Abstract
Objectives
Present evidence has suggested that large tumour suppressor 2 (LATS2) is abnormally expressed in most human cancer. However, the clinical and prognostic value in hepatocellular carcinoma (HCC) is still unknown.
Materials and methods
Large tumour suppressor 2 mRNA and protein expression levels in HCC tissues and cell lines were detected by qRT‐PCR, immunohistochemistry or Western blot. The correlation between LATS2 expression and clinicopathological factors was analysed through immunohistochemistry. The function of LATS2 on HCC cell growth and mobility was explored through MTT, colony formation, Transwell migration and invasion assays. The molecular mechanism of LATS2 was screened and confirmed by qRT‐PCR and Western blot.
Results and conclusion
In this study, LATS2 mRNA and protein expressions were decreased in HCC tissues and cell lines compared with normal hepatic tissues and hepatic cell line. Low LATS2 expression was oppositely corrected with tumour stage, vascular invasion and metastasis. The univariate and multivariate analyses suggested that low LATS2 expression was an independent poor prognostic factor for HCC patients. The in vitro experiments showed that LATS2 regulated HCC cells migration and invasion, but had no effect on HCC cells proliferation. Meanwhile, LATS2 modulated metastasis‐associated genes expression including E‐cadherin, vimentin, snail, slug, MMP2 and MMP9. In conclusion, LATS2 is a prognostic biomarker and a tumour metastasis suppressor in HCC.
Keywords: biomarker, hepatocellular carcinoma, KPM, LATS2
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumour and the third leading cause of cancer death worldwide.1 Hepatitis B virus (HBV) or hepatitis C virus infection, excessive drinking and foodstuff contamination with aflatoxins are the main risk factors for HCC.2, 3, 4, 5 In China, HCC is the most commonly diagnosed cancer and the leading cause of cancer death in men before the age of 60 years.6 The high incidence of HCC in China mainly is attributed to high rate of HBV infection, approximately 5‐8% in the general population.7, 8 Despite the rapid development of targeted therapy, the prognosis of HCC patient remains poor.9 It is urgent for HCC patients to identify more available biomarkers for developing targeted therapies.
Large tumour suppressor 2 (LATS2, also known as KPM) is one member of the novel LATS tumour suppressor gene family and is located in human chromosome 13q11‐12.10 Recently, some studies have suggested that LATS2 is involved in diverse cellular processes, such as growth, migration, invasion apoptosis and angiogenesis.11 LATS2 is generally expressed low in human cancer (non‐small cell lung cancer12 and ovarian cancer13), but is overexpressed in nasopharyngeal carcinoma.14 In HCC, down‐regulation of LATS2 expression could promote tumour cell invasion through decreasing YAP1/TEAD2 phosphorylation and leading to YAP1/TEAD2 transcriptional activation.15 Moreover, LATS2 served as a target of miR‐195 to regulate HCC cell apoptosis.16 However, the clinical significance of LATS2 is still unclear. In our study, we evaluated LATS2 mRNA and protein expressions in HCC tissues and cell lines. Meanwhile, clinical and prognostic values of LATS2 were observed in 106 HCC cases. Furthermore, the biological function and molecular mechanism of LATS2 were explored in HCC cell lines.
2. Materials and methods
2.1. Clinical samples collection
This study was approved by the Ethic Committee of Affiliated Tumour Hospital of Guangxi Medical University and Affiliated Minzu Hospital of Guangxi Medical University. All patients provided written informed consent. A total of 106 primary HCC tissues and 53 matched adjacent non‐cancerous hepatic tissues were collected at our hospital from January 2005 to December 2015. Before surgical therapy, none of HCC patients in this study had received neoadjuvant treatment. Tissues were respectively stored in liquid nitrogen and formaldehyde solution after surgical resection. The histological diagnosis and differentiation were evaluated independently by two pathologists based on the WHO classification system.
2.2. RNA preparation and quantitative real‐time PCR
Total RNA was extracted from tissues cell lines using RNAiso Plus (Takara, Japan), and 1 μg total RNA from each sample was reverse transcribed using the PrimeScript RT Master Mix (Takara, Japan). Real‐time q‐PCR analysis was done using SYBR Premix Ex Taq TM II (Takara, Japan) on a Light Cycler (Roche, USA). The PCR conditions were 30 seconds at 95°C, followed by 40 cycles at 95°C for 5 seconds and 60°C for 20 seconds. Finally, the 2−ΔCt method was performed to calculate the relative expression. All qRT‐PCR reactions were performed in triplicate.
