Abstract
Objectives
Stiffness of bone tissue differs response to its physiological or pathological status, such as osteoporosis or osteosclerosis. Consequently, the function of cells residing in bone tissue including osteoblasts (OBs), osteoclasts and osteocytes will be affected. However, to the best of our knowledge, the detailed mechanism of how extracellular matrix stiffness affects OB function remains unclear.
Materials and methods
We conducted a study that exposed rat primary OBs to polydimethylsiloxane substrates with varied stiffness to investigate the alterations of cell morphology, osteoblastic differentiation and its potential mechanism in mechanotransduction.
Results
Distinctive differences of cell shapes and vinculin expression in rat osteoblasts were detected on different PDMS substrates. As representatives for OB function, expression of alkaline phosphatase, Runx2 and osteocalcin were identified and showed a decrease trend as substrates become soft, which is associated with the Rho/ROCK signalling pathway.
Conclusions
Our results indicated substrate elasticity as a potent regulator in OBs functionalization, which may pave a way for further understanding of bone diseases as well as a potential therapeutic alternative in tissue regeneration.
Keywords: extracellular matrix, mechanotransduction, osteoblasts, substrate stiffness
1. Introduction
In physiological condition, the balance between bone formation and bone resorption is well maintained for metabolic homeostasis. Three critical and vital cell types in bone tissues – osteoblasts (OBs), osteoclasts and osteocytes – form the complex and intricate communication network. However, unexpected OB‐mediated osteogenic activity or osteoclast‐dependent bone resorption leads to pathological alteration in bone quality or/and quantity, resulting in osteosclerosis or osteoporosis. As a load‐bearing tissue, bone function is critically associated with its physical properties. In this regard, tissue stiffness, an inherent physical characteristic of body organs, should be seriously considered. Stiffness of normal tissues varies from 1 kPa (like brain) to stiffer 6 kPa (like articular cartilage), and even 15 MPa (like mineralized bone tissue).1, 2 However, tissues in different physiological situations present varied degrees of stiffness. For instance, stiffness of subchondral bone plate in both osteoporosis and osteoarthritis patients is much lower than that in normal condition.3 In the past decades, matrix stiffness has been proven to be a potent regulator in cell living process including cell adhesion, growth, migration and lineage specification.4, 5, 6, 7 A growing body of evidence has shown that rigid matrices drive mesenchymal stem cells undergoing osteogenic lineage differentiation by enhanced osteogenic marker genes or proteins such as alkaline phosphatase (ALP), osteocalcin, osteopontin and Runx2.8, 9, 10 Therefore, we hypothesize that different degrees of stiffness may contribute to the alteration of OBs functionalization.
Extracellular matrix (ECM) is a collection of various components, which has a close relationship with the cells that reside in. Specifically, OBs mediate extracellular bone matrix formation by calcium phosphate deposition, in which lysyl oxidase undergoes the intra/intermolecular cross‐linking of collagen fibrils and thereby regulates the density and stiffness of bone matrix.11, 12 In turn, ECM presents as a biochemical provider of massive growth factors,13 and also a structural supporter, in which the biophysical cues is a potent regulator for cell living as well as a driving force behind multi‐lineage differentiation.14, 15 Given that the potent effects of biomechanical cues on cell behaviour, studies focusing the natural or artificial replication of ECM are garnering attention, in which substrate stiffness,16 viscoelasticity17 and topography18 are specifically considered. In this regard, research models have been established to examine the effect of biomechanical cues, in particular, matrices stiffness on terminal cells19 or stem cells.5 Optimal functionalization of primary chondrocytes reached as cultured on substrates with stiffness of 5.4 kPa, akin to that existing in vivo.20 Mesenchymal stem cells undergo osteogenic lineage differentiation on comparatively rigid polyacrylamide hydrogels with stiffness of 25‐40 kPa, while showed preference to neural lineage with stiffness 0.1‐1 kPa and muscle of 8‐17 kPa.8 However, the intracellular signalling pathways that mediate the interaction between OBs and the underlying matrices with different degrees of stiffness are poorly understood and are still an attractive field of research.
In the studies focusing on the effects of biophysical cues on cell behaviour, two‐dimensional (2D) culture substrates are continually being improved for better mimicking the biophysical properties of extracellular microenvironments. Based on the priorities of non‐toxic to the attached cells and broad range of stiffness it presents, which can cover the range of those in all natural tissues, polydimethylsiloxane (PDMS) substrates were employed in our study. This kind of material is cross‐linked by base and curing agent, which could form user‐defined different degrees of stiffness. Then rat primary OBs were exposed to PDMS substrates, osteogenic markers and related potential pathway were assessed. The results not only improve our understanding of how OBs response to the biophysical microenvironment changes but also paves a way for the improvement of OBs function in bone reconstruction.
2. Materials and methods
2.1. Isolation and culture of rat primary osteoblasts
Primary calvarial OBs were isolated from rat skulls, as described in our previous study.21 Briefly, cranium was obtained from 3‐ to 5‐day‐old rats. After being washed twice in PBS and cut into small pieces, calvarium fragments were trypsinized for 30 minutes in 0.25% protease solution and replaced by being digested in 0.5% type I collagenase for 1 hour. Then fresh Dulbecco' s modified Eagle's medium (high‐glucose DMEM, 0.1 mmol/L non‐essential amino acids, 4 mmol/L l‐glutamine, 1% penicillin‐streptomycin solution; Hyclone, Logan, UT, USA) containing 10% heat‐activated foetal bovine serum was added at 1:1 (v/v) to cease digestion and the mixture was centrifuged for 5 minutes. After the supernatant was removed, tissues and cells were seeded into flasks and maintained in DMEM growth medium under standard conditions of 37°C and humidified atmosphere of 5% CO2. The second passage of purified OBs at 80%‐90% confluence was used and counted by cytometer in our study. The protocol to obtain animal materials in our study was strictly according to ethical principles and approved by our Institutional Review Board (IRB).
