Abstract
In recent years, increasing evidence has shown the potential role of long non‐coding RNAs (lncRNAs) in multiple cancers. Deregulation of lncRNAs was detected being closely associated with many kinds of tumours where they can act as a tumour suppressor or accelerator. LINC00152 was identified as an oncogene involved in many kinds of cancers, such as gastric cancer, hepatocellular carcinoma, colon cancer, gallbladder cancer and renal cell carcinoma. Moreover, inhibition of LINC00152 can suppress proliferation, migration and invasion of the cancer cells. Increasing evidence has showed that LINC00152 may act as a diagnostic and prognostic biomarker for the above‐mentioned cancers. In our review, we summarize the recent research progress of the expression and role of LINC00152 in various kinds of cancers.
1. Introduction
Over the past decade, there has been a more in‐depth understanding of genomic complexity in eukaryotes, thanks to the huge technological progress in high‐throughput sequencing of DNA and RNA transcripts.1 This has allowed scientists to shed new light on that beyond protein‐coding genes, plenty of transcripts which do not encode for proteins may perform their function through RNA sequences and/or through secondary and tertiary structural modification.1, 2 Among these non‐coding RNAs, microRNAs are well‐known regulators of cellular progresses by modulating gene expression. Additionally, long non‐coding RNAs (lncRNAs) represent a huge family of RNAs with limited protein‐coding potential that are defined by the length of over 200 nucleotides and lack of detectable open reading frame (ORF).3, 4, 5, 6, 7 In the nucleus, lncRNAs regulate gene expression in various biological conditions by binding to transcription factors,8 chromatin‐modifying factors9, 10 or heterogeneous nuclear ribonucleoproteins (hnRNPs),11 or lncRNAs act as a regulator targeting the splicing, stability or translation of host mRNAs by post‐transcriptional mechanisms.12 While in the cytoplasm, lncRNAs are known to act as the endogenous microRNA sponge to regulate the microRNA targets.12, 13, 14 Studies have shown that some lncRNAs are upregulated in multiple cancer tissues and cell lines compared with cancer adjacent tissues and normal cell lines, respectively, and perform pro‐oncogenic capability, such as CDKN2B antisense RNA 1 (ANRIL)15, 16 and metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1),17 while some others have reported low expression and play tumour‐suppressive roles, for example, maternally expressed gene 3 (MEG3)18 and growth arrest‐specific transcript 5 (GAS5).19, 20 Furthermore, certain lncRNAs have shown the developmental and tissue‐specific expression patterns. These characteristics appear critical for their functional analysis and make it possible for application of lncRNAs in diagnosis, prognostic evaluation and treatment of cancer patients. Overall, lncRNA is a major component of epigenetic regulatory networks.
According to GENCODE analysis (www.gencodegenes.org) of the last version (version 25) of the Ensemble human genome annotation (GRch38, version 25 from March 2016), 27692 transcripts transcribing from 15767 genes can be identified as lncRNAs. Multiple studies have reported that lncRNAs participate in various aspects of cell biological conditions and are closely associated with tumour development, but only a small portion of lncRNAs have been functionally characterized and little of their molecular mechanism was confirmed. Here, we shed light on the cancer‐related lncRNA called LINC00152 (long intergenic non‐coding RNA 152), a 828 bp lncRNA that maps to chromosome 2p11.2, which was initially detected as differentially hypomethylated during hepatocarcinogenesis,21 has been demonstrated to regulate genes by various mechanisms, including epigenetic modifications,22 and lncRNA‐miRNA23 and lncRNA‐protein interactions.22
First, LINC00152 is a new lincRNA, which is one of the hot research topics nowadays and in several cancers, it has been verified by large clinical samples. For instance, in colon cancer, large samples of 133 patients were used to verify the conclusion that Linc00152 is increased in human colon cancer tissues and is associated with poor prognosis and in gastric cancer. In addition, analysis in a cohort of 97 gastric cancer patients revealed that LINC00152 expression was positively correlated with tumour invasion depth, lymph node metastasis, higher TNM stage and poor survival. Second, the representative mechanisms by which LINC00152 could regulate target genes expression are researched most deeply and all sidedly nowadays, for example it could act as ceRNA in cytoplasm or bind to PRC2 to epigenetically regulate genes’ expression in nucleus. Third, the relative expression of LINC00152 in cancers was really high, which indicated the importance of LINC00152 in the occurrence and development of tumours, is even comparable to some star molecule, such as HOTAIR, MALAT1. So, it is meaningful and essential to study LINC00152 in more depth.
In our review, we summarized the current studies of functional regulation mechanisms and clinical significance of LINC00152 in the evolution and progression of human cancers (Tables 1 and 2).
Table 1.