2.3. Immunohistochemistry
Immunohistochemical analysis was performed to measure LATS2 protein expression in 106 primary HCC tissues and 53 matched adjacent non‐cancerous hepatic tissues. In brief, slides were baked at 60°C for 1 hour, followed by deparaffinization with xylene, and rehydrated. The sections were submerged in EDTA antigenic retrieval buffer and microwaved for antigen retrieval. They were then treated with 3% hydrogen peroxide in methanol to quench endogenous peroxidase activity, followed by incubation with 5% bovine serum albumin to block non‐specific binding. Sections were incubated with anti‐human anti‐LATS2 antibody (1:160 dilution; Cell Signaling Technology, USA) overnight at 4°C. Negative controls were incubated with negative control antibody under the same condition. After washing, tissue sections were treated with secondary antibody, followed by incubation with conjugated horseradish peroxidase (HRP) streptavidin. Tissue sections were then counterstained with haematoxylin, dehydrated, and mounted. Finally, sections were viewed under a bright‐field microscope.
2.4. Staining evaluation
The tissue sections stained immunohistochemically for LATS2 were reviewed, and scored separately by two pathologists blinded to the clinical parameters. For LATS2 assessment, staining intensity was scored as 0, negative; 1, weak; 2, moderate; or 3, strong. Extent of staining was scored as 0, negative; 1, <10%; 2, 11%‐50%; 3, 51%‐80%; or 4, >80% positive cells. The final score was determined by the combined staining score and proportion score (immunoreactive score=intensity score × proportion score). Low expression of LATS2 was defined as 1‐4 scores and high expression of LATS2 was defined as more than four scores (including four scores).13
2.5. Western blot
Total protein was extracted using cell lysis buffer (Beyotime, China) for Western blot. Equal amounts of protein were denatured and then separated by 8%‐12% SDS‐PAGE. The target proteins were incubated with the following primary antibodies: LATS2, E‐cadherin, vimentin, snail, slug, MMP2, MMP9 (Cell Signaling Technology, USA), β‐actin or GAPDH antibody (CWBIO, China). The proteins were then incubated with homologous secondary antibodies (Cell Signaling Technology, USA). For HRP detection, an ECL chemiluminescence kit (CWBIO, China) was used. Intensity of blots was performed by Quantity One Software (Bio‐Rad, USA).
2.6. Hepatocellular carcinoma cell lines and normal hepatic cell line cultures
The liver cancer cell lines (MHCC97H, MHCC97L, HepG2), and the normal hepatic cell liver cell line (L02) were obtained from the Shanghai Cell Bank (Shanghai, China). MHCC97H, MHCC97L, HepG2 and L02 cells were grown in DMEM (Dulbecco's Modified Eagle Medium) (Invitrogen, USA). All media were supplemented with 10% foetal calf serum (Invitrogen, USA) and 100 IU/mL penicillin (Sigma, USA).
2.7. Plasmid and siRNA transfection
The small‐interfering RNA was synthesized from GenePharma (Shanghai, China). The coding sequence region of human LATS2 gene was amplified from cDNA and cloned into pcDNA3.1 express vector. The plasmid was synthesized from GeneChem (Shanghai, China) and the resulting constructs were confirmed by DNA sequencing. Cells were transfected using lipofectamine TM 3000 reagent (Invitrogen, USA) in Opti‐MEM (Gibco, USA), according to the manufacturer's instructions. The transfection efficiency was tested by qRT‐PCR and Western blot.
2.8. MTT assay
Hepatocellular carcinoma cells were plated in a 96‐well plate. After incubation at 37°C for 24, 48, 72, 96 and 120 h, MTT (10 μL, 5 mg/mL) was added, and then incubated at 37°C for 4 hours. After that, the supernatant was removed, and 100 μL DMSO was added. The absorbance value was detected at 490 nm. Experiments were performed three times.
2.9. Colony formation assay
Hepatocellular carcinoma cells were plated in six‐well culture plates at 100 cells/well. Each cell group had three wells. After incubation for 12 days at 37°C, cells were washed twice with phosphate‐buffered saline and stained with the Giemsa solution (Sigma, USA). The number of colonies containing >50 cells was counted under a microscope.
2.10. Cell migration and invasion assays
Cell migration and invasion experiments were performed using 16 plates. Prior to migration and invasion experiments, cells were starved of FBS for 24 hours. About 2 × 104 cells were suspended in serum‐free medium and seeded in the upper wells. Lower chambers contained media with FBS to assess the migratory behaviour of cultured cells, or without FBS as a negative control accordingly. For the invasion assay, the surface of the upper chamber was covered with a monolayer of 5% growth factor‐reduced matrigel (BD Biosciences, USA). Cells adhering to the lower surface were fixed with methanol and stained with 0.1% crystal violet (Beyotime, China). The number of cells in the membrane was counted from five randomly selected visual fields with a microscope.
2.11. Statistical analysis
All values were represented as mean ± standard deviation (SD). spss 17.0 software was used to perform statistical analysis. The associations of clinicpathological characteristics with LATS2 expression was determined using χ2 tests. Univariate and multivariate Cox regression models were adopted to evaluate prognostic significance. The significance of survival variables in univariate analysis was included into the final multivariable Cox proportional hazards model. Statistical significant difference between groups was measured using the Student's t‐test. P‐values in all experiments were considered statistically significant at ≤.05.
