Abstract
Objectives
The microenvironmental niche plays the key role for maintaining the cell functions. The stem cells from apical papilla (SCAPs) are important for tooth development and regeneration. However, there is limited knowledge about the key factors in niche for maintaining the function of SCAPs. In this study, we analyse the gene expression profiles between apical papilla tissues, SCAPs and SCAPs cell sheet to identify the key genes in SCAPs niche.
Materials and methods
Microarray assays and bioinformatic analysis were performed to screen the differential genes between apical papilla tissues and SCAPs, and SCAPs and SCAPs cell sheet. Recombinant human BMP6 protein was used in SCAPs. Then CCK‐8 assay, CFSE assay, alkaline phosphatase activity, alizarin red staining, quantitative calcium analysis and real‐time reverse transcriptase‐polymerase chain reaction were performed to investigate the cell proliferation and differentiation potentials of SCAPs.
Results
Microarray analysis found that 846 genes were up‐regulated and 1203 genes were down‐regulated in SCAPs compared with apical papilla tissues. While 240 genes were up‐regulated and 50 genes were down‐regulated in SCAPs compared to in SCAPs cell sheet. Moreover, only 31 gene expressions in apical papilla tissues were recovered in cell sheet compared with SCAPs. Bioinformatic analysis identified that TGF‐β, WNT and MAPK signalling pathways may play an important role in SCAPs niche. Based on the analysis, we identified one key growth factor in niche, BMP6, which could enhance the cell proliferation, the osteo/dentinogenic, neurogenic and angiogenic differentiation potentials of SCAPs.
Conclusions
Our results provided insight into the mechanisms of the microenvironmental niche which regulate the function of SCAPs, and identified the key candidate genes in niche to promote mesenchymal stem cells‐mediated dental tissue regeneration.
1. Introduction
In dental clinic, trauma and infection will cause development of immature permanent teeth root to stop. Now apexification and blood capillary regeneration were applied to promote root formation. However, these two kinds of treatment have the limitations and poor prognosis.1, 2 Today, utilizing mesenchymal stem cells (MSCs) and tissue engineering techniques to reconstruct the immature dental root may be the optimistic therapeutic implications. Stem cells derived from dental tissues, such as dental pulp, periodontal ligament, apical papilla and dental follicle, are considered as a new adult stem cells that could be used for tissue engineering and regenerative medicine.3, 4, 5, 6, 7, 8 They are multipotent, destined for osteo/dentinogenic lineages and other lineages such as melanocytes, endothelial cells and functionally active neurons, and capable of self‐renewal.9 The apical papilla is essential for tooth development, and stem cells from the apical papilla (SCAPs) represent a population of early mesenchymal stem/progenitor cells residing in the root apex of immature permanent teeth.10 These postnatal stem cells can generate the calcium nodules in the osteo/odontogenic medium in vitro. Besides, they can bring about the formation of bone‐like tissues and dentin‐like tissues in vivo.11 In addition, recent clinical reports indicate that SCAPs are important to the apexogenesis of developing roots and continuous root maturation in teenagers suffering from the endodontic diseases and periapical lesions.12 Many studies in terms of tooth regeneration are based on SCAPs and have witnessed exciting progress.13 Therefore, stem cells from the apical papilla are a reliable resource for dental tissue regeneration.
Microenvironmental niche supports and maintains the self‐renewal, differentiation and regeneration potentials of MSCs, and now ongoing research is starting to illuminate important aspects of the microenvironmental niche of MSCs. The microenvironmental niche is also an important factor in determining the behaviour of cells and the morphogenesis of teeth. In tooth tissue, stem cells, the growth factors and extracellular matrix (ECM) in niche, and their multiple interactions determine the tooth development, eruption and the biological basis. However, limited by the current methods, the niche cannot be maintained when MSCs are isolated and cultured in vitro. Disruption of the niche may impede the MSC‐mediated tooth regeneration.14, 15, 16 In last decades, tissue regeneration techniques mainly depend on scaffold‐based approaches. In dental root engineering, MSCs usually combine with the scaffold materials to regenerate the dental root. Scaffold‐based methods caused the insufficient cell migration, host inflammatory reactions, limited microscale vascularization, cell proliferation ability compared to degradation of scaffold and the incapability for regenerating functional tissues. Recently, use of continuous cell sheet technology brings more attention of scholars. As endogenous bioactive scaffolds, cell sheet preserve the normal cellular junctions, endogenous ECM, mimicking cellular microenvironments and store the mechanical, chemical and biological properties, which may be beneficial for tissue regeneration.17, 18, 19, 20, 21, 22 Except the structure, the microenvironmental niche also has complex genes regulation, especially the growth factors, which largely affect the function of MSCs. Whether the cell sheet could restore the genes regulation in niche remains unclear.
In this study, we analyse the gene expression profiles between apical papilla tissues, SCAPs and SCAPs cell sheet to identify the key genes in SCAPs niche by microarray and bioinformatic analysis, and investigate whether SCAPs cell sheet recover the genes regulation in niche. Furthermore, we investigate the function of candidate growth factor in niche by in vitro cell proliferation and differentiation assays. Our results identified the key candidate genes in niche to promote MSC‐mediated dental tissue regeneration.