Functional characterization of LINC00152 in various tumours
| Tumour type | Expression | Functional role | Related gene | Protein binding | Role | References |
|---|---|---|---|---|---|---|
| Gastric cancer | Upregulation | Proliferation, apoptosis, migration, invasion, tumorigenesis and epithelial to mesenchymal transition | p15; p21 | EZH2; H3K27me3 | Oncogene | 22, 24, 26 |
| Hepatocellular carcinoma | Upregulation | Proliferation and tumorigenesis | EpCAM | / | Oncogene | 38 |
| Colon cancer | Upregulation | Tumorigenesis and chemoresistance | miR‐193a‐3p/ERBB4 | / | Oncogene | 23 |
| Gallbladder cancer | Upregulation | Proliferation, metastasis, apoptosis and tumorigenesis | / | / | Oncogene | 50 |
| Renal cell carcinoma | Upregulation | Proliferation, invasion, cell cycle and apoptosis | / | / | Oncogene | 57 |
Table 2.
Clinical significance of LINC00152 in various tumours
| Tumour type | Overexpression of LINC00152 | References |
|---|---|---|
| Gastric cancer | Poor survival, invasion depth, lymph node metastasis and higher TNM stage | 22, 24, 26 |
| Hepatocellular carcinoma | Tumour size and Edmondson grade | 38 |
| Colon cancer | Shorter overall survival and recurrence‐free survival | 23 |
| Gallbladder cancer | Tumour progression, lymph node invasion and TNM stage advancement | 50 |
| Renal cell carcinoma | Advanced TNM stages, shorter overall survival | 57 |
2. LINC00152 in Human Cancers
2.1. Gastric cancer
Human gastric cancer, with characteristic of poor prognosis and high death rate, is the second most common cause of cancer‐related death over the world.16, 20, 24 Therefore, it is necessary to find original prognostic markers and develop novel therapeutic strategies in countermeasures for gastric cancer.24, 25
Studies showed that increased expression of LINC00152 was positively correlated with larger tumour size, tumour invasion depth, lymph node metastasis, higher TNM stage and poor survival.22, 24, 26 Interestingly, LINC00152 levels in gastric juice from patients with gastric cancer were observably higher than those from normal controls.26 Moreover, it is in plasma that LINC00152 can be detected as well and the levels of plasma LINC00152 were significantly increased in gastric cancer patients compared with healthy controls, and one of the possible mechanisms of its stable existence in blood is that it is protected by exosomes.27 Unexpectedly, significant joint effect between LINC00152 and H. pylori infection on risk of gastric cancer was also found (OR: 17.49, 95% CI: 4.78‐63.92).28 Therefore, LINC00152 has the possibility to be applied in gastric cancer diagnosis as a novel blood‐based biomarker for diagnosis of gastric cancer, particularly for those with H. pylori infection.
In vitro, LINC00152 in gastric cancer cell lines, BGC‐823, MGC‐803 and SGC‐7901, was significantly higher than those in human normal gastric epithelial cell line GES‐1.26 Moreover, LINC00152 knockdown could inhibit cell proliferation and colony formation, promote cell cycle arrest at G1 phase, trigger late apoptosis, reduce the epithelial to mesenchymal transition (EMT) programme, and suppress cell migration and invasion24 (Figure 1F).
Figure 1.
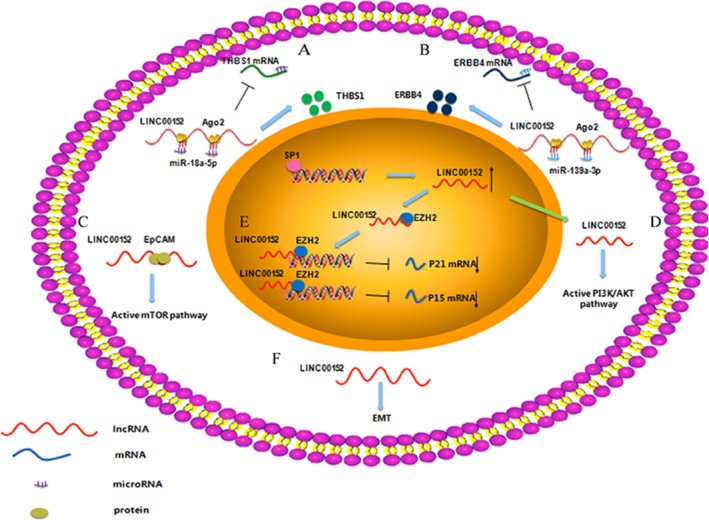
Underlying regulatory mechanisms of LINC00152 in human cancers. (A) LINC00152 may interact with THBS1 mediated by miR‐18a‐5p. (B) LINC00152 may act as a competing endogenous RNA (ceRNA) to regulate the expression of miR‐193a‐3p, and then regulate ERBB4. (C) LINC00152 can activate the mechanistic target of rapamycin (mTOR) pathway by binding to the promoter of EpCAM. (D) LINC00152, which can be activated by SP1, can participate in the phosphatidylinositol 3‐kinase (PI3K)/AKT signalling pathway. (E) LINC00152 can facilitate GC cell proliferation by accelerating the cell cycle by binding to enhancer of zeste homologue 2 (EZH2) and silencing the expression of p15 and p21. (F) LINC00152 can promote the epithelial to mesenchymal transition (EMT) programme
Additionally, subcellular fractionation assay showed that in human gastric cancer cell lines, LINC00152 was present in both the nuclear and the cytoplasmic fractions of BGC‐823 cells and SGC‐7901 cells; however, it was located mainly in the cytoplasm. LINC00152 overexpression facilitated GC cell proliferation by accelerating the cell cycle by binding to enhancer of zeste homologue 2 (EZH2) and silencing the expression of p15 and p21, and this mechanism may be a new molecular signalling link in gastric cancer22 (Figure 1E). P15 and p21, cyclin‐dependent protein kinase inhibitors (CKIs), can contribute to the regulation of both the GC cell cycle and proliferation.29 In addition, Xia, T. et al.30 have found that LINC00152 may interact with THBS1 mediated by miR‐18a‐5p (Figure 1A).