3. Results
3.1. LATS2 mRNA and protein expressions are decreased in hepatocellular carcinoma tissues and cell lines
In order to explore the status of LATS2 HCC tissues, we conducted qRT‐PCR and immunohistochemistry to measure LATS2 mRNA and protein in HCC tissues. Compared with non‐cancerous hepatic tissues, LATS2 mRNA expression was decreased in HCC tissues with an average of 3.27‐fold (P<.001, Figure 1A). We detected LATS2 protein expression in 106 primary HCC tissues and 53 matched adjacent non‐cancerous hepatic tissues using immunohistochemical staining (Figure 2A‐F). Specific LATS2 protein staining was found in the cytoplasm of hepatic cell and tumour cell. We found that LATS2 protein expression was low in HCC tissues in 65.1% (69/106). In comparison, only 22.6% of hepatic tissues showed low expression of LATS2 protein, obviously lower than that in the HCC tissues (P<.001, Table 1).
Figure 1.
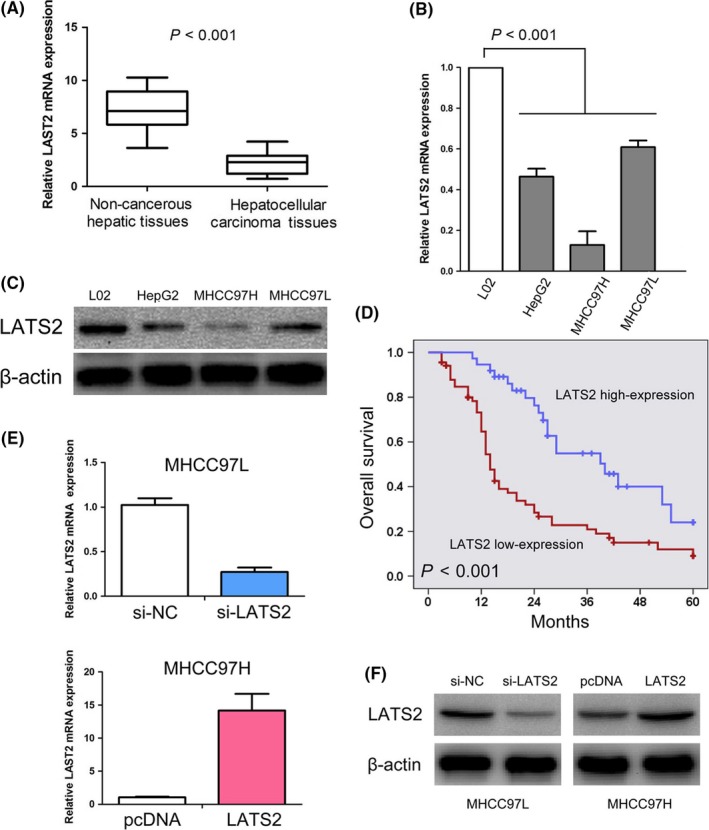
LATS2 expressions in hepatocellular carcinoma tissues and cell lines. (A) Expression of LATS2 is decreased in hepatocellular carcinoma tissues compared with normal hepatic tissues. (B) LATS2 mRNA was markedly lower in hepatocellular carcinoma cell lines (HepG2, MHCC97H and MHCC97L) compared with normal hepatic cell line (L02). (C) LATS2 expression was decreased in hepatocellular carcinoma cell lines (HepG2, MHCC97H and MHCC97L) compared with normal hepatic cell line (L02). (D) Low LATS2 expression predicts an unfavourable prognosis in hepatocellular carcinoma patients. (E, F) MHCC97L cells transfected with si‐LATS2 showed down‐regulation of LATS2 mRNA and protein expressions. MHCC97H cells transfected with pcDNA‐LATS2 resulted in up‐regulation of LATS2 mRNA and protein expressions
Figure 2.
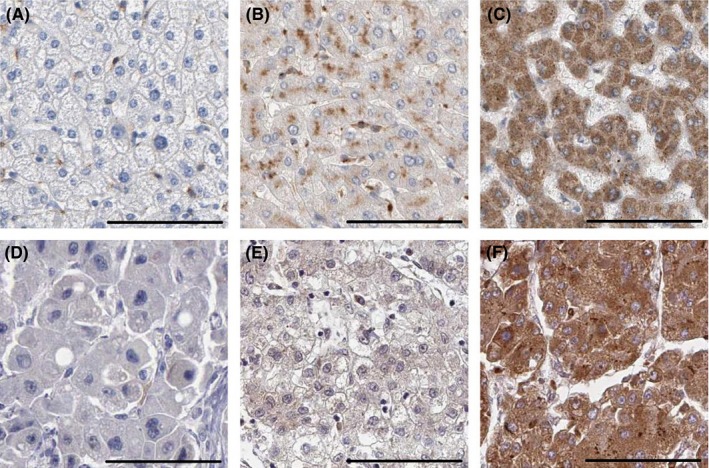
Immunohistochemical staining of LATS2 (the label is 100 μm). (A) Negative expression of LATS2 in normal hepatic tissue. (B) Low expression of LATS2 in normal hepatic tissue. (C) High expression of LATS2 in normal hepatic tissue. (D) Negative expression of LATS2 in hepatocellular carcinoma tissue. (E) Low expression of LATS2 in hepatocellular carcinoma tissue. (F) High expression of LATS2 in hepatocellular carcinoma cancer tissue
Table 1.