2.2. Preparation and characterization of various elastic PDMS substrates
Polydimethylsiloxane substrates (Sylgard 184, Corning, NY, USA) were fabricated via different ratios of base and curing agent, 10:1, 30:1 and 50:1, and were cast on Petri dishes after thoroughly mixing, then cross‐linked in an oven at 60°C for 24 hours. Young modulus of all substrates was measured by universal testing machine (Model 5565; Instron, Norwood, MA, USA) and stiffness for each was evaluated by the following equation:
K – stiffness, E – Young modulus, A – cross‐sectional area (cm2), L – height
For cell adhesion, self‐polymerization of dopamine was performed on substrate surface by being socked in the tris (hydroxymethyl) aminomethane (Adamas) and dopamine solution (pH8) of 24 hours for twice. Before cell seeding, PDMS substrates were sterilized via ultraviolet light for 1 hour.
2.3. Scanning electron microscope
For observation of cell attachment and morphological changes, OBs were plated on stiff (10:1~134 kPa), intermediate (30:1~16 kPa) and soft (50:1~1.4 kPa) PDMS substrates at low density for 3 days, OBs were fixed by 2.5% glutaraldehyde overnight and then dehydrated in a graded ethanol series (50%, 60%, 70%, 80%, 90%, 95% and 100%). After being dried in an exhaust hood, the specimens were mounted on specimen holders, coated with a thin layer of gold, and then examined by scanning electron microscope (SEM).
2.4. Immunofluorescence
After 7 days of incubation, rat primary OBs on different PDMS substrates were fixed in 4% paraformaldehyde and then permeabilized by 0.5% Triton X‐100. After blocking using 5% sheep serum, samples were subjected to immunofluorescence for the primary antibody of vinculin, osteocalcin, RhoA and ROCK‐2 (Abcam, Cambridge, MA, USA). The Alexa Fluor 594 donkey anti‐mouse (A21203; Invitrogen, Carlsbad, CA, USA) were used as secondary antibody and then stained with fluorescent FITC‐phalloidin (Sigma‐Aldrich, St. Louis, MO, USA) conjugate solution. The DAPI (Sigma‐Aldrich) was used for the staining of nuclei. For observing morphologies of OBs, images were captured by CLSM (Leica TCS SP8, Wetzlar, Germany).
2.5. Alkaline phosphatase staining
To assess the ability of osteoblastic differentiation induced by substrate stiffness alone, rat OBs were seeded onto PDMS prepared six‐well inserts at the density of 105 cells/well and cultured in normal DMEM media for 14 days, during which the culture media was changed every 3 days. Then ALP staining was carried out and OBs were fixed in 4% formaldehyde and incubated using staining mixture from cALP Stain Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The detailed protocol was followed by the manufacturer's instruction. After staining, images were captured with microscope.
2.6. Immunoblotting analysis
After 7 days of incubation, OBs were lysed using whole cell lysis assay (KeyGen BioTECH, Nanjing, China) according to the manufacturer's instructions. Total protein samples were prepared by mixing with Bio‐Rad Laemmli Sample Buffer and then separated in 8%‐15% sodium dodecyl sulphate polyacrylamide gel electrophoresis (according to the molecular weights of the GAPDH, RhoA and ROCK‐2). Proteins were transferred onto a polyvinylidene difluoride membrane and then blocked and incubated with primary antibody of GAPDH, RhoA and ROCK‐2 (Abcam) overnight at 4°C. After washing, blots were probed with 1:5000 anti‐goat IgG‐HRP for 1 hour at RT. Blot signals were finally detected with Santa Cruz Western Blotting Luminol Reagent Kit (sc 2048, Santa Cruz, CA, USA) and visualized by Kodak X‐AR (Kodak, Windsor, Colorado, USA). Data quantification for blot images were measured by optical density using Image‐J. The mean value of each group was normalized to that of group 10:1.
2.7. Real‐time PCR analysis
Briefly, total RNA of rat OBs was extracted using TRIzol at day 7. After purification, cDNA was synthesized using a synthesis kit (Mbi, GlenBirnie, MD, USA). The gene transcriptional levels of Ras‐related C3 botulinum toxin substrate 1 (Rac‐1), RhoA, ROCK‐1, ROCK‐2, Runx2 and ALP (sequences of forward and reverse primers of mRNA were shown in Table 1) in OBs were measured using PrimeScript™ RT‐PCR Kit (Takata, Tokyo, Japan) and ABI 7300 (Applied Biosystems, Shanghai, China). RT‐PCR amplification process was according to the manufacturer's instructions. Mean data of each group were first quantified relative to GAPDH, and then normalized to that of group 10:1.
Table 1.