Molecular mechanisms provide additional insight into research of gastric carcinogenesis and strong evidence suggests that LINC00152 may prove a useful biomarker and therapeutic target for GC treatment.
2.2. Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) ranks the third most common cause of cancer‐related death worldwide and the sixth most common cancer malignancies worldwide.31, 32, 33, 34, 35, 36 However, at an advanced stage, hepatocellular carcinoma is often associated with a poor prognosis due to frequent cancer distant metastasis and clinical recurrence.31, 37 Therefore, new targets for hepatocellular carcinoma will be of great help to accelerate research on the molecular pathogenesis of HCC and perform untraditional diagnosis, treatments and prognosis.
Studies showed that LINC00152 could promote cell proliferation in vitro and tumour growth in vivo.38 In hepatocellular carcinoma, the transcript for LINC00152 was located mainly in the nucleus of MHCC‐97H cells. Furthermore, microarray‐based analysis demonstrated that LINC00152 could activate the mechanistic target of rapamycin (mTOR) pathway which plays an essential role in the regulation of cancer cell growth, division and tumorigenesis by binding to the promoter of EpCAM through a cis‐regulation, as confirmed by Gal4‐λN/BoxB reporter system38, 39 (Figure 1C). Thus, LINC00152 might be involved in the oncogenesis of HCC by activating the mTOR signalling pathway and might be a novel biomarker for clinical diagnosis of HCC in the future.
2.3. Colon cancer
Colon cancer, one of the most prevalent cancers in the world, is the third most common cancer and the second leading cause of cancer‐related mortality worldwide.40, 41, 42, 43 Besides surgery, the fundamental countermeasures for CRC patients, traditional cytotoxic chemotherapy and/or targeted therapies are routinely used to treat patients who are at high risk of developing recurrent or metastatic disease.44 However, similar to most other malignant tumours, lack of molecular biomarkers for tumour cell progression is still one of the challenging obstacle for CRC therapy. Therefore, new discovery of the diagnostic and prognostic biomarkers associated with CRC progression and the clinical outcomes would be of great clinical significance and the molecular alterations in CRC have been studied extensively.45
Studies have demonstrated that LINC00152 promoted tumour progression and conferred resistance to oxaliplatin (L‐OHP)‐induced apoptosis in vitro and in vivo. 23 Moreover, it antagonized the sensitivity of chemotherapy through acting as a competing endogenous RNA (ceRNA) to regulate the expression of miR‐193a‐3p, and then regulate ERBB4, which is associated with AKT phosphorylation and can result in L‐OHP resistance23 (Figure 1B).
All the data suggest that LINC00152 is a novel molecule related with progression of colon cancer as well as a therapeutic target for enhancing response to oxaliplatin in the treatment of colon cancer and potential prognostic biomarker.
2.4. Gallbladder cancer
Gallbladder cancer (GBC) is the most common biliary tract cancer and the fifth most common gastrointestinal malignancy.46 Distinctly, gallbladder carcinoma (GBC), which is common in India and Pakistan, but rare in the Western world, has a unique global distribution all over the world.47, 48 Novel therapeutic targets will be of great help in treatments for gallbladder cancer, thanks to the advances in cancer biology, which can lead to better understanding of the mechanism of GBC tumorigenesis.49
Cai et al.50 demonstrated that the high LINC00152 levels correlated positively with tumour status progression, lymph node invasion and TNM stage advancement. Functionally, studies revealed that LINC00152 dramatically promoted cell proliferation, metastasis and inhibited apoptosis in vitro. Moreover, in vivo, LINC00152 overexpression significantly promoted tumour growth. Mechanistic analyses indicated that LINC00152 could participate in the phosphatidylinositol 3‐kinase (PI3K)/AKT signalling pathway. In addition, SP1 directly binds to the LINC00152 promoter and positively regulates its expression in GBC cell (Figure 1D). In summary, the research suggested that LINC00152 contributes to the oncogenic potential of GBC, and SP1/LINC00152/PI3K/AKT may be a potential therapeutic target for GBC.
