Abstract
Objectives
MicroRNAs (miRNAs) are considered as the cellular regulators which post‐transcriptionally modulate gene expression in diverse biological processes including cell development and immunity. In this study, we investigated functions of miR‐181d in dendritic cells (DCs) maturation, and the underlying mechanisms were also explored.
Materials and methods
Here we did the miRNA screening in human DCs in response to lipopolysaccharides (LPS) by quantitative real‐time PCR (qRT‐PCR). The expressions of DCs maturation markers were measured after miRNA mimics transfections. The pharmacological inhibitors of signalling pathways were applied to examine miR‐181d effect on DCs maturation by Western blot. Luciferase assay and mixed lymphocyte reaction (MLR) were also performed to reveal the target gene of miR‐181d and test the viability of T cells treated with miR‐181d transfected DCs.
Results
Overexpression of miR‐181d per se is sufficient to promote DCs maturation, and up‐regulate CD80 and CD83 expressions without LPS. Besides, we showed that miR‐181d activated NF‐κB pathway and also promoted the expression of pro‐inflammatory cytokine IL12 and TNF‐α. Inhibition of NF‐κB pathway suppressed DCs maturation. Luciferase reporter assay and target gene knockdown assay indicated that miR‐181d targets regulator cylindromatosis (CYLD), a primary negative regulator of NF‐κB pathway. MLR assay showed that miR‐181d‐transfected DCs could promote T‐cell proliferation than iDCs in vitro.
Conclusion
Our study demonstrates that miR‐181d is required for DCs maturation through the activation of NF‐κB pathway by targeting CYLD.
Abbreviations
- CYLD
cylindromatosis
- DCs
dendritic cells
- iDCs
immature DCs
- mDC
mature DCs
- miRNA
microRNAs
- qRT‐PCR
quantitative real‐time PCR
1. INTRODUCTION
Dendritic cells (DCs) are the antigen‐presenting cells (APC) that can uptake, process and present antigens to lymphocytes, and effectively stimulate naïve T cells to proliferate.1, 2 DCs‐regulated immune response is largely dependent on maturation status.3 DCs exist in two basic states: immature DCs (iDCs) which could induce tolerance to itself with high phagocytic and antigen‐processing ability, and mature DCs (mDCs) which can initiate immune response to external antigens and elicit a pronounced antigen presentation capacity.4 Recent studies reported the immunological application of dendritic cell (DC)‐based vaccine therapies to target tumours in central nervous system.5, 6, 7, 8 The abundance of DCs in peripheral blood is very low and they are difficult to be isolated from other peripheral leucocytes.9 Although DCs are the most potent APCs, they may fail to mount a robust immune response to tumour antigens due to defective antigen‐processing mechanism or deregulated expression of MHC or other co‐stimulatory molecules for example CD80 and CD83.10 Haematopoietic progenitor cells or peripheral blood monocytes are used to produce mDCs in vitro by co‐culturing with growth factors and cytokines or microbial stimuli to induce maturation and enhance immunological functions.11 The duration of stimulation could regulate DCs maturation status and immunophenotype; 24‐hour stimulation shows a clear alternation of immunophenotype and elevated expression of CD markers compared with the 6‐hour treatment.12
MicroRNAs (miRNAs) are small, non‐coding and conserved RNA molecules around 19‐24 nucleotides, that post‐transcriptionally modulate gene expression and cellular processes, thereby influence cell fate and function.13 Numerous miRNAs have been identified and characterized on the varying genes expression patterns and functions, and the roles of miRNAs are being studied in almost every area of biology. A panel of miRNAs has been reported to regulate DCs development, maturation and function. miR‐155, one of the well‐characterized miRNAs in DCs maturation, is part of a negative feedback loop, which suppresses inflammatory cytokine production in response to stimulus. miR‐155 targets TAB 2 and SOCS1, which are the important transduction molecules in cytokine signalling. In addition, miR‐155 also drives IL‐12p70 production in mDCs, thus potentially promoting the potency of monocyte‐derived DCs in Th1 immune responses.14, 15 miR‐146a overexpression is shown to inhibit the expression of several DC‐specific surface markers such as CD80, CD86, HLA‐DR and CCR7 molecules.16 Overexpression of miR‐301a inhibited IL‐12, IL‐6 and TNFα production in DCs and modulated T‐cell responses; however, there is no effect on MHC class II, CD80, CD86 or CD40 expression.17 Likewise,up‐regulation of the miRNA let‐7i in DCs is required for DCs maturation following lipopolysaccharide (LPS) treatment. Inhibition of let‐7i down‐regulated the expression of the co‐stimulatory molecules CD80 and CD86, as well as pro‐inflammatory cytokine IL6 and TNF‐α production. T‐cell responses to antigen presented by DCs were also compromised.18
In this study, we screened for the candidate miRNAs which control DCs maturation. LPS treatment promoted expression of numerous miRNAs in DCs, highlighting their potential regulatory role for DCs maturation. We demonstrated that the altered miR‐181d level modulates DCs morphology, promotes cytokine release and elevates DCs maturation markers expression. Bioinformatics analysis and luciferase reporter assay indicated cylindromatosis (CYLD) is a target gene of miR‐181d. Our study revealed that miR‐181d regulated DCs maturation and functional status by activating NF‐κB pathway and suppressing CYLD transcription and translation.
2. MATERIALS AND METHODS
2.1. Generation of DCs from human peripheral blood
Human peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll‐Paque (1.077 g/mL; GE Healthcare) density gradient centrifugation from the buffy coat fraction of anticoagulated blood. Monocytes were purified from PBMCs from the middle layer after the centrifugation. About 1 × 106 cells/mL were cultured in 2 mL AIM V containing 10% autologous plasma, 50 ng/mL recombinant human granulocyte‐macrophage colony‐stimulating factor (GM‐CSF; Peprotech), 50 ng/mL recombinant interleukin 4 (IL‐4; Peprotech) and 1% penicillin/streptomycin (P/S; Life technologies) in six‐well flat‐bottomed plates. The adherent cells were selected by overnight culture. Medium was half‐changed every 2 days, the maturation of monocytes derived was induced by 100 ng/mL LPS on day 6, and mDCs were collected on day 7. PBMC were collected from 14 healthy donors (30‐60 years old), and all donors gave their written consent for supplying blood for scientific research. All experiments conducted using human materials were approved by the ethics committee of The Chinese University of Hong Kong.
2.2. Characterization of DC by flow cytometry
For the analysis of surface marker immunophenotype, cells were pre‐incubated for 30 minutes on ice in FACS buffer (1× PBS containing 2% FBS, 0.3% (w/v) NaN3 and 1 mmol/L EDTA) to block non‐specific Ig binding and incubated with the specific monoclonal antibodies (mAb) or control isotype for 30 minutes on ice, washed twice using FACS buffer and fixed with Flow Fixation buffer (1× PBS containing 1 % paraformaldehyde) for 30 minutes. Cell staining was performed using CD83‐PE conjugated mAb, CD80‐PE conjugated mAb, CD‐86‐Alexa conjugated mAb and CD14‐FITC conjugated mAb (Novus Biologicals, Littleton, CO, USA). Flow cytometry was carried out using BD LSR Fortessa (BD Biosciences, San Jose, CA, USA), and the data were analysed using flowing software 2 (Perttu Terho, Turku, Finland).