2. Materials and methods
2.1. Tissue collection, isolation and culture of mesenchymal stem cells and cell sheets
All research involving human stem cells is in compliance with the ISSCR “Guidelines for the Conduct of Human Embryonic Stem Cell Research”. Human impacted third molar with immature roots were collected from 10 healthy female patients (18‐22 years old) under approved guidelines set by the Beijing Stomatological Hospital, Capital Medical University, with informed patient consent (Table S1). Wisdom teeth were first disinfected with 75% ethanol and then washed with phosphate‐buffered saline (PBS). SCAPs were isolated, cultured and identified as previously described.2 Briefly, the tissues were gently separated from the apical papilla of the root, and then digested in a solution of 3 mg/mL collagenase type I (Worthington Biochemical Corp., Lakewood, NJ, USA) and 4 mg/mL dispase (Roche Diagnostics Corp., Indianapolis, IN, USA) for 1 hour at 37°C. Single‐cell suspensions were obtained by passing the cells through a 70 μm strainer (Falcon, BD Labware, Franklin Lakes, NJ, USA). SCAPs were grown in a humidified, 5% CO2 incubator at 37°C in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 15% foetal bovine serum (FBS; Invitrogen), 2 mmol/L glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen). The culture medium was changed every 3 days. Cells from passages 2‐4 were used in further experiments. SCAPs were grown in a normal growth media with 20 μg/mL Vc (Invitrogen) for 2 weeks, then cell sheet was formed and used in further experiments. For recombinant human BMP6 (rhBMP6) (R&D system, Minneapolis, MN, USA) treatment, MSCs were starved for 24 hours to synchronize the cells in DMEM alpha modified Eagle's medium without serum, then changed to routine culture medium or different inducing medium with appropriate concentration of rhBMP6.
2.2. RNA preparation and microarray analysis
Tissues from apical papilla (n=5), SCAPs (n=5) and SCAPs cell sheet (n=5) were briefly rinsed with PBS and lysed in TRIzol reagent (Invitrogen) for RNA extraction. Total RNA was extracted from the tissues and cells using TRIzol and the RNeasy mini kit (Qiagen, Hilden, Germany), and RNA quality and quantity were confirmed by multiImager and spectrophotometer (Meriton, Beijing, China). Gene expression profiles were analysed with the human 1.0ST GeneChip (Affymetrix Santa Clara, CA, USA), strictly following the manufacturer's protocol. Microarray experiments were performed at the Genminix Informatic Ltd (Shanghai, China) with the microarray service certified by Affymetrix.
2.3. Bioinformatic analysis
The differentially expressed genes were selected using TwoClassDif. Gene ontology (GO) analysis was applied to determine the main function of the differentially expressed genes according to GO, which is the key functional classification of the NCBI.23 Pathway analysis of differentially expressed genes was performed based on KEGG and BioCarta database.24, 25 Signal‐net maps were also constructed to present the interaction of molecules based on KEGG.26
2.4. CCK8 assays
Stem cells from apical papilla were seeded at a density of 1.0 × 103 cells/well into 96‐well plates. Cells were grown in 96‐well plates for 1 and 2 days after seeding. A cell counting kit solution (Cell Counting kit‐8; Dojindo, Kumamoto, Japan) was then added to each well of the plate, and absorbance was measured at 450 nm, according to the manufacturer's protocol (Dojindo).
2.5. CFSE assays
Stem cells from apical papilla were stained according to the CellTrace™ CFSE Cell Proliferation Kit Protocol (Invitrogen) for labelling cells in suspension and then seeded at a density of 5.0 × 104 cells/plate in six‐well plates. SCAPs were harvested with 0.25% trypsin after 6 days’ culturing and analysed using a flow cytometer (Calibur; BD Biosciences, Franklin Lakes, NJ, USA) with 488 nm excitation and emission filters appropriate for fluorescein. The proliferation index was calculated by Modfit LT.
2.6. ALP activity assay and alizarin red staining
Stem cells from apical papilla were grown in mineralization‐inducing medium using the StemPro osteogenesis differentiation kit (Invitrogen). Cells were cultured for 3 days, and the alkaline phosphatase (ALP) activity assay was performed with an ALP kit according to the manufacturer's protocol (Sigma‐Aldrich, St. Louis, MO, USA) and normalized based on protein concentrations. The protein concentration was quantitatively determined using Bio‐Rad protein assay solution (Bio‐Rad Laboratories Hercules, CA, USA). For detecting mineralization, cells were induced for 2 weeks, fixed with 70% ethanol and stained with 2% alizarin red (Sigma‐Aldrich). To quantitatively determine calcium mineral content, alizarin red was destained with 10% cetylpyridinium chloride in 10 mmol/L sodium phosphate for 60 minutes at room temperature. The concentration was determined by absorbance measurement at 562 nm on a multiplate reader using a standard calcium curve in the same solution. The final calcium level in each group was normalized with the total protein concentrations prepared from a duplicate plate.
2.7. Neurogenic differentiation induction
Stem cells from apical papilla were seeded at a density of 2.0 × 105 cells/well into low adhesion plates. SCAPs were grown in modified neurogenic differentiation medium (Neurobasal‐A; Invitrogen) with 2 mmol/L glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen), 20 ng/mL EGF, 40 ng/mL bFGF and 20 μg/mL B27 (R&D system). After induction, neuron‐like cells were observed by using microscope. The real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) was used to detect the neurogenic differentiation markers.
2.8. Angiogenic differentiation induction
Stem cells from apical papilla were seeded at a density of 1.0 × 105 cells/well into six‐well plates. SCAPs were grown in vascular differentiation medium (M199; Invitrogen) with 10% FBS (Invitrogen), 2 mmol/L glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen), 10 ng/mL VEGF and 5 ng/mL bFGF (R&D system). After induction, real‐time RT‐PCR was used to detect the angiogenic differentiation markers.