Sequences of forward and reverse primers of selected genes designed for qPCR
| mRNA | Product length | Primer pairs | |
|---|---|---|---|
| GAPDH | 233 bp | Forward | ACAGCAACAGGGTGGTGGAC |
| Reverse | TTTGAGGGTGCAGCGAACTT | ||
| Runx2 | 106 bp | Forward | CCTCTGACTTCTGCCTCTGG |
| Reverse | GATGAAATGCCTGGGAACTG | ||
| ALP | 101 bp | Forward | CCTGACTGACCCTTCCCTCT |
| Reverse | CAATCCTGCCTCCTTCCACT | ||
| Rac‐1 | 200 bp | Forward | GAGAGTACATCCCCACCGTC |
| Reverse | AACACGTCTGTTTGCGGGTA | ||
| RhoA | 132 bp | Forward | AACAGGATTGGCGCTTTTGG |
| Reverse | GATGAGGCACCCCGACTTTT | ||
| ROCK‐1 | 108 bp | Forward | TTGGTTGGGACGTACAGTAAAA |
| Reverse | CGTAAGGAAGGCACAAATGAGA | ||
| ROCK‐2 | 135 bp | Forward | GACATTGAACAGCTTCGGTCGGA |
| Reverse | CATTGAACAGCTTCGGTCG | ||
2.8. Statistical analysis
All experiments were conducted in triplicate and reproduced at least three independent times. All statistical analysis was performed with spss 21.0 using one‐way ANOVA. Data were considered significantly different if the two‐tailed P value was <.05.
3. Results
3.1. Polydimethylsiloxane substrates with varied stiffness induce osteoblasts to form distinct morphologies
Fabrication of PDMS substrates was achieved by mixing different proportions of base and curing agent (Figure 1A). In our study, mechanical test of substrates showed a decrease in stiffness with increasing base to curing ratios. The stiffness of 10:1, 30:1 and 50:1 groups was approximately 134, 16, and 1.4 kPa, respectively, which in our study termed as rigid, intermediate and soft substrates (Figure 1B). To dissect the effect of stiffness on OBs living process, well–cross‐linked substrates with hydrophilic modification by dopamine treatment facilitate the adhesion of primary OBs (Figure 1C). From SEM images, OBs displayed distinct cell morphologies. Rigid substrate (~134 kPa) directed cells to spread widely with a polygonal shape, consistent with nature conditions. On intermediate substrate with the stiffness of 16kPa, the majority of OBs reshaped to fusiform fibroblasts‐like cells. However, cells cultured on soft one (~1.4kPa) yielded to quite small and round morphologies, as shrank and lay down in the soft matrix. These data demonstrated that the modified elastic substrates had the ability to facilitate the attachment of OBs for further study.
Figure 1.
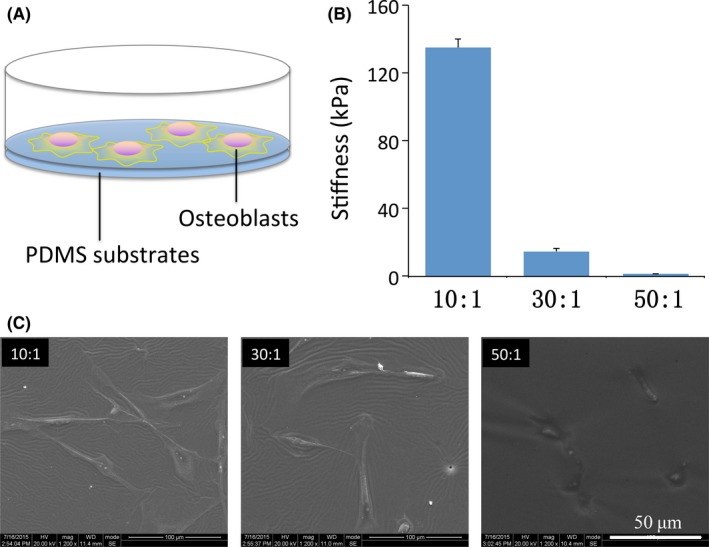
Polydimethylsiloxane (PDMS) substrates with varied stiffness induce osteoblasts to form distinct morphologies. (A) Schematic draft of osteoblasts exposed to PDMS substrates. (B) Stiff, intermediate and soft PDMS substrates fabricated by different proportions of base and curing agents (10:1, 30:1 and 50:1) displayed stiffness of 134, 16 and 1.4 kPa, respectively. The presented results are representative of three different samples (n=3). Data are presented as means ± SD. (C) SEM images of osteoblasts plated on varying elastic PDMS materials. Scale bars are 100 μm
3.2. Rigid substrates promote osteoblastic differentiation
Given the successful fabrication of PDMS elastic matrices to set a friendly platform for cell‐matrix interaction study, we then carried out three tests for osteoblastic differentiation‐extracellular ALP activity, transcription levels of Runx2 and ALP, and immunofluorescence expression of osteocalcin (Figures 2 and 3). Low magnification images clearly showed that formation of ALP, a specific marker for osteoblastic differentiation, increased in OBs with increasing substrate stiffness (Figure 2A), and further verified by close‐up images of each samples (Figure 2B). For corroboration at transcription level, we assessed the mRNA expressions of ALP and Runx2 in OBs at 7‐day post‐seeding (Figure 2C,D). The same trend of distinctively increased ALP expression was observed on rigid substrates. Statistically differences were found between expressions in rigid group and those in intermediate and soft ones (P<.05) (Figure 2D). Similarly, expression level of Runx2, the prime control gene for osteoblastogenesis, significantly increased in rigid substrate compared to softer ones (Figure 2C). Additionally, immunofluorescent staining of osteocalcin, a preliminary biomarker for bone mineralization was carried out and stronger expression was observed on rigid matrix, confirming that the functional level of OBs was enhanced by matrix stiffness (Figure 3).