2.3. Cytokine analysis
Dendritic cell supernatants were collected on day 0, 2, 4 and 6, and frozen at −20◦C until ELISA analysis. Quantification measurements of cytokines and chemokines were performed using IL12 SEH00544A, Single‐Analyte ELIS Array Kits (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
2.4. Quantitative real‐time PCR
Total RNA, including miRNA, was extracted from cells using the miRCURY RNA isolation kit (Exiqon, Denmark) following the manufacturer's instruction. RNA concentrations were measured with NanoDrop instrument (Thermo Scientific, Waltham, MA, USA). miRNA expression was assessed by qRT‐PCR analysis using miScript SYBR Green PCR Kit (Qiagen). miRNA‐specific primers were designed by miRprimer software and the cDNA synthesis was followed as previously reported.19 All the oligo primers were synthesized by Tech Dragon Ltd. For mRNA expression, 1 μg of total RNA was reverse transcribed using High‐Capacity Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) as specified by the manufacturer, qRT‐PCR amplification was done using SYBR Select Master Mix (Applied Biosystems, Foster City, CA, USA). All qRT‐PCR analyses were performed using abi prism Vii 7 Sequence Detection System (PerkinElmer Biosystems, Waltham, MA, USA). Relative miRNA and mRNA levels were calculated using the 2△△Ct method; GAPDH and U6 were used for mRNA and miRNA normalization respectively, and primer sequences are detailed in Tables S2and S3.
2.5. miRNA mimics and siRNA transfection
Cells were added at 2×105 to 5×105/mL in 12‐well tissue culture plates for transfection. miRNA (Genepharm, Shanghai, China) mimics and negative control (N.C.) were transfected into the cells at the recommended concentration using Lipofectamine 3000 (Invitrogen). For miRNA transfections, a final concentration of 50 nmol/L miRNA was used. For siRNA transfections, CYLD siRNA (siCYLD, #sc‐37326, Santa Cruz, CA, USA), ESR1 siRNA (siESR1, #sc‐44204, Santa Cruz) and DUSP10 siRNA (siDUSP10, #sc‐61048, Santa Cruz) were used at the final concentration of 20 nmol/L. iDCs were then grown in AIM V medium containing 10% autologous plasma without antibiotic for 24 hours before harvesting. The transfection efficiency was assessed by the FAM‐labelled mimics‐transfected cells. Total RNA and protein were harvested 48 hours post‐transfection for qRT‐PCR and Western blot assay respectively.
2.6. Qlucore analysis
miRNA mimics were transfected into iDCs, and CD80 and CD83 expression levels were detected by qRT‐PCR. Data generated by these assays were analysed using Qlucore Omics Explorer Analysis (http://www.qlucore.com/) to determine similarity among samples within and between groups, and measure differentially expressed miRNAs unique to each phenotype. To identify the candidate miRNAs for DCs maturation, qRT‐PCR results were analysed in the flexible and easy‐to‐use heatmap with hierarchical clustering.
2.7. Mixed lymphocyte reaction
PBMCs were isolated from healthy donors using Ficoll gradient centrifugation, and were suspended in AIM V with 10% autologous serum for 2 hours; the non‐adherent cells were collected and cultured in AIM V with 10% autologous serum in the presence of IL‐2 (10g/mL) for further T‐cell expansion. iDCs‐ and DCs‐transfected miRNA mimics were harvested and treated with 50 μg/mL mitomycin (#M0503, Sigma, St. Louis, MO, USA) for 30 minutes. T cells served as the responding cell cocultured with stimulator cells DCs 10:1 and 50:1 ratio in 96‐well plate for 72 hours. MTS assay (#G3580, Promega, San Luis Obispo, CA, USA) was performed to detect T‐cell proliferation, and absorbance at OD of 490 nm was measured by a microplate reader.
2.8. Identification of miRNAs target
Genome‐wide mRNA expression analysis was performed to compare the gene expression profile between miRNA mimics‐transfected DCs and scrambled mimics‐transfected cells. mRNA transcripts with more than 2‐fold down‐regulated were selected and compared with the in silico targets from four publicly available databases (miRanda, PITA, miR‐Walk and TargetScan 6.2), generating a list of candidate target mRNAs. Meanwhile, the target genes and signalling‐related genes were analysed by Ingenuity® Pathway Analysis (IPA).
2.9. Ingenuity pathway analysis (IPA)
Data sets representing a gene list or gene expression profile obtained from qRT‐PCR analyses were submitted to the Ingenuity Pathway Analysis Tool (IPA Tool; Ingenuity H Systems, Redwood City, CA, USA; http://www.ingenuity.com) for core analysis and miRNA targets prediction.
2.10. Signal transduction in DCs maturation
To identify the key signalling pathways involved in DCs maturation, two or three key genes were selected and validated by the qRT‐PCR. On day 6, iDCs were transfected with the miRNA mimics at 50 nmol/L. In some experiments, iDCs were pre‐treated with PD98059 (25 μmol/L, Tocris, Bristol, UK) and Bay 11‐7821 (20 μmol/L, Tocris), the pharmacological inhibitors of ERK pathway and NF‐κB pathway respectively. Control group received an equivalent amount of vehicle (DMSO). Cell maturation was characterized by the CD80 and CD83 expressions using qRT‐PCR.
2.11. Luciferase reporter assays
The sequences of the target genes at 3′ UTR were retrieved from http://genome.ucsc.edu and fragments (~600 bp) of 3′UTR were cloned into pmirGLO vector (Promega), downstream of luciferase open reading frame. The primer sequences were shown as follows: ESR1 F, 5′‐ GCGAGCTCGACAATTTTATGTATCTGTG‐3′; ESR1 R, 5′‐ GCTCTAGAATTACATCGTCTAGTCCTAG‐3′; CYLD F, 5′‐ GCGAGCTCAAGAAGTCTAAATGAAGTTA‐3′; CYLD R, 5′‐ GCTCTAGATGTTAGCTGACAGTAACTAC‐3′; DUSP10 F, 5′‐ GCGAGCTCGATTTTGT GTGTTTCATGCT‐3′; DUSP10 R, 5′‐ GCTCTAGACCAGTTTCATTTATATAATG‐3′; Bold italic characters were for restriction sites. The sequences of the whole constructs were confirmed by DNA sequencing. 293T cells (Clontech, Mountain View, CA, USA) were plated at 1×105 cells per well in 24‐well plate and transfected 24 hours later using Lipofectamine 3000 (Invitrogen); each transfection contains 200 ng pmirGLO reporter vector (Promega) with the 3′UTR of putative target genes together with the human miRNA mimics or a control scramble at a final concentration 20 nmol/L. Firefly and Renilla luciferase activities were measured 48 hours post‐transfection using the Dual‐Glo Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to the Renilla luciferase activity to control for cell number variation. Results were further normalized with respect to values obtained with an empty pmirGLO reporter vector in triplicates.