Expression of LATS2 protein between hepatocellular carcinoma tissues and non‐cancerous hepatic tissues
| Group | Cases | LATS2 | P | |
|---|---|---|---|---|
| Low expression (%) | High expression (%) | |||
| Hepatocellular carcinoma tissues | 106 | 69 (65.1) | 37 (34.9) | <.001 |
| Hepatic tissues | 53 | 12 (22.6) | 41 (77.4) | |
The status of LATS2 mRNA and protein expressions in HCC lines was detected by qRT‐PCR and Western blot. We found LATS2 mRNA was markedly lower in HCC cell lines compared with normal hepatic cell line (P<.001, Figure 1B). Similarly, LATS2 protein expression was low in HCC cell lines compared with normal hepatic cell line (Figure 1C).
3.2. LATS2 expression is corrected with malignant status of hepatocellular carcinoma patients
In order to explore the clinical significance of LATS2 in HCC patients, LATS2 protein expression was measured in 106 primary HCC samples through immunohistochemistry (Figure 2A‐F). The correlation between clinicopathological characteristics and LATS2 expression in HCC patients is summarized in Table 2. We indicated that low LATS2 expression is oppositely corrected with tumour stage (I‐II vs III‐IV, P=.007), vascular invasion (absent vs present, P<.001) and metastasis (absent vs present, P=.001). However, there were no significant relationships between LATS2 expression and age (<50 years vs ≥50 years, P=.783), gender (female vs male, P=.245), tumour size (<5 cm vs ≥5 cm, P=.127), HBV infection (absent vs present, P=.362) and histological differentiation (well vs moderate/poor, P=.261).
Table 2.
Correlations between LATS2 expression and clinicopathological characteristics in hepatocellular carcinoma
| Characteristics | n | Low expression (%) | High expression (%) | P |
|---|---|---|---|---|
| Age (years) | ||||
| 50 | 42 | 28 (66.7) | 14 (33.3) | .783 |
| ≥50 | 64 | 41 (64.1) | 23 (35.9) | |
| Gender | ||||
| Female | 38 | 22 (57.9) | 16 (42.1) | .245 |
| Male | 68 | 47 (69.1) | 21 (30.9) | |
| Tumour stage | ||||
| I‐II | 34 | 16 (47.1) | 18 (52.9) | .007 |
| III‐IV | 72 | 53 (73.6) | 19 (26.4) | |
| Tumour size | ||||
| 5 cm | 64 | 38 (59.4) | 26 (40.6) | .127 |
| ≥5 cm | 42 | 31 (73.8) | 11 (26.2) | |
| Vascular invasion | ||||
| Absent | 54 | 26 (48.1) | 28 (51.9) | <.001 |
| Present | 52 | 43 (82.7) | 9 (17.3) | |
| Metastasis | ||||
| Absent | 89 | 52 (58.4) | 37 (41.6) | .001 |
| Present | 17 | 17 (100) | 0 (0) | |
| HBV infection | ||||
| Absent | 80 | 54 (67.5) | 26 (32.5) | .362 |
| Present | 26 | 15 (57.7) | 11 (42.3) | |
| Histological differentiation | ||||
| Well | 65 | 45 (69.2) | 20 (30.8) | .261 |
| Moderate/Poor | 41 | 24 (58.5) | 17 (41.5) | |
3.3. Low LATS2 expression predicts an unfavourable prognosis in hepatocellular carcinoma patients
We further explored the prognostic significance of LATS2 in HCC patients. Kaplan‐Meier survival analysis suggested that HCC patients who expressed low level of LATS2 had lower overall survival compared to patients with high expression of LATS2 (P<.001, Figure 1D). Moreover, we performed univariate analysis and identified five prognostic parameters: tumour stage, tumour size, vascular invasion, metastasis and LATS2 expression. Furthermore, we found that low expression of LATS2 was an independent poor prognostic factor for HCC patients through multivariate analysis (HR=0.459, 95% CI: 0.247‐0.851, P=.012, Table 3).
Table 3.
Univariate and multivariate Cox regression of prognostic factors for overall survival in hepatocellular carcinoma patients
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (<50 vs ≥50) | 0.810 | 0.507‐1.295 | .379 | |||
| Gender (female vs male) | 1.258 | 0.759‐2.084 | .373 | |||
| Tumour stage (I‐II vs III‐IV) | 2.620 | 1.516‐4.530 | .001 | 2.798 | 1.133‐6.911 | .026 |
| Tumour size (<5 cm vs ≥5 cm) | 2.087 | 1.298‐3.354 | .002 | 0.963 | 0.521‐1.781 | .905 |
| Vascular invasion (absent vs present) | 1.966 | 1.222‐3.164 | .005 | 0.490 | 0.228‐1.050 | .067 |
| Metastasis (absent vs present) | 7.038 | 3.642‐13.602 | <.001 | 5.822 | 2.709‐12.512 | <.001 |
| HBV infection (absent vs present) | 0.785 | 0.459‐1.344 | .378 | |||
| Histological differentiation (well vs moderate/poor) | 0.944 | 0.582‐1.532 | .817 | |||
| LATS2 expression (low vs high) | 0.365 | 0.215‐0.620 | <.001 | 0.459 | 0.247‐0.851 | .013 |
HR, hazard ratio; 95% CI, 95% confidence interval.