Figure 2.
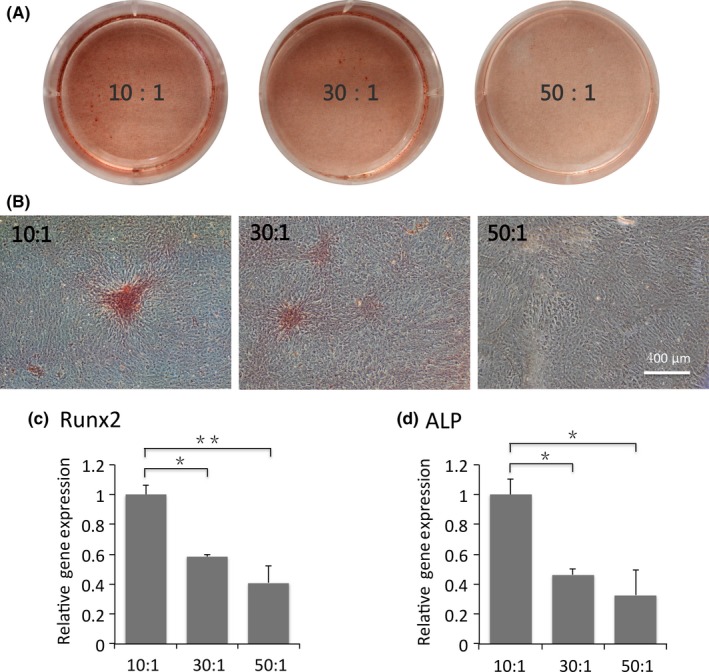
Rigid substrates promote osteoblastic differentiation. Macroscopic (A) and microscopic (B) images of alkaline phosphatase (ALP) staining in osteoblasts plated on stiff, intermediate and soft substrates after 14 days. Scale bars are 400 μm. (C) Transcriptional levels of Runx2 and ALP in osteoblasts at 7‐day post‐seeding. Data have been firstly normalized to GAPDH, and then normalized to that of group 10:1. The results shown are representative of three different samples (n=3). Data are presented as means ± SD, *P<.05, **P<.01
Figure 3.
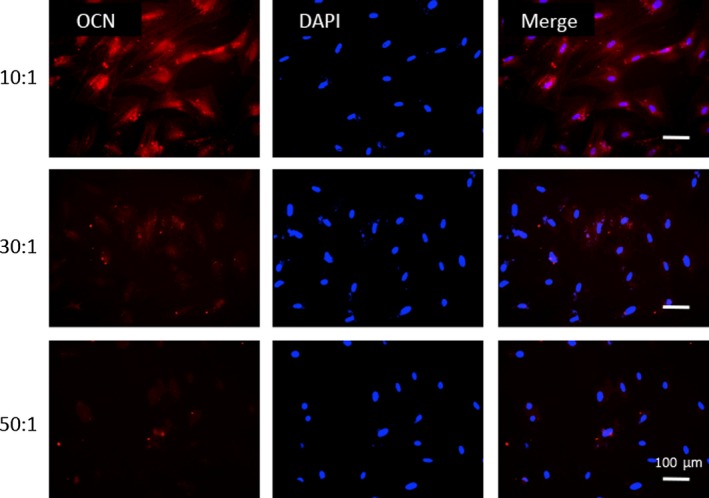
More osteocalcin expressed on stiffer substrates. Immunofluorescence images of osteocalcin expressed in osteoblasts cultured on stiff, intermediate and soft polydimethylsiloxane substrates. Osteocalcin (red) and nucleus (blue). Scale bars are 100 μm
3.3. Substrates stiffness modulates the assembly of actin fibres and distribution of vinculin in osteoblasts
Consistent with the consensus that cell reshaping and focal adhesion reorganization exhibited on elastic substrates, assembly of actin stress fibres and vinculin, a main recruited factor in the cell‐matrix interaction process, were also assessed in our study. Fluorescent images revealed that primary OBs cultured on rigid matrix had broad spreading area along with a high‐organized F‐actin network, while those cultured on softer matrices (eg, intermediate group of 16 kPa and soft group of 1.4 kPa) exhibited shrinking morphologies and shaped into round with poorly defined and fuzzy filaments (Figure 4). Similar alterations in cell morphologies were observed in SEM images (Figure 1C). Distribution of vinculin displayed obvious differences. Rod‐like expression of vinculin arranged along with cell axis was observed in fibroblast‐like OBs as the underlying substrates were rigid. However, cells cultured on compliant and soft matrices assembled ill‐defined actin fibres that terminated in decreased and fuzzy vinculin (Figure 4).
Figure 4.