2.12. Western blot assay
Total protein was extracted using N‐PER Neuronal Protein Extraction Reagents (#87792; Thermo Scientific), and protein concentration was measured by Bradford assay (Bio‐Rad, CA, USA). Ten microgram proteins from each sample was separated on Tris‐polyacrylamide gel by electrophoresis and blotted onto nitrocellulose membranes (GE Healthcare, Chicago, IL, USA). Membranes were incubated with primary antibodies at a 1:1000 dilution overnight at 4°C and an appropriate secondary antibody at a 1:2000 dilution for 1 hour at room temperature. Primary antibodies used are: CYLD (D1A10) (#8462), phospho‐p65 (Ser‐536) (#3033), phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (#9106), NF‐κB‐P65 (#4764), p44/42 MAPK (ERK 1/2) (#9102) AKT (#4691) phospho‐AKT (Thr308) (#5106) (Cell Signaling Technology, MA, USA) and GAPDH antibodies (#sc‐25778; Santa Cruz Technology).
2.13. Statistical analysis
Data presented in the graphics are from at least three independent experiments. Statistical difference between groups was analysed by two‐tailed Student's t tests. Value of P<.05 was considered significant (Excel; GraphPad Prism 6).
3. RESULTS
3.1. LPS promotes DCs maturation
Monocytes isolated from peripheral blood of healthy human donors were differentiated into iDCs with GM‐CSF, IL4 growth factors and a classical maturation inducer lipopolysaccharide (LPS) for DCs maturation. The mDCs displayed the characteristics of typical mature dendritic cells with a clustering of protruding veils (Figure 1A). The drug treatment promoted cell growth measured by the Scepter Cell Counter; the mDCs were bigger than iDC (Figure 1B). The expression of DC surface markers including CD80, CD83 and CD86 were elevated after DCs maturation (Figure 1C). Meanwhile, the expressions of these cell surface markers were assessed by qRT‐PCR over a 7‐day culture; CD80 and CD83 were strongly induced after exposure to LPS (Figure 1D). Nearly 2‐fold induction of IL‐12p70 cytokine secretion was detected in the culture supernatant after 24 hours post‐LPS treatment (Fig. S1a). Consistently, the expression of CD83, CD80 and CD86 were significantly higher in LPS‐treated group compared with the iDCs by flow cytometry; however, CD14 expression remained unchanged (Fig. S1b)
Figure 1.
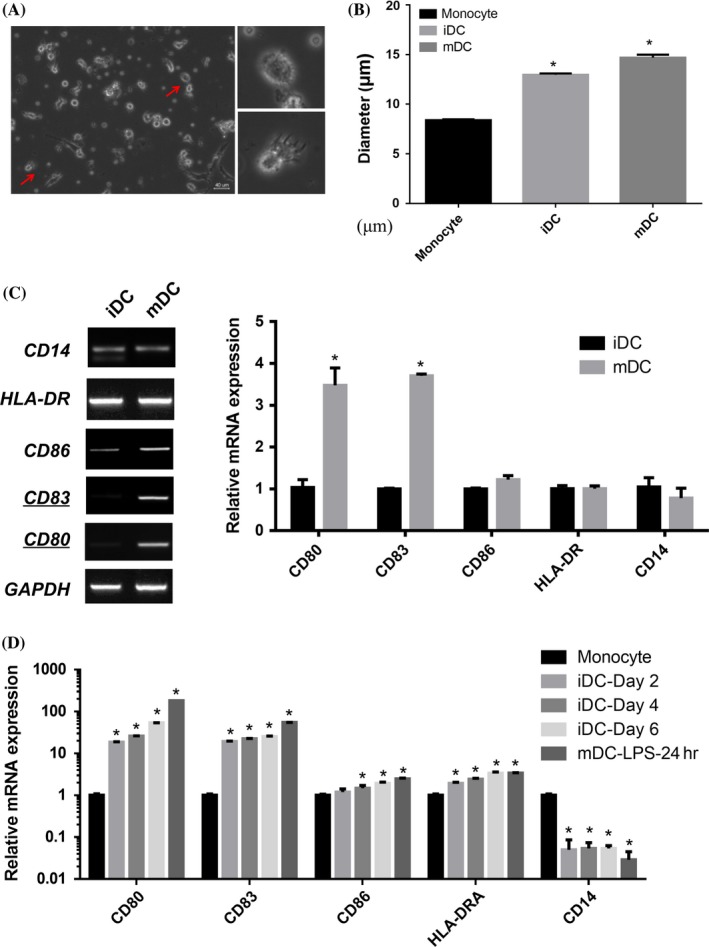
Characterization of dendritic cells (DC) morphology and cell surface markers expression in different stages. (A) Cell morphology was evaluated by phase contrast microscopy. Images were captured with a Nikon Ti‐U inverted microscope, processed with Adobe Photoshop 7.0 software (Adobe, San Jose, CA; row, original magnification ×200 and ×400 respectively). Representative images are shown for the gross DC morphology. Scale bar=40 μm. (B) Monocytes cultured in the presence of GM‐CSF and IL‐4 for 6 days and then LPS for another day show clustered and protruding veils that resemble mature DCs. The diameter of cells in monocyte, iDC and mDC is measured by Scepter cell counter (n=6). Data represent the means±SEM. (C) The expression of maturation‐related markers including CD86, CD83, CD80 and HLA‐DR were all detected by PCR and qRT‐PCR. qRT‐PCR were performed in three replicates from three independent experiments (n=6). (D) iDCs generated from monocytes by incubation with GM‐CSF/IL‐4 for 6 days, then the DCs were collected on day 2, day 4 and day 6, while mDCs were treated by LPS on day 6 and collected on day 7. CD86, CD83, CD80, HLA‐DR and CD14 were detected by qRT‐PCR in different stages respectively These results are representative of one experiment out of three independent experiments. (Student's t tests; *P<.05)
3.2. Identification of differentially expressed miRNAs between iDCs and mDCs
We attempted to identify miRNAs required for DC differentiation and maturation by combining the multiple microarray data sets. The raw CEL files of mDCs (accession number GSE23371) and (accession number GSE21708) data sets were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database.15, 20 The blast analysis revealed that a total of 36 miRNAs were differentially expressed (Figure 2A). Then we measured the expression of these miRNAs by qRT‐PCR in both mDC and iDC samples (Figure 2B). The expression profiles of miRNAs were then submitted to IPA software for analysis. The IPA analysis relates the differentially expressed miRNAs to their associated biological functions in inflammatory diseases, inflammatory response and other diseases (Figure 2C). In the associated network analysis, cancer, haematological diseases and hereditary disorder were enriched as the top network functions (Figure 2D). We further characterized the top 12 miRNAs showing greatest differential expression which may play an important role in DCs maturation. These miRNAs were individually transfected into the iDCs on day 6, and miR‐155 served as the positive control. We confirmed the overexpression of individual miRNAs mimics compared with the N.C.‐transfected DCs (Fig. S2a). The morphology of the transfected cells was examined 24 hours post‐transfection, and the cellular protrusions were calculated as the percentage of that observed in the N.C. As shown in Fig. S2b, 8 of the 13 miRNA mimics namely miR‐181d, miR‐575, miR‐629, miR‐422a, miR‐451a, miR‐134, miR‐612 and miR‐155 promoted the development of protruding veils in the iDC compared to the negative control. It was proposed that the iDC may undergo a conversion from an immature to a mature phenotype by the miRNA introduction, and morphological change resembling in iDCs confirming their role in DCs maturation.