2.9. Reverse transcriptase‐polymerase chain reaction and real‐time RT‐PCR
Total RNA was isolated from SCAPs with Trizol reagents (Invitrogen). We synthesized cDNA from 2 μg aliquots of RNA, random hexamers or oligo(dT), and reverse transcriptase, according to the manufacturer's protocol (Invitrogen). Real‐time PCR reactions were performed with the QuantiTect SYBR Green PCR kit (Qiagen) and an IcycleriQ Multi‐color Real‐time PCR Detection System. The primers for specific genes were shown in Table S2.
2.10. Statistics
All statistical calculations were performed using spss 10 statistical software. Student's t test was performed to determine statistical significance. A P‐value ≤.05 was considered statistically significant.
3. Results
3.1. Comparison of gene expression profiles between apical papilla tissues and SCAPs, and SCAPs and SCAPs cell sheet
First, we screened the gene expression patterns between apical papilla tissues and SCAPs using the human GeneChip. From the microarray data, 2049 genes were differently expressed in SCAPs compared with apical papilla tissues, with 846 genes up‐regulated and 1203 genes down‐regulated in SCAPs compared to in apical papilla tissues (Table S3). Furthermore, we screened the gene expression patterns between SCAPs and SCAPs cell sheet. From the microarray data, 290 genes were differently expressed in SCAPs cell sheet compared with SCAPs, with 240 genes up‐regulated and 50 genes down‐regulated in SCAPs compared to in SCAPs cell sheet (Table S4). To confirm the reliability of the microarray data, three up‐regulated genes (S100A4, FOXM1, FGF5) and three down‐regulated genes (CXCL14, IGF2, BMP6) in SCAPs were chosen, and the expressions of these genes were detected by real‐time RT‐PCR. The results showed that the expressions of these six genes were consistent with the microarray results in apical papilla tissues, SCAPs and SCAPs cell sheet, confirming the reliability of the microarray data (Table 1). Furthermore, based on data statistics, we found that only 31 genes expressions in apical papilla tissues were recovered in cell sheet compared with SCAPs (Table S5).
Table 1.
Gene expression detected by real‐time RT‐PCR in apical papilla tissues, SCAPs and SCAPs cell sheets
| Genes | Tissues | SCAPs | Cell sheets | Microarray results |
|---|---|---|---|---|
| BMP6a | 1.00 ± 0.39 | 0.002 ± 0.006** | 0.016 ± 0.024** | Down‐regulated in SCAPs |
| CXCL14 | 1.00 ± 0.27 | 0.00003 ± 0.004** | 0.000016 ± 0.003** | Down‐regulated in SCAPs |
| IGF2 | 1.00 ± 0.073 | 0.139 ± 0.034** | 0.245 ± 0.087** | Down‐regulated in SCAPs |
| FGF5a | 1.00 ± 0.95 | 651.40 ± 99.56** | 119.02 ± 40.04** | Up‐regulated in SCAPs |
| HOXM1a | 1.00 ± 0.54 | 92.45 ± 57.47** | 27.33 ± 12.86** | Up‐regulated in SCAPs |
| S100A4 | 1.00 ± 0.32 | 4.76 ± 4.68** | 4.912 ± 3.23** | Up‐regulated in SCAPs |
RT‐PCR, reverse transcriptase‐polymerase chain reaction; SCAPs, stem cells from apical papilla.
GAPDH was used as internal control. The results represent mean ± standard deviation from five independent experiments (mean value of gene expression/GAPDH in apical papilla tissues was set as 1). Student's t test was performed to determine statistical significance.
**P<.01.
aExpressions that were significantly different in SCAPs and SCAP cell sheets.
3.2. Bioinformatic analysis of microarray data
Finally, bioinformatic analysis was executed to discover the key factors that controlled the SCAP functions. First, GO analysis was applied to analyse the main function of the differentially expressed genes according to the gene ontology, which is the key functional classification of NCBI. The result showed that the top‐five GOs were responsible for cell adhesion, cell division, regulation of cell proliferation, extracellular structure organization and cell cycle process (Fig. S1). Similarly, pathway and path‐net analyses were used to determine the significant pathway of the differentially expressed genes according to the KEGG, and Biocarta and Reatome databases. We identified that the TGF‐β signalling pathway may play an important role in SCAPs, and that WNT and MAPK signalling pathways are also likely involved (Fig. S2). To further investigate the global network, we computationally identified the most important nodes by signal‐net analysis. ITGA9, PIK3R1, PIK3CG, FOS and ENTPD1 were determined to be the important genes by high betweenness centrality calculation (Fig. S3).