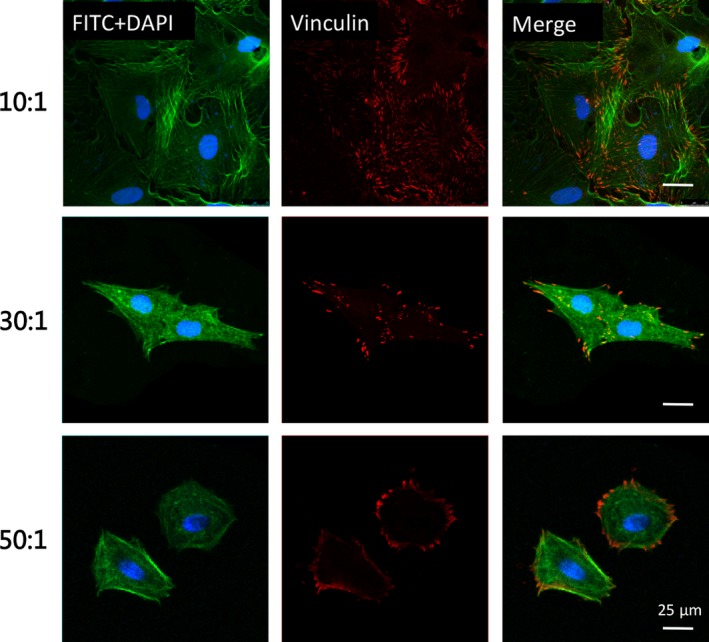
Substrates stiffness modulated the assembly of actin fibres and distribution of vinculin in osteoblasts. Immunofluorescence analysis of cytoskeleton staining by FITC‐phalloidin (green), vinculin (red) and DAPI (blue) in osteoblasts. Scale bars are 25 μm
3.4. Regulation of osteoblasts functionalization by matrix stiffness may be related to Rho/ROCK pathway
To probe the mechanism by which matrices stiffness modulates the osteoblastic differentiation, RhoA/ROCK signal, a well‐known pathway in mechanotransduction, was detected visually by immunofluorescence staining of RhoA and ROCK‐2 and qualitatively by protein blotting and transcriptional level measurements. From immunofluorescence images, RhoA and ROCK‐2 expressed stronger on rigid matrix (10:1) (Figure 5A). Similarly, expressions of the two proteins were increased with the increase in stiffness identified by Western blots (Figure 5B). Additionally, transcription levels of Rac‐1, RhoA, ROCK‐1 and ROCK‐2 genes showed statistical increase on stiff matrices (Figure 5C). However, expression of Rac‐1, ROCK‐1 and ROCK‐2 remained no statistically different to one another in intermediate and soft groups, indicating that primary OBs may not be sensitive to relatively small changes in stiffness, from 16 to 1.4 kPa.
Figure 5.
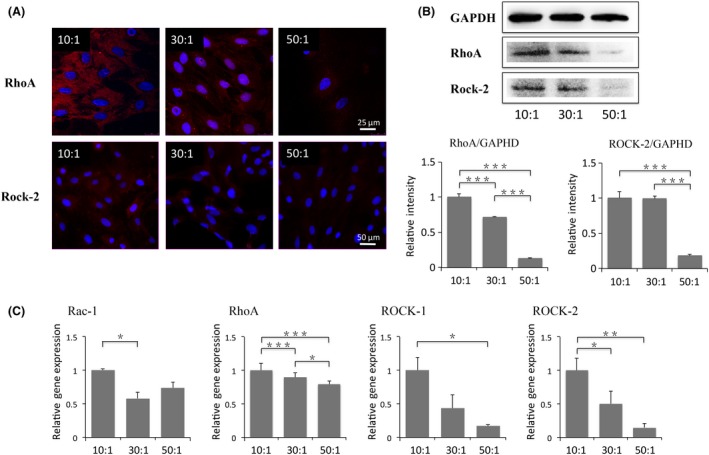
Regulation of osteoblasts functionalization by matrices stiffness may be related to Rho/ROCK pathway. (A) Immunofluorescence images of RhoA (red in the first row) and ROCK‐2 (red in the second row)) with DAPI (blue) in osteoblasts cultured on stiff, intermediate and soft substrates. Scale bars are 25 and 50 μm, respectively. (B) Protein expressions of RhoA and ROCK‐2 in osteoblasts plated on different elastic substrates. (C) Transcriptional levels of Rac‐1, RhoA, ROCK‐1 and ROCK‐2 in osteoblasts. Gene expressions are relative to that of GAPDH, and then normalized to those in 10:1 group. The results shown are representative of three different samples (n=3). Data are presented as means ± SD, *P<.05, **P<.01, ***P<.001
4. Discussion
The results of our study showed that the osteoblastic differentiation in primary OBs was more favoured when cultured on rigid substrates. Specifically, based on the successful employment of 2D PDMS materials with the superiorities of non‐toxicity and broad range of stiffness, we observed the distinct alteration of OBs morphologies and higher ALP formation and osteocalcin as well as enhanced transcription levels of Runx2 and ALP on stiffer matrices with stiffness of 134 kPa, compared to intermediate (~16 kPa) and soft ones (~1.4 kPa). Potential mechanism of stiffness‐dependent osteoblastic differentiation was identified to be related to Rho/ROCK pathway, which may be a promising approach not only for the understanding of pathological mechanism of bone diseases but also for biomaterial designing in bone tissue engineering.
In physiological or pathological conditions, the stiffness of bone tissue differs. For instance, reduction of mineralized bone matrix may lead to decreased bone density resulting in osteoporosis. A recent study reported that stiffness of subchondral bone plate in either osteoporosis or osteoarthritis patients were much lower than that of normal condition.3 Regarding cell level, the well‐known consensus goes to the breakdown of the balance between OB‐derived bone formation and osteoclast‐dependent bone resorption. As cells reside in a complex and elaborate three‐dimensional (3D) communication network, ECM, the components in ECM or their spatial arrangement which serves as a supporter for the network microstructure have biochemical and biophysical effects on cell living process. Previous studies have shown that one of the physical properties of ECM, stiffness, is recognized as a vital regulator of a variety of cell behaviours such as proliferation,22 migration,23 multiple differentiation8 as well as phenotype maintenance.20 Therefore, a speculation regarding the correlation between bone ECM stiffness and function of the embedded OBs is easily raised.