Figure 2.
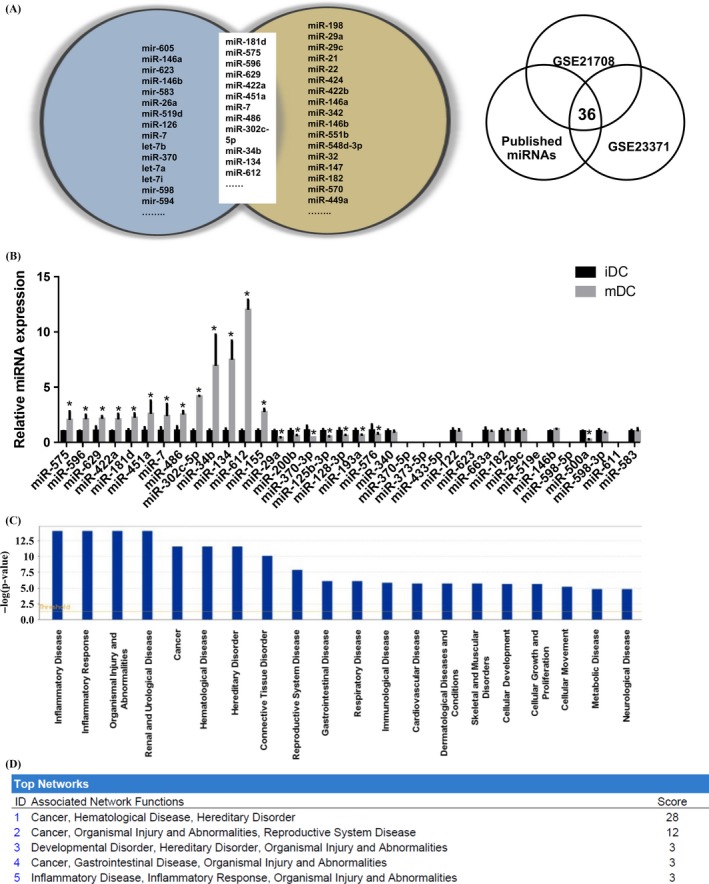
Identification of differentially expressed miRNAs in iDC and mDC (A) Venn diagram showing differential miRNA expression in different databases. (B) miRNA expression levels were measured using qRT‐PCR in mDC, normalized to U6 and expressed relative to iDC control. For miRNAs designated (fold change >1.5), induction with LPS was determined by qRT‐PCR. qRT‐PCR measurements were performed in triplicate (n=3). Data represent the means±SEM. (C) The bar chart showed the top predicted diseases and disorders with their respective scores obtained from Ingenuity Pathway Analysis Software (IPA). (D) The list of top‐five networks and biofunctions with their respective scores obtained from IPA. (Student's t tests; *P<.05)
3.3. miR‐181d regulates DCs maturation and modulates cell surface markers expression and cytokine release
Dendritic cell maturation status is important for antigen presentation and we measured the expression of maturation markers (CD80 and CD83) in the miRNA‐transfected iDCs by qRT‐PCR (Figure 3A). To identify the miRNAs required for CD marker expression, we drew the hierarchically clustered heatmap according to the expression profiles of CD80 and CD83 using Qlucore software. As shown in Figure 3B, compared with iDC stimulated by LPS, iDCs transfected with miR‐181d mimics showed the strongest induction of CD markers, suggesting that miR‐181d is required for driving DCs maturation. Overexpression of miR‐181d in DCs also enhances transcription of Th1 cytokine IL12 and pro‐inflammation cytokine TNF‐α by semi‐RT‐PCR (Fig. S3a). The secretion of IL12 in miR‐181d‐transfected DCs showed an increased level compared with the negative control (Fig. S3b). Furthermore, miR‐181d markedly promoted T‐cell proliferation compared with N.C.‐treated DCs in a dose‐dependent fashion with a stronger effect at 1:10 ratio (Figure 3C). A gene list was generated by the IPA software according to experimentally observed selection, which means the observation was linked with at least one reference (Table S1). To identify the potential miR‐181d targets, the gene list was imported into IPA software, and the top associated signalling pathways involved in the network were identified including NF‐κB, MAPK and Akt pathways (red colour‐filled circle) (Figure 3D). This network was associated with haematological disease and immunological disease (figure was not shown).
Figure 3.
