical‐induced squamous cell tumorigenesis that is associating with Ku80 repression. Int J Cancer. 2015;136:1263–1275. doi: 10.1002/ijc.29106. [DOI] [PubMed] [Google Scholar]
- 11. Ahmad A, Sethi S, Chen W, Ali‐Fehmi R, Mittal S, Sarkar FH. Up‐regulation of microRNA‐10b is associated with the development of breast cancer brain metastasis. Am J Transl Res. 2014;6:384–390. [PMC free article] [PubMed] [Google Scholar]
- 12. Hu A, Huang JJ, Xu WH, et al. miR‐21 and miR‐375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am J Transl Res. 2014;6:604–613. [PMC free article] [PubMed] [Google Scholar]
- 13. Huang K, Dong X, Sui C, et al. MiR‐223 suppresses endometrial carcinoma cells proliferation by targeting IGF‐1R. Am J Transl Res. 2014;6:841–849. [PMC free article] [PubMed] [Google Scholar]
- 14. Jiao Y, Zhu M, Mao X, et al. MicroRNA‐130a expression is decreased in Xinjiang Uygur patients with type 2 diabetes mellitus. Am J Transl Res. 2015;7:1984–1991. [PMC free article] [PubMed] [Google Scholar]
- 15. Kang M, Ren MP, Zhao L, Li CP, Deng MM. miR‐485‐5p acts as a negative regulator in gastric cancer progression by targeting flotillin‐1. Am J Transl Res. 2015;7:2212–2222. [PMC free article] [PubMed] [Google Scholar]
- 16. Niu G, Li B, Sun J, Sun L. miR‐454 is down‐regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c‐Met. Cell Prolif. 2015;48:348–355. doi: 10.1111/cpr.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR‐99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–595. doi: 10.1111/cpr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denoyelle C, Lambert B, Meryet‐Figuiere M, et al. miR‐491‐5p‐induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL‐XL and EGFR leading to BIM activation. Cell Death Dis. 2014;5:e1445. doi: 10.1038/cddis.2014.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su Z, Chen D, Li Y, et al. microRNA‐184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9:961–966. doi: 10.3892/etm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song KB, Liu WJ, Jia SS. miR‐219 inhibits the growth and metastasis of TSCC cells by targeting PRKCI. Int J Clin Exp Med. 2014;7:2957–2965. [PMC free article] [PubMed] [Google Scholar]
- 21. Yu X, Li Z, Chen G, Wu WK. MicroRNA‐10b induces vascular muscle cell proliferation through Akt pathway by targeting TIP30. Curr Vasc Pharmacol. 2015;13:679–686. [DOI] [PubMed] [Google Scholar]
- 22. Yu X, Li Z, Shen J, et al. MicroRNA‐10b promotes nucleus pulposus cell proliferation through RhoC‐Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS ONE. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Li Z, Yu X, Wang Y, et al. By downregulating TIAM1 expression, microRNA‐329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui Y, Xiao Z, Chen T, et al. The miR‐7 identified from collagen biomaterial‐based three‐dimensional cultured cells regulates neural stem cell differentiation. Stem Cells Dev. 2014;23:393–405. doi: 10.1089/scd.2013.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurashige J, Mima K, Sawada G, et al. Epigenetic modulation and repression of miR‐200b by cancer‐associated fibroblasts contribute to cancer invasion and peritoneal dissemination in gastric cancer. Carcinogenesis. 2015;36:133–141. doi: 10.1093/carcin/bgu232. [DOI] [PubMed] [Google Scholar]
- 26. Yuan F, Wang W. MicroRNA‐802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep. 2015;12:4647–4651. doi: 10.3892/mmr.2015.3921. [DOI] [PubMed] [Google Scholar]
- 27. Li N, Qin ZB. Inflammation‐induced miR‐802 promotes cell proliferation in cholesteatoma. Biotechnol Lett. 2014;36:1753–1759. doi: 10.1007/s10529-014-1545-y. [DOI] [PubMed] [Google Scholar]
- 28. Cao ZQ, Shen Z, Huang WY. MicroRNA‐802 promotes osteosarcoma cell proliferation by targeting p27. Asian Pac J Cancer Prev. 2013;14:7081–7084. [DOI] [PubMed] [Google Scholar]
- 29. Pavese JM, Ogden IM, Voll EA, et al. Mitogen‐activated protein kinase kinase 4 (MAP2K4) promotes human prostate cancer metastasis. PLoS ONE. 2014;9:e102289. doi: 10.1371/journal.pone.0102289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tesser‐Gamba F, Petrilli AS, de Seixas Alves MT, Filho RJ, Juliano Y, Toledo SR. MAPK7 and MAP2K4 as prognostic markers in osteosarcoma. Hum Pathol. 2012;43:994–1002. doi: 10.1016/j.humpath.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 31. Pazzaglia L, Chiechi A, Conti A, et al. Genetic and molecular alterations in rhabdomyosarcoma: mRNA overexpression of MCL1 and MAP2K4 genes. Histol Histopathol. 2009;24:61–67. [DOI] [PubMed] [Google Scholar]
- 32. Su GH, Song JJ, Repasky EA, Schutte M, Kern SE. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- 33. Xin W, Yun KJ, Ricci F, et al. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 34. Cunningham SC, Gallmeier E, Hucl T, et al. Theoretical proposal: allele dosage of MAP2K4/MKK4 could rationalize frequent 17p loss in diverse human cancers. Cell Cycle. 2006;5:1090–1093. doi: 10.4161/cc.5.10.2805. [DOI] [PubMed] [Google Scholar]
- 35. Lee JW, Soung YH, Kim SY, et al. Kinase domain mutation of MAP2K4 is rare in gastric, colorectal and lung carcinomas. Pathology. 2006;38:263–264. doi: 10.1080/00313020600699219. [DOI] [PubMed] [Google Scholar]
- 36. Davis SJ, Choong DY, Ramakrishna M, Ryland GL, Campbell IG, Gorringe KL. Analysis of the mitogen‐activated protein kinase kinase 4 (MAP2K4) tumor suppressor gene in ovarian cancer. BMC Cancer. 2011;11:173. doi: 10.1186/1471-2407-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahn YH, Yang Y, Gibbons DL, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator‐activated receptor gamma2 expression. Mol Cell Biol. 2011;31:4270–4285. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan W, Wang H, Jianwei R, Ye Z. MicroRNA‐27a promotes proliferation, migration and invasion by targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem. 2014;33:402–412. doi: 10.1159/000356679. [DOI] [PubMed] [Google Scholar]
- 39. Wan X, Huang W, Yang S, et al. Androgen‐induced miR‐27A acted as a tumor suppressor by targeting MAP2K4 and mediated prostate cancer progression. Int J Biochem Cell Biol. 2016;79:249–260. doi: 10.1016/j.biocel.2016.08.043. [DOI] [PubMed] [Google Scholar]