3.3. BMP6 enhanced the osteo/dentinogenic differentiation potential of SCAPs
Stem cells from apical papilla were cultured in mineralization‐inducing medium with 0, 5, 20 or 50 ng/mL rhBMP6. After 3 days, ALP activity results showed that 20 or 50 ng/mL rhBMP6 could increase ALP activity of SCAPs, while 5 ng/mL rhBMP6 had no effect (Figure 1A). Then 20 ng/mL of rhBMP6 was used for the further experiments. After culturing SCAPs in mineralization‐inducing medium for 2 weeks, alizarin red staining and calcium quantitative assay results revealed that mineralization was also significantly stronger in SCAPs with 20 ng/mL of rhBMP6 treatments than untreated SCAPs (Figure 1B,C). We examined the osteo/dentinogenic markers including DSPP, DMP1, BSP, OPN and OCN. The real‐time RT‐PCR results showed that DSPP and DMP1 were highly expressed in rhBMP6‐treated SCAPs compared with untreated group at 3, 7, 10 and 14 days after mineralization induction (Figure 1D,E). And BSP, which encodes ECM proteins of bone and dentin, was more strongly induced in rhBMP6‐treated SCAPs compared with untreated group at 10 days after mineralization induction (Figure 1F). Two other markers, OPN and OCN, were increased in rhBMP6‐treated SCAPs compared with untreated group at 14 days after mineralization induction (Figure 1G,H). We further examined the key transcription factors involved in osteo/dentinogenic differentiation including RUNX2 and OSX. Real‐time RT‐PCR results showed that OSX was significantly increased in rhBMP6‐treated SCAPs compared with untreated SCAPs at 3 and 7 days after mineralization induction (Figure 1I), and RUNX2 was significantly increased in BMP6‐treated SCAPs compared with untreated SCAPs at 3 days after mineralization induction (Figure 1J).
Figure 1.
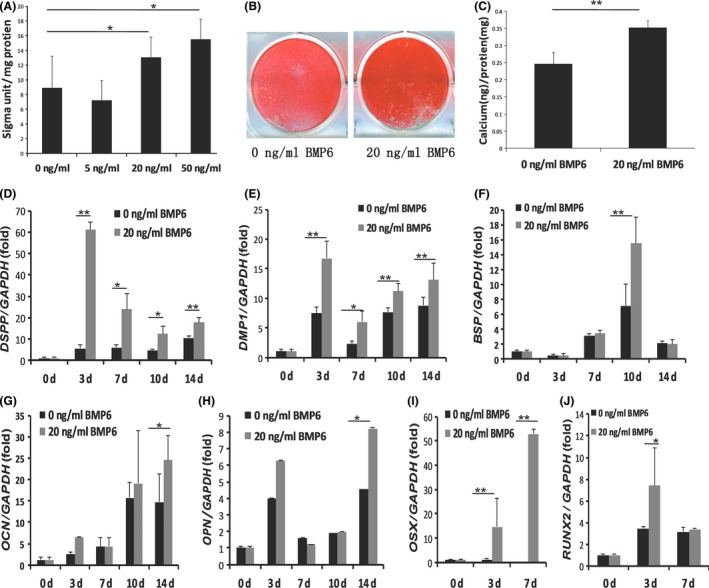
BMP6 enhanced the osteo/dentinogenic differentiation potential in stem cells from apical papilla (SCAPs). (A) Alkaline phosphatase (ALP) activity assays showed that 20 or 50 ng/mL BMP6 could increase ALP activity of SCAPs. (B, C) Alizarin Red staining (B) and calcium quantitative analysis (C) showed that 20 ng/mL BMP6 significantly enhanced the mineralization of SCAPs. (D‐J) Real‐time RT‐PCR results showed that DSPP (D), DMP1 (E), BSP (F), OPN (G), OCN (H), OSX (I), and RUNX2 (J) were highly expressed in 20 ng/mL of BMP6‐treated SCAPs compared with untreated SCAPs after mineralization induction. GAPDH was used as an internal control. Student's t test was performed to determine statistical significance. All error bars represent SD (n=5). *P<.05, **P<.01
3.4. BMP6 prompted the expressions of neurogenic and angiogenic differentiation markers in SCAPs
Stem cells from apical papilla were culturing with neurogenic‐inducing medium with 20 ng/mL of rhBMP6 or without rhBMP6 for 10 days, we examined the neurogenic differentiation markers including TH‐1, NeuroD, NCAM and βIII tubulin. The real‐time RT‐PCR results showed that TH‐1, as the key marker of neuroblast, was highly expressed in BMP6‐treated SCAPs compared with untreated SCAPs at 10 days after neurogenic induction (Figure 2A). And the neural markers, NeuroD and NCAM were more strongly induced in BMP6‐treated SCAPs compared with untreated group at 10 days after neurogenic induction (Figure 2B,C). However, the expression level of βIII tubulin was not significantly different after BMP6 treatment (data not shown). Then SCAPs were culturing with angiogenic‐inducing medium with 20 ng/mL of rhBMP6 or without rhBMP6 for 10 days, we examined the angiogenic differentiation markers including ANG1, PDGF, VEGF and HGF. The real‐time RT‐PCR results showed that the mRNA level of ANG1 was significantly increased in BMP6‐treated SCAPs compared with untreated group at 10 days after angiogenic induction (Figure 2D). However, the expressions of PDGF, VEGF and HGF were not significantly different after BMP6 treatment (data not shown).
Figure 2.