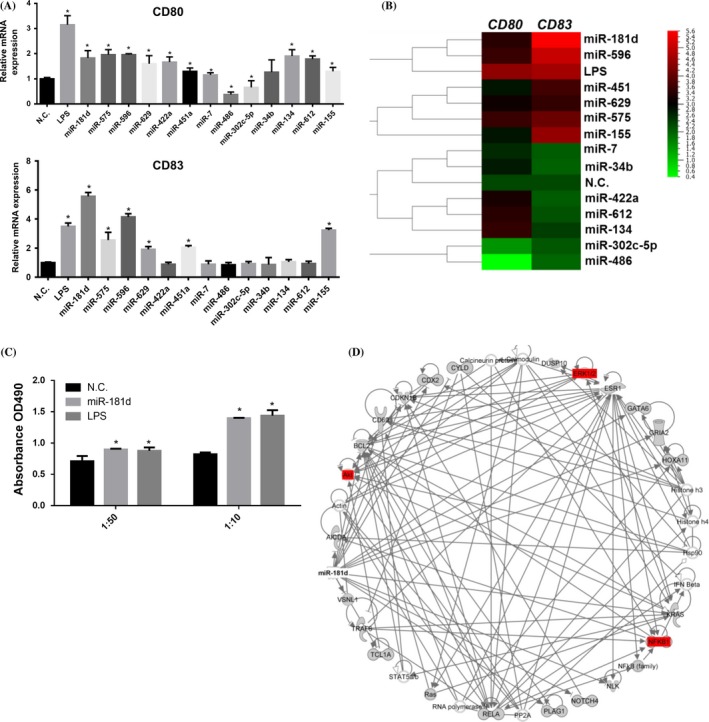
miR‐181d affects the dendritic cells (DCs) maturation by regulating the expression of cell surface markers and cytokine release (A) iDCs were transfected with 12 miRNA mimics, plus miR‐155 as the positive control. CD80 and CD83 expression levels were detected by qRT‐PCR. qRT‐PCR measurements were performed in triplicate (n=3). Data represent the means±SEM. (B) Heatmap of the miRNA expression profile by Qlucore analysis (http://www.qlucore.com/). (C) DCs were cocultured with responding T cells as different ratios, 1:10 and 1:50, in 96‐well plate for 72 hours. After MTS one solution incubation, the ability of DCs to interact T cell proliferation was measuring the absorbance at 490 nm. (D) The represented network show the most highly ranked based on the IPA analysis. The genes that are blanked were determined to be highly rated in this network from the statistical analysis. A solid line represents a direct interaction between the two gene products and an arrow line means acting‐in, and the solid line ended with vertical bar means inhibition. (Student's t tests; *P<.05)
3.4. NF‐kB signalling pathway is modulated by miR‐181d in mDCs
miR‐181d is shown to be a key regulator of cellular differentiation in human mammary epithelial MCF‐10A cells and associated with the inflammation response.21 However, the functional specificity of miR‐181d in mDCs remains largely unexplored. Thus, we screened for the signalling pathways altered by miR‐181d during DCs maturation by monitoring the expression of the downstream components. As shown in Figure 4A, overexpression of miR‐181d activated two signalling pathways including NF‐κB and MAPK. Besides, we evaluated the abundance of phosphorylated forms of p65, Akt and ERK1/2 after miR‐181d transfection. Western blotting showed that the miR‐181d overexpression strongly promoted phosphorylation of p65 and ERK in DCs and 293T cell line, however there is only a little change for Akt pathway (Figure 4B). In contrast, supplementation of Bay 11‐7821 (a NF‐κB pathway inhibitor) and PD‐98059 (a MAPK pathway inhibitor) strongly suppressed phosphorylation of p65 and ERK induced by miR‐181d respectively (Figure 4C). We also qPCR‐measured the expression of two DCs maturation markers, CD80 and CD83, with the pathway inhibitors in miR‐181d‐overexpressing DCs. As shown in Figure 4D, blockade of NF‐κB pathway inhibited the up‐regulation of both maturation markers induced by miR‐181d; inhibition of MAPK pathway selectively suppressed CD80 expression. To further test the role of miR‐181d on DCs maturation, we evaluated the effect of the expression levels of CD80 and CD83 as well as the abundance of phosphorylated p65 after depletion of miR‐181d. We confirmed that transfection of anti‐miR‐181d pronouncedly inhibited its endogenous expression (Figure 4E). Expression of the cell surface markers CD80 and CD83 revealed mild but insignificant reduction in anti‐miR‐181d‐transfected DCs (Figure 4F). Western blotting showed that inhibition of miR‐181d could not promote phosphorylation of p65 compared to N.C.‐transfected DCs (Figure 4G). Taken together, these results showed that miR‐181d could activate NF‐κB pathway, and DCs maturation marker up‐regulation is mainly dependent on NF‐κB pathway, rather than MAPK pathway.
Figure 4.
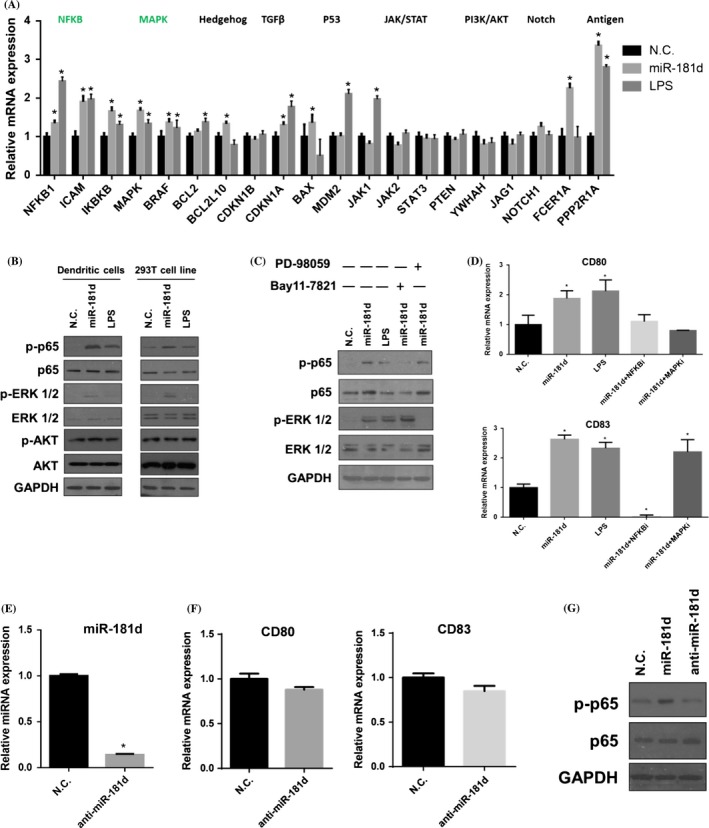
NF‐κB signalling pathway is modulated by miR‐181d in mDCs (A) qRT‐PCR was performed to examine the expressions of the key components which involved in the signalling pathways. qRT‐PCR measurements were performed in triplicate (n=3). Data represent the means±SEM. (B) Western blotting of p‐p65, p‐ERK 1/2 and p‐Akt in the miR‐181d, LPS and N.C.‐treated dendritic cells (DCs). The same treatment was done in 293T cell line. Data are representative of three independent experiments. (C) DCs were treated with signalling inhibitor PD 98059 and Bay 11‐7821, combined the treatment with miR‐181d transfection, N.C. and LPS. Western blotting showed the p‐p65 and p‐ERK 1/2 expression after the treatment. Data are representative of three independent experiments. (D) CD80 and CD83 mRNA expressions were detected by qRT‐PCR with PD 98059 and Bay 11‐7821, and miR‐181d mimics transfection. qRT‐PCR measurements were performed in triplicate (n=3). (E) DCs were transfected with anti‐miR‐181d (50 nmol/L) for 24 hours on day 6 culture. Expression of miR‐181d was assessed by qRT‐PCR. (F) CD80 and CD83 expression were detected by qRT‐PCR in iDC and anti‐miR‐181d‐transfected iDC. (G) Expression of p‐p65 and p‐65 proteins was detected by Western blotting in DCs transfected with miR‐181d, anti‐miR‐181d or N.C. respectively. GAPDH was used as the internal control. Data represent the means±SEM (Student's t tests; *P<.05)
3.5. CYLD was targeted by miR‐181d
Given that miR‐181d induced the NF‐κB pathway, we attempted to identify its downstream targets for DCs maturation. In the murine forebrain, HtrA1 were identified as the target of miR‐181d.22 CDX2, a known positive regulator of hepatocyte differentiation, was regulated by miR‐181d and targeted SOAT2 in hepatic cells.23 Then we qPCR‐measured the expression levels of the putative miR‐181d direct targets generated by IPA. As shown in Figure 5A, overexpression of miR‐181d down‐regulated the expression of ESR1, DUSP10 and CYLD during DCs maturation. TargetScan analysis predicts a putative miR‐181d binding site on the 3′UTR of the ESR1, DUSP10 and CYLD, suggesting they are the direct targets of miR‐181d (Figure 5B). Among the four isomiRs in the miR‐181 family, miR‐181d showed an elevated expression by LPS stimulation in mDCs (Figure 5B). We tested whether miR‐181d could directly repress the identified mRNA targets by interacting with their 3′UTR regions. The 3′UTRs of ESR1, DUSP10 and CYLD were cloned into pmirGLO reporter vector, downstream of firefly luciferase element. The luciferase reporters and miR‐181d mimics were co‐transfected into 293T cells, and the luciferase activity was measured by the bioluminescence analyser in 48 hours post‐transfection. As shown in Figure 5C, overexpression of miR‐181d significantly reduced CYLD and ESR1 luciferase reporter activity by 30% and 20% respectively, and also influence DUSP10 luciferase reporter assay slightly. Next, we examined the role of ESR1, DUSP10 and CYLD in CD80 and CD83 expressions. We first confirmed that silencing of CYLD, ESR1 or DUSP10 by siRNA effectively down‐regulated their endogenous expression level in DCs by qRT‐PCR (Figure 5D). CD80 and CD83 expressions in DCs transfected with siESR1, siDUSP10 and siCYLD were then determined by qRT‐PCR. The results showed that silencing of CYLD significantly up‐regulated the expression levels of CD80 and CD83; however, there was no effect upon silencing of ESR1 and DUSP10 (Figure 5E). Immunoblot consistently demonstrated that CYLD protein expression was strongly inhibited upon miR‐181d overexpression. A growing number of dendrites were observed in the miR‐181d‐transfected group (Figure 5F). Moreover, we found that silencing of CYLD by siRNA transfection (siCYLD) elevated the expression of p‐p65 measured using Western blotting (Figure 5G). Taken together, we demonstrate that miR‐181d could regulate human DCs maturation and cytokine production, and it can activate NF‐κB pathway potentially by targeting CYLD (Figure 5H).