3.4. LATS2 has no effect on hepatocellular carcinoma cells proliferation
We analysed the expression level of LATS2 in a panel of HCC cell lines including HepG2, MHCC97H (high metastatic ability) and MHCC97L (low metastatic ability). We observed that LATS2 expression was relatively lower in MHCC97H cell line (high metastatic ability) than in MHCC97L cell line (low metastatic ability), suggesting that LATS2 expression may be associated with metastatic ability (Figure 1B,C). Based on this expression pattern, we therefore chose MHCC97L for loss‐of‐function studies and MHCC97H for gain‐of‐function studies. LATS2 mRNA and protein expressions after transfected with si‐LATS2 or pcDNA‐LATS2 in HCC lines was detected, respectively, by qRT‐PCR and Western blot (Figure 1E,F). Meanwhile, we provided evidence that up‐/down‐regulation of LATS2 has no effect on the expression of β‐actin in HCC lines (Figure S1).
Subsequently, we explored the effect of LATS2 on HCC cell growth in vitro. The growth curves determined by MTT assay showed that up‐/low‐regulation of LATS2 expression has no effect on HCC cell viability (Figure 3A, P>.05). The results of colony formation assay was similar to MTT assay; up‐/down‐regulation of LATS2 expression has no effect on the number of colonies of HCC cell (Figure 3B, P>.05).
Figure 3.
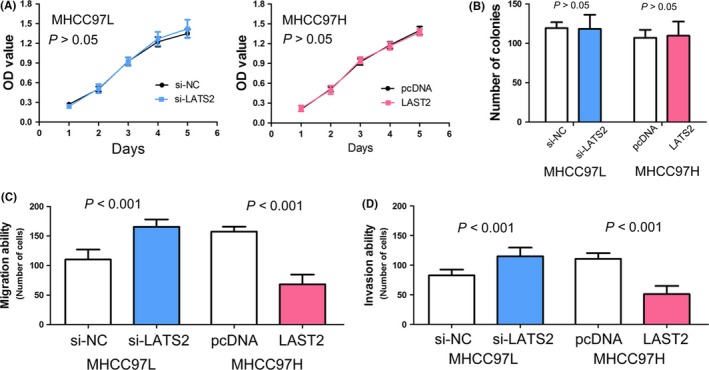
The biological function of LATS2 in hepatocellular carcinoma. (A) In vitro viability of MHCC97L and MHCC97H cells did not affected respectively by si‐LATS2 and pcDNA‐LATS2 through MTT assay. (B) Down‐regulated or up‐regulated LATS2 expression has no effect on hepatocellular carcinoma cell proliferative ability of MHCC97L and MHCC97H cells respectively through colony formation assay. (C) Down‐regulated LATS2 expression decreased the ability of MHCC97L cells migration in vitro, and up‐regulated LATS2 expression increased the ability of MHCC97H cells migration in vitro. (D) Suppressed LATS2 expression inhibited invasiveness of MHCC97L cells, and LATS2 overexpression promoted invasiveness of MHCC97H cells
3.5. LATS2 regulates migration and invasion of hepatocellular carcinoma cells
Because LATS2 expression was associated with vascular invasion and metastasis in HCC clinical samples, and metastatic ability in cell lines, we were interested in the function of LATS2 in regulating tumour cell migration and invasion. We found that low regulation of LATS2 expression could accelerate HCC cells migration, and up‐regulation of LATS2 expression obviously inhibited HCC cells migration (both P<.001, Figure 3C). Consistent with results of cell migration assay, matrigel invasion assay showed that knocking down LATS2 expression markedly enhanced HCC cells invasion, and LATS2 overexpression significantly suppressed HCC cells invasion (both P<.001, Figure 3D).
3.6. LATS2 regulates EMT‐associated genes expression in hepatocellular carcinoma cells
To explore the molecular mechanisms by which LATS2 contributes to cellular metastasis in HCC, we performed qRT‐PCR to screen the mRNA expression of metastasis‐associated molecules including E‐cadherin, vimentin, ZEB1, ZEB2, β‐catenin, snail, slug, MT1‐MMP, MMP2 and MMP9. Among them, we found E‐cadherin, vimentin, snail, slug, MMP2 and MMP9 could be regulated by LATS2 (all P<.001, Figure 4A). We further confirmed these genes protein expression by Western blot. Our results suggested that down‐regulation of LATS2 expression obviously increased the expression of vimentin, snail, slug, MMP2 and MMP9, and decreased E‐cadherin expression. On the contrary, up‐regulation of LATS2 expression obviously suppressed vimentin, snail, slug, MMP2 and MMP9 expressions, and elevated E‐cadherin expression (Figure 4B).