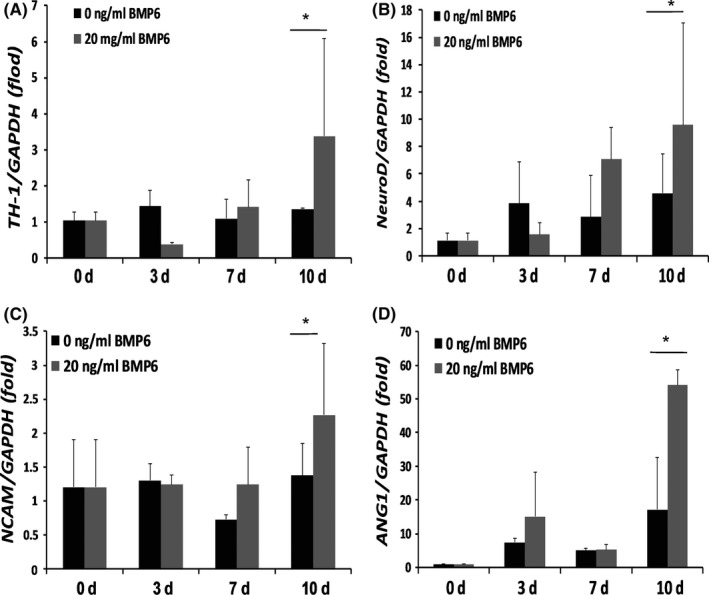
BMP6 prompted the expressions of neurogenic and angiogenic differentiation markers in stem cells from apical papilla (SCAPs). (A‐C) Real‐time RT‐PCR results showed that neurogenic differentiation markers, TH‐1 (A), NeuroD (B) and NCAM (C) were highly expressed in 20 ng/mL of BMP6‐treated SCAPs compared with untreated SCAPs after neurogenic induction. (D) Real‐time RT‐PCR results showed that angiogenic differentiation marker, ANG1, was highly expressed in 20 ng/mL of BMP6‐treated SCAPs compared with untreated SCAPs after angiogenic induction. GAPDH was used as an internal control. Student's t test was performed to determine statistical significance. All error bars represent SD (n=5). *P<.05
3.5. BMP6 increased the cell proliferation ability of SCAPs
CCK‐8 assay was used to evaluate cell proliferation of SCAPs with 20 ng/mL of rhBMP6 or without rhBMP6 for 2 days, and we found that the OD value of BMP6‐treated SCAPs was significantly higher than the untreated group at 1 and 2 days (Figure 3A). To further confirm the results of cell proliferation, we detected the cell proliferation indexes by CFSE cell proliferation assays. The result also showed that the cell indexes of SCAPs with BMP6 treatments were much higher than SCAPs without treatments (Figure 3B,C).
Figure 3.
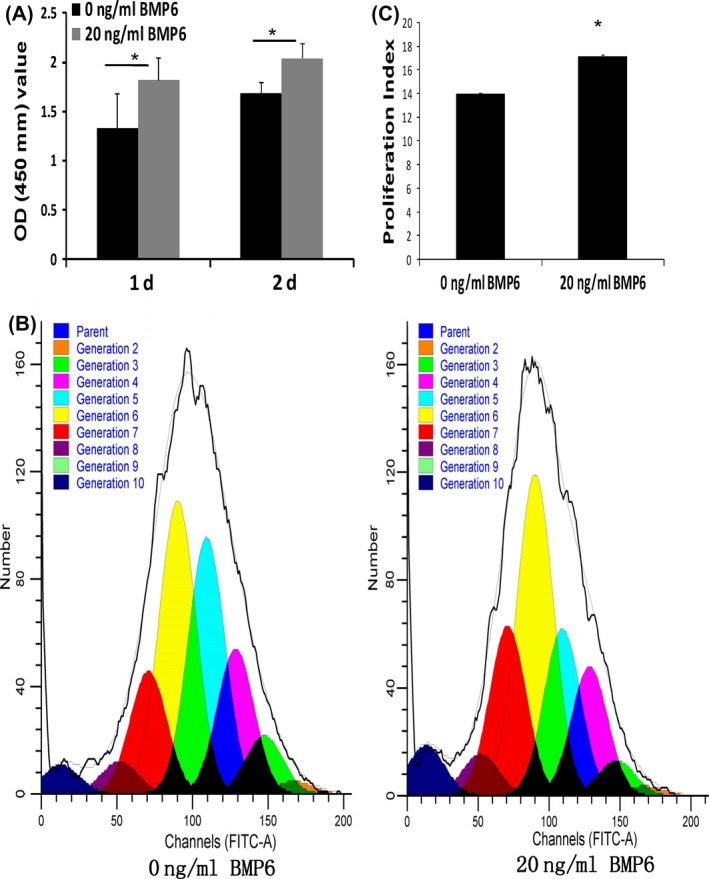
BMP6 increased the cell proliferation of stem cells from apical papilla (SCAPs). (A) CCK‐8 assay results showed that 20 ng/mL BMP6 could increase the cell proliferation of SCAPs. (B,C) CFSE cell proliferation assays showed that 20 ng/mL BMP6 prompted the cell proliferation indexes of SCAPs. Student's t test was performed to determine statistical significance. All error bars represent SD (n=5). *P<.05
4. Discussion
Stem cells from apical papilla are important for tooth development and regeneration. A key variable for successful tissue regeneration is the microenvironmental niche in which cells and tissues grow.27 In present study, in order to illuminate important aspects of the microenvironmental niche of SCAPs, we identify the key genes in SCAPs niche by microarray and bioinformatic analysis based on the analysis of the gene expression profiles between apical papilla tissues and SCAPs, and SCAPs and SCAPs cell sheet. And then we investigated whether SCAPs cell sheet could recover the genes regulation in niche. By Gene Chip analysis and further data statistics, only 31 gene expressions were recovered in cell sheet compared with SCAPs niche. These results indicated that cell sheet could preserve the normal cellular junctions, endogenous ECM, and mimic the mechanical, chemical and biological properties in niche, while the cell sheet could not restore the comprehensive gene regulation in SCAPs niche. Some modulator should be supplied into the cell sheet to mimic the MSCs niche. So the key genes must be identified in microenvironmental niche to support the MSCs functions.