Figure 5.
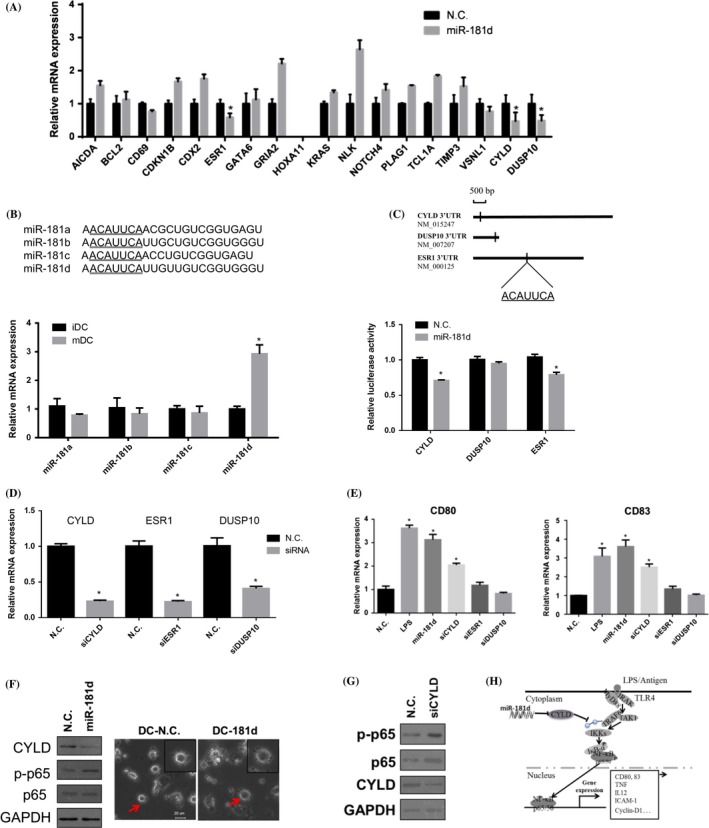
Cylindromatosis (CYLD) was targeted by miR‐181d (A) iDCs at day 6 were transfected with miRNA mimics and scrambled control (50 nmol/L), followed by LPS (100 ng/mL) for 24 hours. The targets of miR‐181d were predicted by the bioinformatics tool and were screened by qRT‐PCR. GAPDH was served as the internal control. qRT‐PCR measurements were performed in triplicate (n=3). Data represent the means±SEM (B) Sequence alignment is shown for miR‐181 isomiRs including miR‐181a, b, c and d. Their normalized expression levels iDCs were detected by qRT‐PCR. Data are representative of four independent donors. Data represent the means±SEM. (C) The 3′UTR of CYLD, DUSP10, ESR1 inserts into pmirGLO Dual‐Luciferase miRNA Target Expression Vector. The direct binding between miR‐181d and target genes was detected by Dual‐Glo luciferases reporter assay, and the mimics and luciferase vectors were co‐transfected by transient transfection in 293T cells. Data are representative of three independent experiments. Data represent the means±SEM. (D) Dendritic cells (DCs) were transfected with siCYLD, siESR1 or siDUSP10 (20 nmol/L) for 24 hours on day 6 culture. Expressions of CYLD, ESR1 and DUSP10 were assessed by qRT‐PCR respectively. (E) CD80 and CD83 expressions were detected by qRT‐PCR in LPS‐stimulated DCs, siCYLD, siESR1 or siDUSP10‐treated DCs, compared with N.C.‐treated DCs. (F) Expression of cylindromatosis (CYLD), p‐p65 and p‐65 proteins were detected by Western blotting in DCs transfected with miR‐181d mimics or N.C. respectively. GAPDH was used as the internal control. Morphology of the DCs transfected with N.C. and miR‐181d was examined by Nikon Ti‐U inverted microscope. Data are representative of three independent experiments. Scale bar=20 μm. Data represent the means±SEM. (Student's t tests; *P<.05) (G) Expression of CYLD, p‐p65 and p‐65 proteins were detected by Western blotting in DCs transfected with siCYLD or N.C. respectively. GAPDH was used as the internal control. (H) A schematic representation of miR‐181d regulation of DCs maturation
4. DISCUSSION
Dendritic cells maturation upon stimulation is essential for the initiation of allergic reactions, and then become highly efficient APC for naïve T lymphocytes.24 In this study, we provide a mechanistic insight underlying the miR‐181d‐mediated DC differentiation and maturation for immunity. First, a specific miR‐181d variant was up‐regulated during human monocyte differentiation from iDCs and mDCs. We also demonstrated that overexpression of miR‐181d enhanced DCs maturation markers including CD80 and CD83 transcription during human DC differentiation. Second, miR‐181d is a critical regulator of DCs maturation via NF‐κB signalling pathway and cytokine production. Third, mechanistically, miR‐181d activates NF‐κB pathway and suppresses CYLD (a negative regulator of NF‐κB) transcription and translation during DCs maturation.