Fabrication or replication of culture materials is being developed to investigate cell‐matrix interaction for optimizing biophysical properties on cell behaviour. Examples are 2D culture formats24 where cells are adhesive to the surfaces, and 3D materials9, 25, 26 where cells are encapsulated in hydrogels. Although better mimicking in vivo microenvironment, it is hard to control its rigidity as well as replicating high degree of stiffness for 3D materials. Therefore, in our work, the well‐known PDMS material usually being involved in 2D cell‐matrix study was used. By mixing different proportions of base and curing agent, stiffness of 134, 16 and 1.4 kPa were achieved. Our previous study has confirmed that the surface roughness of different PDMS substrates was under 20 nm, which had been proved no statistically differences on cell behaviour.20 Water contact angle of 55° achieved by dopamine modification gave the priority for cell attachment, also see Figure 1C, which allowed us to focus on the sole effect of stiffness on OBs function. Our results showed distinct alteration in OBs morphologies cultured on rigid, intermediate and soft substrates. From SEM and immunofluorescence images, broad spreading area and polygonal fibroblast‐like morphology of OBs were observed on rigid substrate with stiffness of 134 kPa. While in the 1.4 kPa soft group, OBs seemed to sink into the base matrix. It can be explained that as traction force of cells on the surrounding environment is greater than the constrain stress given by matrices, OBs preferred round morphologies.
In our study, ALP, Runx2 and osteocalcin were chosen for the measurement of osteoblastic differentiation. Functional level represented by ALP formation and transcription level of marker genes in OBs showed the highest expression on rigid PDMS material with the stiffness of 134 kPa, indicating the well‐accepted consensus that polygonal shapes of OBs afford more mineralized matrix formation. As reduction in stiffness, a decreased trend was observed in marker gene or protein expressions. Our findings have corroborated the enhanced OBs functionalization on stiffer matrix, which was in accordance with previous studies.27, 28 It should be noted that, however, no significant changes in ALP activity and Runx2 gene expression induced by substrate stiffness were detected in human bone‐derived OB precursor cells, in contrast to mesenchymal stem cells isolated from umbilical cord in the same study, which showed a stiffness‐dependent response.29 A possible explanation may refer to the fact that MSCs are more sensitive to mechanical stimuli during OB differentiation. Interestingly, another research reported the possibility that osteogenic differentiation assessed by calcium staining and ALP activity detection of differentiated human dental follicle cells (DFCs) was initiated by comparatively soft matrix with a dexamethasone treatment,30 which was totally opposite to current opinion in stiffness‐mediated differentiation. The fact may be attributed to tissue specification and characteristic of periodontal development, in which a higher amount of periodontal ligament with increased softness‐directed DFCs differentiated into cementoblasts or OBs to maintain a balance in periodontium.
In current knowledge, the optimum stiffness for different cell types to form their functions is varied. The best stiffness for the phenotypic maintenance of chondrocytes is 5.6 kPa when cultured on 2D PDMS substrate,20 while for cell differentiation, MSCs preferred neuron lineage specification on stiffness of 0.1‐1 kPa and favoured an osteogenic differentiation on 25‐40 kPa matrix, all of which showed a similar stiffness in vivo.8 The ECM stiffness for OBs varies during its living process. From the osteogenic differentiation of MSCs to pre‐OBs and OBs, the ECM is relatively lower compared to that in the transformation process from OBs to osteocytes. It is reported that the stiffness of soft collagenous bone is around 100 kPa.8 As maturation, OBs undergo matrix secretion into the ECM which is termed as osteoid, and then followed by the mineralization which shows a higher stiffness of ECM,31, 32 up to 1000 kPa of mineralized bone.33 Thereby, the ECM stiffness for OBs is dynamic. Although our results showed the highest expression of markers on 134 kPa matrix, no solid conclusion of the best stiffness for OBs function can be made. Given that stiffness of bone tissue is tunable, studies focusing on replication of its natural physical property in vivo have been paid much attention, in which 2D matrices34 or 3D hydrogel biomaterials28 with gradient modulus are notable examples. These materials not only set a platform to understand cell‐material physical interaction but also give prospective to engineer seamless tissue interfaces for integration of hard tissues like mineralized bone and soft tissues like the adjacent articular cartilage or tendons.28
Substrate stiffness clearly activated vinculin expression and formed distinctively cell morphologies that transmit physical information of the surrounding microenvironment to intracellular signalling pathways in our work. As cells being adhesive to the underlying substrate, transmembrane integrin responded to extracellular stimuli immediately, and then transmitted the mechanical signal to integrin linked kinase ILK and focal adhesion to modulate cell motility. A clear confirmation has been made that matrix rigidity was bound up with enhanced integrin signalling35 as well as ILK,36 which was proved by the effects of depleting ILK by short hairpin RNA and ectopically overexpression using a recombinant adenovirus encoding for ILK on cancer stem cells behaviour. In our study, immunofluorescent staining of vinculin demonstrated clearly different distribution among varying stiffness groups, from strong and rod‐like expression on 134 kPa matrix to diffuse and spot‐like images on 1.4 kPa. For further quantitative analysis and corroboration, protein blotting and gene knockout experiments could be a point of future investigation. Although the lack of literature regarding the relationship between stiffness and vinculin limits our understanding of the detailed mechanism of stiffness‐related biophysical transmission, our results do underscore the importance of vinculin that had been recruited and showed an enhanced high‐organized expression on rigid matrix.