By bioinformatic analysis, we identified several important signalling pathways and genes which might regulate the SCAPs functions, and found that the TGF‐β, WNT and MAPK signalling pathway may play an important role in SCAPs. TGF‐β signalling pathway could promote osteoblastic differentiation and maturation in early stage and obstruct osteoblastic differentiation in late stage and mineralization.28 Wnt signalling pathway is involved in cell differentiation, bone homeostasis and bone formation.29, 30 The inductive and inhibitory effect of Wnt signalling pathway on osteogenic differentiation were both reported depending on microenvironment, cell types and other supplements.31 The MAPK signalling pathway transmits signals from the cell membrane into nucleus, exerting an important role in regulation of cell differentiation.32, 33 Furthermore, ITGA9, PIK3R1, PIK3CG, Fos and ENTPD1 were determined to be the important genes by high betweenness centrality calculation. ITGA9 is one of the integrin subunits which mediate cell‐cell and cell‐matrix adhesion,34 could facilitate cell migration and regulate diverse biological functions such as angiogenesis, lymphangiogenesis, cancer cell proliferation and migration.35 PIK3R1 encodes the p85, p55 and p50 regulatory subunits of class IA phosphatidylinositol 3 kinases (PI3Ks), which are known to play a key role in insulin signalling.36The PIK3CG gene is located in a commonly deleted segment of chromosome 7. This gene plays an important role in maintenance of the structural and functional integrity of epithelia.37 Fos is a trans‐acting factor that is capable of stimulating gene expression not by direct binding to DNA but by interaction with the sequence‐specific transcription factor AP‐1.38 ENTPD1, also known as CD39, is the dominant vascular ectonucleotidase. By hydrolysing ATP and ADP to AMP, ENTPD1 regulates ligand availability to a large family of purinergic receptors, and is an important factor in several acute and subacute models of vascular injury by modulation of extracellular nucleotide metabolism.39
Growth factors induce intracellular signalling pathways, which result in the activation of genes that change cellular activity and phenotype.40, 41 Recent in vitro and in vivo studies have confirmed that growth factors can improve the capacity of tissues to regenerate, and improve cellular chemoattraction, differentiation and proliferation. In present study, we identified that several down‐regulated growth factors in SCAPs may be associated with cell proliferation and differentiation functions, such as BMPs (BMP2, BMP5, BMP6 and BMP7), IGF2 and PDGF were more than 5‐fold down‐regulated in SCAPs compared to apical papilla tissues. BMPs are a well‐studied growth factors involved in the processes of bone healing.42 BMPs could induce the formation of both bone and cartilage by stimulating the cellular events of mesenchymal progenitor cells. Studies involving mutations of BMP ligands, receptors and signalling molecules have shown important roles of BMPs in embryonic and postnatal development. Severe skeletal deformation, development of osteoporosis, reduction in bone mineral density and bone volume are all aberrations associated with disrupted and dysregulated BMP signalling.43, 44 IGF2 is one of the key regulators of differentiation and exerts a beneficial effect on the osteogenic differentiation of MSCs.45, 46 PDGF showed the strong mitogenic effect on MSCs, and could regulate the proliferation, differentiation and migration of MSCs.47, 48
In order to identify the function of candidate growth factor in niche, we choose one BMPs protein, BMP6, which was 12 times highly expressed in apical papilla tissues than that in SCAPs, to investigate the function on the directed differentiation and cell proliferation potentials of SCAPs. BMP6 was more potent and consistent than BMP2 and BMP7 in inducing osteoblast differentiation in primary MSCs.49 In addition, another study proved that BMP6 may be unique among the BMP family in mediating terminal osteoblast differentiation in human‐derived cells.50 Growth factor's effect on cell proliferation and differentiation will be various, depending on the cell species, the culture conditions and the concentration of the growth factor.51 First, we investigate the effective dose of BMP6 and found that 20 ng/mL might be the optimal concentrations of rhBMP6 by ALP activity assay. Alizarin red staining and quantitative calcium analysis confirmed that 20 ng/mL of rhBMP6 could effectively enhance the mineralization of SCAPs in vitro. Real‐time RT‐PCR results showed that 20 ng/mL rhBMP6 could enhance the expressions of osteo/dentinogenic differentiation markers, including DSPP, DMP1, BSP, OCN and OPN, and the key transcript factors OSX and RUNX2. Therefore, we concluded that BMP6 can enhance the osteo/dentinogenic differentiation potential of SCAPs, which were consistent with previous reports.52 We also investigate the effect of BMP6 on the neurogenic and angiogenic differentiation potentials of SCAPs. Real‐time RT‐PCR results showed that neurogenic differentiation markers, including TH‐1, NeuroD, NCAM, and angiogenic differentiation marker, ANG1, were more strongly induced in 20 ng/mL rhBMP6‐treated SCAPs compared with untreated group. In addition, we investigated the proliferation potentials of SCAPs. CCK‐8 and CFSE assays showed that 20 ng/mL rhBMP6 could increase the proliferation ability of SCAPs. Taken together, these results suggest that BMP6 could promote the proliferation and osteo/dentinogenic, neurogenic and angiogenic differentiation potentials of SCAPs.
In summary, our study provides insight into the mechanisms of stem cells niche to regulate the stem cells function, provide the key target genes and certain theoretical basis to maintain the stem cell characteristics and promote stem cell‐mediated dental tissue regeneration. Our results also demonstrated that one candidate growth factor, BMP6 significantly enhanced the cell proliferation and directed differentiation abilities of SCAPs, indicating that BMP6 might be the potential mediator in microenvironmental niche to promote SCAPs‐mediated dental tissue regeneration.