Dendritic cells maturation could be characterized by cell surface markers. It was previously reported that CD83 was the most specific marker in mature DCs.25 For the cell surface markers, CD80 is expressed on activated B and T cells, macrophages and dendritic cells. CD80 could interact with CD28 to provide a potent co‐stimulatory signal for T‐cell activation through the CD3 complex. CD83 is expressed on a subset of dendritic cells, Langerhans cells and weakly on activated lymphocytes. CD83 was also reported to be stably expressed on mature dendritic cells and lack of expression on immature DCs.26 CD86 is expressed on monocytes/macrophages, dendritic cells, activated B and T cells, and astrocytes; however, CD86 showed an earlier expression in immune response than CD80. CD80 could also bind to CD28 to signal T‐cell activation 27. For HLA‐DR, it is expressed in B cells, activated T cells, monocytes/macrophages and dendritic cells, which plays a critical role to present peptide presentation to CD4+ T cells. Previous study showed that multiple cell surface markers including HLA‐DR, CD80, CD86, CD83, CD40 and CD11c showed increased expression upon the maturation of DCs.28 According to the previous reports and our results, we mainly focus on CD80 and CD83 to characterize DCs maturation in our study.
Recent characterization and understanding of miRNA‐mRNA regulatory networks substantially facilitate the development of immune therapy for human diseases.29 In addition, whole transcriptome expression analysis and miRNA profiling also provide mechanistic insight into DC developmental biology and help identify biomarkers for DC potency.14 miR‐181d was first identified in B‐lymphoid cells in mouse bone marrow, and its ectopic expression could increase the fraction of B‐lineage cells in vitro and in vivo.30 Previous study showed that miR‐181a and miR‐181b induced the apoptosis of glioma cells and function as potential tumour suppressors.31 Subsequently, miR‐181d was reported to act as a tumour suppressor in glioma by targeting K‐ras and Bcl‐2.32 However, miR‐181b/d was shown to be up‐regulated in early stage of hepatocarcinogenesis induced by CDAA diet, and miR‐181b is required to promote hepatocarcinogenesis.33
Our study indicates a direct role of NF‐κB pathway in regulating miR‐181d‐mediated DCs maturation. NF‐κB is an important transcription factor of the nuclear factor κ B (NF‐κB)/Rel family, which play a pivotal role in inflammatory and immune responses.34 There are five family members in mammals: RelA (p65), c‐Rel, RelB, NF‐κB1 (p105/p50) and NF‐κB2 (p100/p52).35 The p65/p50 heterodimer is the major form of NF‐κB in most cells, and it regulates a number of genes which play a pivotal role in the immune response, inflammation and development.36 NF‐κB signalling pathway could affect DCs maturation marker expression and functional characteristics dependent on different stimuli.37 NF‐κB was reported to regulate DCs maturation, immune response and protection of DCs from apoptosis.38, 39 Mutant Escherichia coli (EPEC) was shown to activate NF‐kB signalling pathway, secrete large amounts of pro‐inflammatory cytokines and promote CD80 and CD83 expression when incubated with myeloid DCs.40 However, no reports address the role of NF‐κB signalling pathway in DCs maturation induced by miR‐181d, which could target CYLD in DCs. A recent report has shown that miR‐21 could involve in a positive feedback loop to activate NF‐κB signalling pathway, by targeting PTEN expression.21
CYLD is a deubiquitination enzyme, which contains a deubiquitinase domain and three glycine‐rich domains. The deubiquitinase domain can regulate signal transduction pathways by deubiquitinating signal molecules.41 CYLD has been widely considered as a negative regulator for both canonical and non‐canonical NF‐κB pathways.42 CYLD could inhibit tumour cell proliferation by blocking Bcl‐3‐dependent NF‐κB pathway.43 CYLD was also reported to be associated with multiple types of human cancers, including melanoma, colon, hepatocellular carcinomas and kidney.44, 45, 46 Recently, CYLD was demonstrated to regulate the function of murine regulatory T cells.47 However, the role of CYLD in DCs maturation and activation remains elusive.
Our study revealed that miR‐181d regulates DCs maturation and functional state by targeting CYLD. Overexpressing miR‐181d can promote DCs undergo maturation, exhibit higher surface expression of co‐stimulatory molecules CD80 and CD83, and activate NF‐κB pathway followed by the shift of cytokine profile from pro‐inflammatory mediators. In this study, we propose that miR‐181d could serve as an adjuvant to induce DCs maturation and enhance immunity.
CONFLICT OF INTEREST
The authors declare no commercial or financial conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by the CUHK direct grant (#4054139) and the Otto Wong Brain Tumour Centre.
AUTHOR CONTRIBUTIONS
W.S.P. and G.L. conceived and designed the study, contributed to manuscript writing; X.W.S performed experiments, analysed data, wrote the manuscript; C.K. L. contributed to analyse data, manuscript editing; K.S.T. and Q.L. contributed to flow cytometry analysis. Y.L. and D.T.M.C. contributed to sample collection. S.D.Z. contributed to bioinformatics analysis; H.F.K. provided the technology support for this study.
Su XW, Lu G, Leung CK, et al. miR‐181d regulates human dendritic cell maturation through NF‐κB pathway. Cell Prolif. 2017;50:e12358 10.1111/cpr.12358
Contributor Information
Gang Lu, Email: lugang@cuhk.edu.hk.
Wai Sang Poon, Email: wpoon@surgery.cuhk.edu.hk.