Although signalling pathways involved in mechanotransduction transferring from vinculin to phenotype alterations have gained much attention, no clear confirmation has been made yet. Recently, Ras superfamily of small guanosine triphosphatases‐Rho GTPase, of which the major members are Rho, Rac and Cdc42, has been found as a key regulator in cell morphology and migration.37, 38 Rho‐associated protein kinase (ROCK) acts as a downstream effector of small Rho GTPase and is a mediator in rearrangement of cell cytoskeleton.39 Based on that, stiffness‐dependent regulation in cell morphology and actin filaments was detected in our study, and the report on the relationship between Rho/ROCK pathway and MSC differentiation,40 thereby major proteins involved in this pathway such as RhoA, ROCK‐1, ROCK‐2 and Rac1 expressed in rat primary OBs have also been studied. Blotting and transcription levels of RhoA and ROCK‐2 were enhanced with the increase in matrix stiffness. Additionally, the same trend had been found in the transcription levels of Rac‐1 and ROCK‐1 that the expressions decreased as the matrix stiffness became soft. Consequently, a conclusion can be made that the Rho/ROCK pathway showed a stiffness‐dependent manner in mechanotransduction.
Limitations should also be noted in our study. Firstly, given that three major types of cells (OBs, osteoclasts and osteocytes) reside in bone tissue, the interaction and cross‐talk need to be carefully addressed when a comprehensive understanding of the precisely organized niche is called.41 Although the preference of soft matrix for osteoclast differentiation and specific favour of ECM stiffness on the effect of RANK/RANK‐L/OPG axis in osteocoupling process27 as well as morphological changes and phenotypic transferring in OB‐osteocyte differentiation31, 42 have been recently reported, more well‐designed model for cell‐matrix study in bone biology should also be further conducted. Secondly, the downstream molecules of Rho/ROCK pathway in the regulation of osteoblastic differentiation are still needed. In summary, this work presents a study on the understanding of cell‐matrix interactions in 2D format that mimics the stiffness experienced by OBs in vivo. Results presented demonstrated that rigid substrate promoted natural OB morphology maintenance and osteoblastic differentiation with a potential mechanism by which vinculin and Rho/ROCK signalling pathway were involved. Physical properties like stiffness of surrounding microenvironment should be considered in future aetiological study as well as an alternative to biochemical cues in optimizing cell‐material interaction in tissue engineering.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (81471803) and Sichuan Science and Technology Innovation Team (2014TD0001).
Zhang T, Lin S, Shao X, et al. Effect of matrix stiffness on osteoblast functionalization. Cell Prolif. 2017;50:e12338 10.1111/cpr.12338
References
- 1. Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30:6867–6878. [DOI] [PubMed] [Google Scholar]
- 2. Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93:4453–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li B, Aspden RM. Mechanical and material properties of the subchondral bone plate from the femoral head of patients with osteoarthritis or osteoporosis. Ann Rheum Dis. 1997;56:247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Assoian RK, Klein EA. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008;18:347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banks JM, Mozdzen LC, Harley BA, Bailey RC. The combined effects of matrix stiffness and growth factor immobilization on the bioactivity and differentiation capabilities of adipose‐derived stem cells. Biomaterials. 2014;35:8951–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaudhuri O, Koshy ST, Branco da Cunha C, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13:970–978. [DOI] [PubMed] [Google Scholar]
- 7. Guo Q, Liu C, Li J, Zhu C, Yang H, Li B. Gene expression modulation in TGF‐beta3‐mediated rabbit bone marrow stem cells using electrospun scaffolds of various stiffness. J Cell Mol Med. 2015;19:1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. [DOI] [PubMed] [Google Scholar]
- 9. Chen G, Dong C, Yang L, Lv Y. 3D scaffolds with different stiffness but the same microstructure for bone tissue engineering. ACS Appl Mater Interfaces. 2015;7:15790–15802. [DOI] [PubMed] [Google Scholar]
- 10. Jiang P, Mao Z, Gao C. Combinational effect of matrix elasticity and alendronate density on differentiation of rat mesenchymal stem cells. Acta Biomater. 2015;19:76–84. [DOI] [PubMed] [Google Scholar]
- 11. Bailey AJ, Wotton SF, Sims TJ, Thompson PW. Biochemical changes in the collagen of human osteoporotic bone matrix. Connect Tissue Res. 1993;29:119–132. [DOI] [PubMed] [Google Scholar]
- 12. Knott L, Bailey AJ. Collagen cross‐links in mineralizing tissues: a review of their chemistry, function, and clinical relevance. Bone. 1998;22:181–187. [DOI] [PubMed] [Google Scholar]
- 13. Zhu J, Clark RA. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM‐GF paradigm. J Invest Dermatol. 2014;134:895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye K, Wang X, Cao L, et al. Matrix stiffness and nanoscale spatial organization of cell‐adhesive ligands direct stem cell fate. Nano Lett. 2015;15:4720–4729. [DOI] [PubMed] [Google Scholar]