Disclosure of potential conflicts of interest
The authors deny any conflicts of interest related to this research.
Author contributions
SD: data analysis and interpretation, manuscript writing financial support; XL: collection and/or assembly of data; LW: collection and/or assembly of data; RD: conception and design, collection and/or assembly of data; JD: collection and/or assembly of data, data analysis and interpretation; DMY: collection and/or assembly of data, data analysis and interpretation, ZF: conception and design, data analysis and interpretation, financial support, final approval of manuscript.
Supporting information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81625005 to Z.P.F.), and High‐level Talents of the Beijing Health System (2013‐3‐035 to Z.P.F.), the Program for “Hundred‐Thousand‐Ten thousand” Talents in Beijing (2014006 to Z.P.F.), Chinese Medicine Science and Technology Foundation of Beijing (JJ2015‐10 to Z.P.F.), Discipline Construction Foundation of Beijing Stomatological Hospital (16‐09‐05 to S.D.).
Diao S, Lin X, Liping W. Analysis of gene expression profiles between apical papilla tissues, stem cells from apical papilla and cell sheet to identify the key modulators in MSCs niche. Cell Prolif. 2017;50:e12337 10.1111/cpr.12337
Contributor Information
Juan Du, Email: dujuang16@163.com.
Dongmei Yang, Email: yangdm10042@gmail.com.
Zhipeng Fan, Email: zpfan@ccmu.edu.cn.
References
- 1. Sujesh M, Rangarajan V, Ravi Kumar C, Sunil Kumar G. Stem cell mediated tooth regeneration: new vistas in dentistry. J Prosthodont Res. 2012;12:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2. Sonoyama W, Liu Y, Yamaza T, et al. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008;34:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yen AH, Yelick PC. Dental tissue regeneration – a mini‐review. Gerontology. 2011;57:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci USA. 2011;108:6503–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang GTJ, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miura M, Gronthos S, Zhao M, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. [DOI] [PubMed] [Google Scholar]
- 9. Zuba‐Surma EK, Kucia M, Ratajczak J, Ratajczak MZ. “Small stem cells” in adult tissues: very small embryonic‐ like stem cells stand up! Cytometry A. 2009;75:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang S, Mu J, Fan Z, et al. Insulin‐like growth factor 1 can promote the osteogenic differentiation and osteogenesis of stem cells from apical papilla. Stem Cell Res. 2012;8:346–356. [DOI] [PubMed] [Google Scholar]
- 11. Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. [DOI] [PubMed] [Google Scholar]
- 12. Govindasamy V, Ronald VS, Totey S, et al. Micromanipulation of culture niche permits long‐term expansion of dental pulp stem cells–an economic and commercial angle. In Vitro Cell Dev Biol Anim. 2010;46:764–773. [DOI] [PubMed] [Google Scholar]
- 13. Volponi AA, Pang Y, Sharpe PT. Stem cell‐based biological tooth repair and regeneration. Trends Cell Biol. 2010;20:715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5‐mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol. 2011;31:2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palosaari H, Wahlgren J, Larmas M, et al. The expression of MMP‐8 in human odontoblasts and dental pulp cells is down‐regulated by TGF‐beta1. J Dent Res. 2000;79:77–84. [DOI] [PubMed] [Google Scholar]
- 16. Iohara K, Zheng L, Ito M, Tomokiyo A, Matsushita K, Nakashima M. Side population cells isolated from porcine dental pulp tissue with self‐renewal and multipotency for dentinogenesis, chondrogenesis, adipogenesis, and neurogenesis. Stem Cells. 2006;24:2493–2503. [DOI] [PubMed] [Google Scholar]
- 17. Catelas I, Sese N, Wu BM, Dunn JC, Helgerson S, Tawil B. Human mesenchymal stem cell proliferation and osteogenic differentiation in fibrin gels in vitro. Tissue Eng. 2006;12:2385–2396. [DOI] [PubMed] [Google Scholar]
- 18. Bemardo ME, Avanzini MA, Perotti C, et al. Optimization of in vitro expansion of human multipotent mesenchymal stromal cells for cell‐therapy approaches: further insights in the search for a fetal calfserum substitute. J Cell Physiol. 2007;211:121–130. [DOI] [PubMed] [Google Scholar]
- 19. Urist MR. Bone: formation by autoinduction. Science. 2005;150:893–899. [DOI] [PubMed] [Google Scholar]
- 20. Rubio D, Garcia‐Castro J, Martin MC, et al. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035–3039. [DOI] [PubMed] [Google Scholar]
- 21. Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self‐renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. [DOI] [PubMed] [Google Scholar]
- 22. Rolf HJ, Niebert S, Niebert M, Gaus L, Schliephake H, Wiese KG. Intercellular transport of Oct4 in mammalian cells: a basic principle to expand a stem cell niche? PLoS ONE. 2012;7:e32287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlitt T, Palin K, Rung J, et al. From gene networks to gene function. Genome Res. 2003;13:2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaigler D, Cirelli JA, Giannobile WV. Growth factor delivery for oral and periodontal tissue engineering. Expert Opin Drug Deliv. 2006;3:647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houshmand B, Behnia H, Khoshzaban A, et al. Osteoblastic differentiation of human stem cells derived from bone marrow and periodontal ligament under the effect of enamel matrix derivative and transforming growth factor‐beta. Int J Oral Maxillofac Implants. 2013;8:440–450. [DOI] [PubMed] [Google Scholar]