Figure 4.
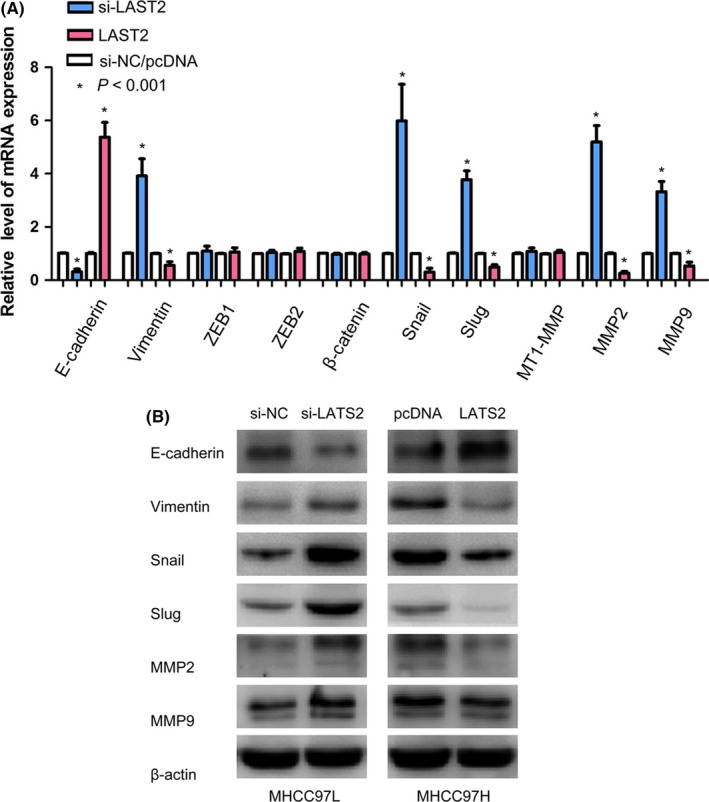
LATS2 regulates metastasis‐associated genes expression in hepatocellular carcinoma. (A) The mRNA levels of cell metastasis‐related molecules were detected by qTR‐PCR in MHCC97L and MHCC97H cells transfected with by si‐LATS2 or pcDNA‐LATS2. (B) E‐cadherin, vimentin, snail, slug, MMP2 and MMP9 protein levels were determined by Western blot in MHCC97L and MHCC97H cells transfected with si‐LATS2 or pcDNA‐LATS2
4. Discussion
Large tumour suppressor 2, also known as KPM, is one of the two human homologues of Drosophila. LATS2 is a tumour suppressor gene located on 13q11‐12 which exhibits the loss of heterozygosity in primary cancers of lung, breast and liver.10, 17 Recently, LATS2 has been confirmed as miRNAs target to regulate human cancer cells growth, motility, apoptosis and cell cycle.18, 19, 20, 21, 22, 23 Meanwhile, LATS2 has been showed dysregulated in human cancer. Nowadays, there are more evidence indicating that LATS2 expression has decreased in tumour tissues such as lung cancer,24, 25 breast cancer,26 colorectal cancer,27 ovarian cancer,13, 23 and oesophageal cancer.28 On the contrary, LATS2 was found to be overexpressed in nasopharyngeal carcinoma14 and acute myeloid leukaemia.29 The status of LATS2 expression in HCC is still unknown. In our study, we found LATS2 mRNA and protein expressions were decreased in HCC tissues and cell lines compared with normal hepatic tissues and hepatic cell line.