- 15. Wen JH, Vincent LG, Fuhrmann A, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lv H, Li L, Sun M, et al. Mechanism of regulation of stem cell differentiation by matrix stiffness. Stem Cell Res Ther. 2015;6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar S, Maxwell IZ, Heisterkamp A, et al. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys J. 2006;90:3762–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim HS, Kim YJ, Jang JH, Park JW. Surface engineering of nanostructured titanium implants with bioactive ions. J Dent Res. 2016;95:558–565. [DOI] [PubMed] [Google Scholar]
- 19. Schuh E, Hofmann S, Stok KS, Notbohm H, Muller R, Rotter N. The influence of matrix elasticity on chondrocyte behavior in 3D. J Tissue Eng Regen Med. 2012;6:e31–e42. [DOI] [PubMed] [Google Scholar]
- 20. Zhang T, Gong T, Xie J, et al. Softening substrates promote chondrocytes phenotype via RhoA/ROCK pathway. ACS Appl Mater Interfaces. 2016;8:22884–22891. [DOI] [PubMed] [Google Scholar]
- 21. Zhang T, Xie J, Sun K, et al. Physiological oxygen tension modulates soluble growth factor profile after crosstalk between chondrocytes and osteoblasts. Cell Prolif. 2016;49:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hadjipanayi E, Mudera V, Brown RA. Close dependence of fibroblast proliferation on collagen scaffold matrix stiffness. J Tissue Eng Regen Med. 2009;3:77–84. [DOI] [PubMed] [Google Scholar]
- 23. Zaman MH, Trapani LM, Sieminski AL, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell‐matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schellenberg A, Joussen S, Moser K, et al. Matrix elasticity, replicative senescence and DNA methylation patterns of mesenchymal stem cells. Biomaterials. 2014;35:6351–6358. [DOI] [PubMed] [Google Scholar]
- 25. Gholipourmalekabadi M, Sameni M, Radenkovic D, Mozafari M, Mossahebi‐Mohammadi M, Seifalian A. Decellularized human amniotic membrane: how viable is it as a delivery system for human adipose tissue‐derived stromal cells? Cell Prolif. 2016;49:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ardeshirylajimi A, Farhadian S, Adegani FJ, et al. Enhanced osteoconductivity of polyethersulphone nanofibres loaded with bioactive glass nanoparticles in in vitro and in vivo models. Cell Prolif. 2015;48:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hwang MP, Subbiah R, Kim IG, et al. Approximating bone ECM: crosslinking directs individual and coupled osteoblast/osteoclast behavior. Biomaterials. 2016;103:22–32. [DOI] [PubMed] [Google Scholar]
- 28. Chatterjee K, Lin‐Gibson S, Wallace WE, et al. The effect of 3D hydrogel scaffold modulus on osteoblast differentiation and mineralization revealed by combinatorial screening. Biomaterials. 2010;31:5051–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Witkowska‐Zimny M, Wrobel E, Mrowka P. Alpha2beta1 integrin‐mediated mechanical signals during osteodifferentiation of stem cells from the Wharton's jelly of the umbilical cord. Folia Histochem Cytobiol. 2014;52:297–307. [DOI] [PubMed] [Google Scholar]
- 30. Viale‐Bouroncle S, Vollner F, Mohl C, et al. Soft matrix supports osteogenic differentiation of human dental follicle cells. Biochem Biophys Res Commun. 2011;410:587–592. [DOI] [PubMed] [Google Scholar]
- 31. Mc Garrigle MJ, Mullen CA, Haugh MG, Voisin MC, McNamara LM. Osteocyte differentiation and the formation of an interconnected cellular network in vitro. Eur Cell Mater. 2016;31:323–340. [DOI] [PubMed] [Google Scholar]
- 32. Barragan‐Adjemian C, Nicolella D, Dusevich V, Dallas MR, Eick JD, Bonewald LF. Mechanism by which MLO‐A5 late osteoblasts/early osteocytes mineralize in culture: similarities with mineralization of lamellar bone. Calcif Tissue Int. 2006;79:340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moffat KL, Sun WH, Pena PE, et al. Characterization of the structure‐function relationship at the ligament‐to‐bone interface. Proc Natl Acad Sci USA. 2008;105:7947–7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rao N, Grover GN, Vincent LG, et al. A co‐culture device with a tunable stiffness to understand combinatorial cell‐cell and cell‐matrix interactions. Integr Biol (Camb). 2013;5:1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levental KR, Yu H, Kass L, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hannigan G, Troussard AA, Dedhar S. Integrin‐linked kinase: a cancer therapeutic target unique among its ILK. Nat Rev Cancer. 2005;5:51–63. [DOI] [PubMed] [Google Scholar]
- 37. Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. [DOI] [PubMed] [Google Scholar]
- 38. Evers EE, Zondag GC, Malliri A, et al. Rho family proteins in cell adhesion and cell migration. Eur J Cancer. 2000;36:1269–1274. [DOI] [PubMed] [Google Scholar]
- 39. Amano M, Nakayama M, Kaibuchi K. Rho‐kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken). 2010;67:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maharam E, Yaport M, Villanueva NL, et al. Rho/Rock signal transduction pathway is required for MSC tenogenic differentiation. Bone Res. 2015;3:15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Q, Gu Y, Zhang W, Dziopa L, Zilberberg J, Lee W. Ex vivo 3D osteocyte network construction with primary murine bone cells. Bone Res. 2015;3:15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mullen CA, Haugh MG, Schaffler MB, Majeska RJ, McNamara LM. Osteocyte differentiation is regulated by extracellular matrix stiffness and intercellular separation. J Mech Behav Biomed Mater. 2013;28:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]