REFERENCES
- 1. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245‐252. [DOI] [PubMed] [Google Scholar]
- 2. Randolph GJ, Jakubzick C, Qu C. Antigen presentation by monocytes and monocyte‐derived cells. Curr Opin Immunol. 2008;20:52‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steinman RM. The control of immunity and tolerance by dendritic cell. Pathol Biol (Paris). 2003;51:59‐60. [DOI] [PubMed] [Google Scholar]
- 4. Reis e Sousa C. Dendritic cells in a mature age. Nat Rev Immunol 2006;6:476‐483. [DOI] [PubMed] [Google Scholar]
- 5. Kim W, Liau LM. Dendritic cell vaccines for brain tumors. Neurosurg Clin N Am. 2010;21:139‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liau LM, Prins RM, Kiertscher SM, et al. Dendritic cell vaccination in glioblastoma patients induces systemic and intracranial T‐cell responses modulated by the local central nervous system tumor microenvironment. Clin Cancer Res. 2005;11:5515‐5525. [DOI] [PubMed] [Google Scholar]
- 7. Prins RM, Soto H, Konkankit V, et al. Gene expression profile correlates with T‐cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. 2011;17:1603‐1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wheeler CJ, Black KL, Liu G, et al. Vaccination elicits correlated immune and clinical responses in glioblastoma multiforme patients. Cancer Res. 2008;68:5955‐5964. [DOI] [PubMed] [Google Scholar]
- 9. Hasskamp J, Zapas JL, Elias EG. Dendritic cell counts in the peripheral blood of healthy adults. Am J Hematol. 2005;78:314‐315. [DOI] [PubMed] [Google Scholar]
- 10. Sadun RE, Sachsman SM, Chen X, et al. Immune signatures of murine and human cancers reveal unique mechanisms of tumor escape and new targets for cancer immunotherapy. Clin Cancer Res. 2007;13:4016‐4025. [DOI] [PubMed] [Google Scholar]
- 11. Castiello L, Sabatino M, Jin P, et al. Monocyte‐derived DC maturation strategies and related pathways: a transcriptional view. Cancer Immunol Immunother. 2011;60:457‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malanga D, Barba P, Harris PE, Maffei A, Del Pozzo G. The active translation of MHCII mRNA during dendritic cells maturation supplies new molecules to the cell surface pool. Cell Immunol. 2007;246:75‐80. [DOI] [PubMed] [Google Scholar]
- 13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 14. Ceppi M, Pereira PM, Dunand‐Sauthier I, et al. MicroRNA‐155 modulates the interleukin‐1 signaling pathway in activated human monocyte‐derived dendritic cells. Proc Natl Acad Sci U S A. 2009;106:2735‐2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu C, Huang X, Zhang X, et al. miR‐221 and miR‐155 regulate human dendritic cell development, apoptosis, and IL‐12 production through targeting of p27kip1, KPC1, and SOCS‐1. Blood. 2011;117:4293‐4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karrich JJ, Jachimowski LC, Libouban M, et al. MicroRNA‐146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood. 2013;122:3001‐3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pyfferoen L, Mestdagh P, Vergote K, et al. Lung tumours reprogram pulmonary dendritic cell immunogenicity at the microRNA level. Int J Cancer. 2014;135:2868‐2877. [DOI] [PubMed] [Google Scholar]
- 18. Zhang M, Liu F, Jia H, et al. Inhibition of microRNA let‐7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol. 2011;187:1674‐1683. [DOI] [PubMed] [Google Scholar]
- 19. Balcells I, Cirera S, Busk PK. Specific and sensitive quantitative RT‐PCR of miRNAs with DNA primers. BMC Biotechnol. 2011;11:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jansen BJ, Sama IE, Eleveld‐Trancikova D, et al. MicroRNA genes preferentially expressed in dendritic cells contain sites for conserved transcription factor binding motifs in their promoters. BMC Genom. 2011;12:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR‐21 and miR‐181b‐1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nigro A, Menon R, Bergamaschi A, et al. MiR‐30e and miR‐181d control radial glia cell proliferation via HtrA1 modulation. Cell Death Dis 2012;3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yap CS, Sinha RA, Ota S, Katsuki M, Yen PM. Thyroid hormone negatively regulates CDX2 and SOAT2 mRNA expression via induction of miRNA‐181d in hepatic cells. Biochem Biophys Res Commun. 2013;440:635‐639. [DOI] [PubMed] [Google Scholar]
- 24. Bousso P. T‐cell activation by dendritic cells in the lymph node: lessons from the movies. Nat Rev Immunol. 2008;8:675‐684. [DOI] [PubMed] [Google Scholar]
- 25. Iwamoto M, Shinohara H, Miyamoto A, et al. Prognostic value of tumor‐infiltrating dendritic cells expressing CD83 in human breast carcinomas. Int J Cancer. 2003;104:92‐97. [DOI] [PubMed] [Google Scholar]
- 26. Cao W, Lee SH, Lu J. CD83 is preformed inside monocytes, macrophages and dendritic cells, but it is only stably expressed on activated dendritic cells. Biochem J. 2005;385:85‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacPhee IA, Turner DR, Yagita H, Oliveira DB. CD80(B7.1) and CD86(B7.2) do not have distinct roles in setting the Th1/Th2 balance in autoimmunity in rats. Scand J Immunol 2001;54:486‐494. [DOI] [PubMed] [Google Scholar]
- 28. Cloosen S, Thio M, Vanclée A, et al. Mucin‐1 is expressed on dendritic cells, both in vitro and in vivo. Int Immunol. 2004;16:1561‐1571. [DOI] [PubMed] [Google Scholar]
- 29. Jin P, Han TH, Ren J, et al. Molecular signatures of maturing dendritic cells: implications for testing the quality of dendritic cell therapies. J Transl Med 2010;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83‐86. [DOI] [PubMed] [Google Scholar]
- 31. Shi L, Cheng Z, Zhang J, et al. hsa‐mir‐181a and hsa‐mir‐181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185‐193. [DOI] [PubMed] [Google Scholar]
- 32. Wang XF, Shi ZM, Wang XR, et al. MiR‐181d acts as a tumor suppressor in glioma by targeting K‐ras and Bcl‐2. J Cancer Res Clin Oncol. 2012;138:573‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang B, Hsu SH, Majumder S, et al. TGFbeta‐mediated upregulation of hepatic miR‐181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787‐1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoesel B, Schmid JA. The complexity of NF‐kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghosh S, May MJ, Kopp EB. NF‐kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225‐260. [DOI] [PubMed] [Google Scholar]
- 36. Tak PP, Firestein GS. NF‐kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ade N, Antonios D, Kerdine‐Romer S, Boisleve F, Rousset F, Pallardy M. NF‐kappaB plays a major role in the maturation of human dendritic cells induced by NiSO(4) but not by DNCB. Toxicol Sci. 2007;99:488‐501. [DOI] [PubMed] [Google Scholar]
- 38. Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816‐825. [DOI] [PubMed] [Google Scholar]
- 39. van de Laar L, van den Bosch A, van der Kooij SW, et al. A nonredundant role for canonical NF‐kappaB in human myeloid dendritic cell development and function. J Immunol. 2010;185:7252‐7261. [DOI] [PubMed] [Google Scholar]
- 40. Vossenkamper A, Marches O, Fairclough PD, et al. Inhibition of NF‐kappaB signaling in human dendritic cells by the enteropathogenic Escherichia coli effector protein NleE. J Immunol. 2010;185:4118‐4127. [DOI] [PubMed] [Google Scholar]
- 41. Kovalenko A, Chable‐Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF‐kappaB signalling by deubiquitination. Nature. 2003;424:801‐805. [DOI] [PubMed] [Google Scholar]
- 42. Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF‐kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl‐3‐dependent NF‐kappaB signaling. Cell. 2006;125:665‐677. [DOI] [PubMed] [Google Scholar]
- 44. Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21‐27. [DOI] [PubMed] [Google Scholar]
- 45. Massoumi R, Kuphal S, Hellerbrand C, et al. Down‐regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med. 2009;206:221‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strobel P, Zettl A, Ren Z, et al. Spiradenocylindroma of the kidney: clinical and genetic findings suggesting a role of somatic mutation of the CYLD1 gene in the oncogenesis of an unusual renal neoplasm. Am J Surg Pathol. 2002;26:119‐124. [DOI] [PubMed] [Google Scholar]
- 47. Reissig S, Hovelmeyer N, Weigmann B, et al. The tumor suppressor CYLD controls the function of murine regulatory T cells. J Immunol. 2012;189:4770‐4776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