The clinical and prognostic significance of LATS2 has been reported in several types of human cancer. In non‐small cell lung cancer, Wu25 found that low levels of LATS2 protein were inversely associated with the T classification, N classification and clinical stage. Moreover, Luo30 showed that aberrant LATS2 gene expression correlated with EGFR (Epidermal Growth Factor Receptor) mutation in lung adenocarcinoma. Strazisar12 demonstrated that LATS2 mutation in non‐small cell lung cancer often harboured a P53 but not K‐RAS gene mutation and was mostly in an advanced stage or with regional lymph node metastasis. In colorectal cancer, Li27 suggested that LATS2 expression was obviously decreased in metastatic tumours from the lymph node and liver compared with primary colorectal tumours. Takahashi31 reported that low expression of LATS2 was corrected with large tumour size, high lymph node metastasis, and oestrogen receptor and progesterone receptor negativity. In HCC patients, we first suggested that low expression of LATS2 –was oppositely corrected with tumour stage, vascular invasion and metastasis. Meanwhile, we further explored the prognostic significance of LATS2 in HCC patients, and found that patients with low LATS2 expression had lower overall survival compared to patients with high LATS2 expression. According to univariate and multivariate analyses, low LATS2 expression was an independent poor prognostic factor for HCC patients. Similar to results in lung cancer reported by Wu25, patients with lower levels of LATS2 protein expression had poorer survival rates than those with higher levels of LATS2 expression, and low expression of LATS2 protein was an independent prognostic marker for non‐small cell lung cancer patients. Interestingly, Luo30 also showed that LATS2 mRNA level was found to be a significant independent predictor for disease‐free survival and overall survival in lung adenocarcinoma patients. In addition, LATS2 expression was inversely associated with recurrence‐free survival times and disease‐specific survival in patients with colorectal cancer.27 Contrarily, only one study showed that LATS2 overexpression was an independent unfavourable prognosis factor in nasopharyngeal carcinoma patients.14 Besides, survival time was not significantly different between patients with the high and low LATS2 expression in ovarian cancer and breast cancer.13, 31
The biological function of LATS2 had been shown to involve in diverse cellular processes, such as growth, migration, invasion apoptosis and angiogenesis. Fang26 found that down‐regulation of LATS2 expression accelerated breast cancer cell proliferation, tube formation and invasion, while up‐regulation of LATS2 expression could suppress cell survival and invasion in breast cancer cell. Meanwhile, LATS2 serve as a negative factor to control cell proliferation through regulating G1/S or G2/M transition and inducing apoptosis.32, 33 In lung cancer, LATS2 acted as a tumour suppressor to regulate lung cancer cells mobility by MMP2 and MMP9.25 Moreover, Zhang34 found that LATS2 acted as a positive modulator of Snail protein level and regulated cellular EMT (Epithelial‐Mesenchymal Transition) and tumour cell invasion. In our study, we found that LATS2 could significantly regulate HCC cells migration and invasion, but has no effect on HCC cells proliferation. Similarly, Guo15 reported that down‐regulation of LATS2 expression could promote tumour cell invasion. These cytological results were consistent with clinical significance of LATS2 in HCC. In order to explore the molecular mechanisms by which LATS2 contributes to cellular metastasis in HCC, we performed qRT‐PCR to screen the mRNA expression of metastasis‐associated molecules. Then, we found that E‐cadherin, vimentin, snail, slug, MMP2 and MMP9 could be regulated by LATS2 in mRNA level, and confirmed by Western blot that these genes also could be controlled by LATS2 in protein level. Meanwhile, Guo15 found that knock‐down of LATS2 increased MMP2/9 activities by LATS2‐mediated YAP1 phosphorylation.
In conclusion, LATS2 expression is decreased in HCC tissues and cell lines, and corrected with malignant status and prognosis of HCC patients. LATS2 controls HCC cells migration and invasion through regulating EMT‐associated genes.
Conflict of interest
The authors declare no conflict of interest.
Ethics statement
This study was approved by The Research Ethics Committee of Affiliated Tumour Hospital of Guangxi Medical University and Affiliated Minzu Hospital of Guangxi Medical University. The informed written consents were collected from all eligible patients and the entire study was performed based on the Declaration of Helsinki.
Author's contribution
RL analysed the data and wrote the manuscript; JZY and HHY designed the study and revised the manuscript; YL, C‐LY, Z‐HL and YQL performed all the experiments and collected all the data; X‐LL analysed the data.
Supporting information
Acknowledgements
This research was supported by Regional science fund project of China natural science foundation (Grant No. 81660498), China Postdoctoral Science Foundation, the 60th grant funding of general program for the post‐doctoral funding program in the western region (Grant No. 2016M602919XB), Guangxi Natural Science Foundation (Grant No. 2016GXNSFBA380090 and 2015GXNSFAA139128), Self‐raised Scientific Research Funds of Ministry of Health of Guangxi Province(Grant No. Z2015605 and Z2016480).
Liang R, Lin Y, Yuan C‐L, et al. The clinical significance and biological function of large tumour suppressor 2 in hepatocellular carcinoma. Cell Prolif. 2017;50:e12340 10.1111/cpr.12340
Contributor Information
Jia‐Zhou Ye, Email: yejiazhou@hainan.com.
Hai‐Hong Ye, Email: yehong139@hainan.com.