- 29. Zhang H, Wang J, Deng F, et al. Canonical Wnt signaling acts synergistically on BMP9‐induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs). Biomaterials. 2015;39:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu W, Konermann A, Guo T, Jäger A, Zhang L, Jin Y. Canonical Wnt signaling differently modulates osteogenic differentiation of mesenchymal stem cells derived from bone marrow and from periodontal ligament under inflammatory conditions. Biochim Biophys Acta. 2014;1840:125–134. [DOI] [PubMed] [Google Scholar]
- 31. De Boer J, Wang HJ, Van Blitterswijk C. Effects of Wnt signaling on proliferation and differentiation of human mesenchymal stem cells. Tissue Eng. 2004;10:393–401. [DOI] [PubMed] [Google Scholar]
- 32. Greenblatt MB, Shim JH, Glimcher LH. Mitogen‐activated protein kinase pathways in osteoblasts. Annu Rev Cell Dev Biol. 2013;29:63–79. [DOI] [PubMed] [Google Scholar]
- 33. James AW. Review of signaling pathways governing MSC osteogenic and adipogenicdifferentiation. Scientifica (Cairo). 2013;2013:684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Y, Enis D, Zheng H, et al. Cell adhesion mediated by VCAM‐ITGa9 interactions enables lymphatic development. Arterioscler Thromb Vasc Biol. 2015;35:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mostovich LA, Prudnikova TY, Kondratov AG, et al. Integrin alpha9 (ITGA9) expression and epigenetic silencing in human breast tumors. Cell Adh Migr. 2011;5:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thauvin‐Robinet C, Auclair M, Duplomb L, et al. PIK3R1, mutations cause syndromic insulin resistance with lipoatrophy. Am J Hum Genet. 2013;93:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sawczuk M, Maciejewska‐Karłowska A, Skotarczak B, Pawlik A. Association between single nucleotide polymorphism rs342286 near PIK3CG gene and acute coronary syndromes. Pol Arch Med Wewn. 2014;124:210–212. [DOI] [PubMed] [Google Scholar]
- 38. Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c‐Fos protein interacts with c‐Jun/AP‐1 to stimulate transcription of AP‐1 responsive genes. Cell. 1998;54:541–552. [DOI] [PubMed] [Google Scholar]
- 39. Behdad A, Sun X, Khalpey Z, et al. Vascular smooth muscle cell expression of ectonucleotidase CD39 (ENTPD1) is required for neointimal formation in mice. Purinergic Signal. 2009;5:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giannobile WV. Periodontal tissue engineering by growth factors. Bone. 1996;19:23S–37S. [DOI] [PubMed] [Google Scholar]
- 41. Anusaksathien O, Giannobile WV. Growth factor delivery to re‐engineer periodontal tissues. Curr Pharm Biotechnol. 2002;3:129–139. [DOI] [PubMed] [Google Scholar]
- 42. Blumenthal NM, Koh‐Kunst G, Alves ME, et al. Effect of surgical implantation of recombinant human bone morphogenetic protein‐2 in a bioabsorbable collagen sponge or calcium phosphate putty carrier in intrabony periodontal defects in the baboon. J Periodontol. 2002;73:1494–1506. [DOI] [PubMed] [Google Scholar]
- 43. Cheng H, Jiang W, Phillips FM, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J Bone Joint Surg Am. 2003;85:1544–1552. [DOI] [PubMed] [Google Scholar]
- 44. Wu XB, Li Y, Schneider A, et al. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin‐overexpressing mice. J Clin Invest. 2003;112:924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding W, Li J, Singh J, et al. miR‐30e targets IGF2‐regulated osteogenesis in bone marrow‐derived mesenchymal stem cells, aortic smooth muscle cells, and ApoE‐/‐ mice. Cardiovasc Res. 2015;106:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hamidouche Z, Fromigué O, Ringe J, Häupl T, Marie PJ. Crosstalks between integrin alpha 5 and IGF2/IGFBP2 signalling trigger human bone marrow‐derived mesenchymal stromal osteogenicdifferentiation. BMC Cell Biol. 2010;11:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen W, Baylink DJ, Brier‐Jones J, et al. PDGFB‐based stem cell gene therapy increases bone strength in the mouse. Proc Natl Acad Sci USA. 2015;112:E3893–E3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hye Kim J, Gyu Park S, Kim WK, Song SU, Sung JH. Functional regulation of adipose‐derived stem cells by PDGF‐D. Stem Cells. 2015;33:542–556. [DOI] [PubMed] [Google Scholar]
- 49. Kalamajski S, Aspberg A, Lindblom K, Heinegård D, Oldberg A. Asporin competes with decorin for collagen binding, binds calcium and promotes osteoblast collagen mineralization. Biochem J. 2009;423:53–59. [DOI] [PubMed] [Google Scholar]
- 50. Zhu F, Friedman MS, Luo W, Woolf P, Hankenson KD. The transcription factor osterix (SP7) regulates BMP6‐induced human osteoblast differentiation. J Cell Physiol. 2012;227:2677–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang L, Huang Y, Pan K, Jiang X, Liu C. Osteogenic responses to different concentrations/ratios of BMP‐2 and bFGF in bone formation. Ann Biomed Eng. 2010;38:77–87. [DOI] [PubMed] [Google Scholar]
- 52. Shen R, Jia R, Liu W, Lin Q, Hai Y, He Z. The function and regulation of BMP6 in various kinds of stem cells. Curr Pharm Des. 2015;21:3634–3643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