References
- 1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Walter SR, Thein HH, Gidding HF, et al. Risk factors for hepatocellular carcinoma in a cohort infected with hepatitis B or C. J Gastroenterol Hepatol. 2011;26:1757–1764. [DOI] [PubMed] [Google Scholar]
- 3. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chuang SC, La Vecchia C, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer Lett. 2009;286:9–14. [DOI] [PubMed] [Google Scholar]
- 5. Chacko S, Samanta S. Hepatocellular carcinoma: a life‐threatening disease. Biomed Pharmacother. 2016;84:1679–1688. [DOI] [PubMed] [Google Scholar]
- 6. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Jia J. Control of hepatitis B in China: prevention and treatment. Expert Rev Anti Infect Ther. 2011;9:21–25. [DOI] [PubMed] [Google Scholar]
- 8. Zhang S, Li RT, Wang Y, et al. Seroprevalence of hepatitis B surface antigen among pregnant women in Jiangsu, China, 17 years after introduction of hepatitis B vaccine. Int J Gynaecol Obstet. 2010;109:194–197. [DOI] [PubMed] [Google Scholar]
- 9. Zhu ZX, Huang JW, Liao MH, et al. Treatment strategy for hepatocellular carcinoma in China: radiofrequency ablation versus liver resection. Jpn J Clin Oncol. 2016;46:1075–1080. [DOI] [PubMed] [Google Scholar]
- 10. Yabuta N, Fujii T, Copeland NG, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–270. [DOI] [PubMed] [Google Scholar]
- 11. Yu T, Bachman J, Lai ZC. Evidence for a tumor suppressor role for the large tumor suppressor genes LATS1 and LATS2 in human cancer. Genetics. 2013;195:1193–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strazisar M, Mlakar V, Glavac D. LATS2 tumour specific mutations and down‐regulation of the gene in non‐small cell carcinoma. Lung Cancer. 2009;64:257–262. [DOI] [PubMed] [Google Scholar]
- 13. Xu B, Sun D, Wang Z, et al. Expression of LATS family proteins in ovarian tumors and its significance. Hum Pathol. 2015;46:858–867. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Hu CF, Chen J, et al. LATS2 is de‐methylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC Cancer. 2010;10:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo C, Wang X, Liang L. LATS2‐mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int J Clin Exp Pathol. 2015;8:1690–1697. [PMC free article] [PubMed] [Google Scholar]
- 16. Yang X, Yu J, Yin J, et al. MiR‐195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012;67:645–651. [PubMed] [Google Scholar]
- 17. Li Y, Pei J, Xia H, et al. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. [DOI] [PubMed] [Google Scholar]
- 18. Zheng YB, Xiao K, Xiao GC, et al. MicroRNA‐103 promotes tumor growth and metastasis in colorectal cancer by directly targeting LATS2. Oncol Lett. 2016;12:2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao H, Feng Y, Chen L. Repression of MicroRNA‐372 by arsenic sulfide inhibits prostate cancer cell proliferation and migration through regulation of LATS2. Basic Clin Pharmacol Toxicol. 2016;120:256–263. [DOI] [PubMed] [Google Scholar]
- 20. Hua K, Jin J, Zhao J, et al. miR‐135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int J Oncol. 2016;48:1997–2006. [DOI] [PubMed] [Google Scholar]
- 21. Zhang M, Wang X, Li W, et al. miR‐107 and miR‐25 simultaneously target LATS2 and regulate proliferation and invasion of gastric adenocarcinoma (GAC) cells. Biochem Biophys Res Commun. 2015;460:806–812. [DOI] [PubMed] [Google Scholar]
- 22. He Y, Wang J, Wang J, et al. MicroRNA‐135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am J Cancer Res. 2015;5:1382–1395. [PMC free article] [PubMed] [Google Scholar]
- 23. Feng S, Pan W, Jin Y, et al. MiR‐25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. [DOI] [PubMed] [Google Scholar]
- 24. Yao F, Liu H, Li Z, et al. Down‐regulation of LATS2 in non‐small cell lung cancer promoted the growth and motility of cancer cells. Tumour Biol. 2015;36:2049–2057. [DOI] [PubMed] [Google Scholar]
- 25. Wu A, Li J, Wu K, et al. LATS2 as a poor prognostic marker regulates non‐small cell lung cancer invasion by modulating MMPs expression. Biomed Pharmacother. 2016;82:290–297. [DOI] [PubMed] [Google Scholar]
- 26. Fang L, Du WW, Yang W, et al. MiR‐93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Chen X, Ding X, et al. LATS2 suppresses oncogenic Wnt signaling by disrupting beta‐catenin/BCL9 interaction. Cell Rep. 2013;5:1650–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee KH, Goan YG, Hsiao M, et al. MicroRNA‐373 (miR‐373) post‐transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. [DOI] [PubMed] [Google Scholar]
- 29. Gholami M, Mirfakhraie R, Movafagh A, et al. The expression analysis of LATS2 gene in de novo AML patients. Med Oncol. 2014;31:961. [DOI] [PubMed] [Google Scholar]
- 30. Luo SY, Sit KY, Sihoe AD, et al. Aberrant large tumor suppressor 2 (LATS2) gene expression correlates with EGFR mutation and survival in lung adenocarcinomas. Lung Cancer. 2014;85:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takahashi Y, Miyoshi Y, Takahata C, et al. Down‐regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. [DOI] [PubMed] [Google Scholar]
- 32. Ke H, Pei J, Ni Z, et al. Putative tumor suppressor Lats2 induces apoptosis through downregulation of Bcl‐2 and Bcl‐x(L). Exp Cell Res. 2004;298:329–338. [DOI] [PubMed] [Google Scholar]
- 33. Murakami H, Mizuno T, Taniguchi T, et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–883. [DOI] [PubMed] [Google Scholar]
- 34. Zhang K, Rodriguez‐Aznar E, Yabuta N, et al. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012;31:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