2.5. Renal cell carcinoma
Renal cell carcinoma (RCC) is one of the most common kidney cancers worldwide and the morbidity and lethality remain increasing although great advantages have been made in the past decades.24 Clinically, RCC is refractory to conventional cytotoxic agents. With therapeutic approaches, immunotherapy or targeted therapies such as tyrosine kinase inhibitors, mTOR inhibitors instead of favouring surgery, a significant proportion of patients will yet undergo disease recurrence and metastases.51, 52, 53, 54 It is therefore critical to develop novel therapeutic tactics in countermeasures for renal cell carcinoma by finding particularly biomarkers for the early detection and prognosis of RCC, for prioritization of asymptomatic individuals and for the identification of patients with greater risk for progression.55, 56
Wu et al.57 found that LINC00152 expression was significantly upregulated in renal cancerous tissues compared with normal counterparts, and high LINC00152 expression was closely associated with advanced TNM stage. Moreover, LINC00152 was found to be able to serve as an independent predictor of overall survival. Further experiments verified that LINC00152 expression was obviously upregulated in renal cancer cell lines and overexpression of LINC00152 can promote cell proliferation and invasion, inhibit cell cycle arrest in G1 phase and dramatically decrease apoptosis in both Caki‐2 and 786O cell lines, whereas the opposite results were observed with down‐expression of LINC00152. Overall, the data suggest that LINC00152 is a novel molecule related with progression of renal cell carcinoma as well as a potential prognostic biomarker and therapeutic target.
2.6. Future directions
Recently, an increasing number of lncRNAs are deregulated in diverse human cancers and associated with disease progression. For instance, prostate cancer antigen 3 (PCA3), which is highly expressed in prostate cancer can promote cancer cell proliferation by modulating the androgen receptor signal pathway.58 A highly conversed lncRNA named metastasis associated in lung adenocarcinoma transcript 1 (MALAT1), also referred to as NEAT2 (nuclear‐enriched abundant transcript 2), which is overexpressed in many kinds of cancers, especially in non‐small cell lung carcinoma (NSCLC), can promote cancer cell migration by regulating the expression of targeted genes.17 These aberrantly expressed lncRNAs are expected to become novel molecular biomarkers for developing novel therapeutic strategies in treatments for diagnosis and prognosis of various kinds of cancers.
There is no doubt that upregulation of LINC00152 affects various cancer‐related aggressive phenotypes, signalling pathways and has a critical role in cancer development, and the dysregulated LINC00152 found in tumours suggests that it may represent an effective target for diagnostic, prognostic and therapeutic purposes.22, 23, 24, 26, 28, 57 However, while we know that LINC00152 knockdown could suppress cancer cell proliferation and colony formation, promote cell cycle arrest at G1 phase, trigger late apoptosis, reduce the epithelial to mesenchymal transition (EMT) programme and suppress cell migration and invasion,24 the molecular mechanism of LINC00152 remains to be further explored. Most interesting is that the location of LINC00152 in part of cells was specific in different cell lines and it is hard to conclude that where is LINC00152 locate primarily. For example, in human gastric cancer cell lines, LINC00152 was present in both the nuclear and the cytoplasmic fractions of BGC‐823 cells and SGC‐7901 cells; however, it was located mainly in the cytoplasm. In hepatocellular carcinoma, the transcript for LINC00152 was located mainly in the nucleus of MHCC‐97H cells. So, further more researches are needed to explore the location in different cell lines. In addition, current understanding of the role of LINC00152 in tumour biology is still in an early stage, and these discoveries have not been applied for tumour patients in clinical practice. Hence, further more large‐scale researches are needed to expound the exact molecular mechanisms by which LINC00152 is involved in tumorigenesis.
In conclusion, LINC00152 is expected to become molecular markers for cancer diagnosis and prognosis in the future with the detailed molecular mechanism explored, and large case‐control studies are needed to prove the sensitivity and specificity of LINC00152 as a serum marker to monitor disease biological behaviour and response to treatment.
Competing Interests
The authors have declared that no competing interest exists.
Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: A pivotal oncogenic long non‐coding RNA in human cancers. Cell Prolif. 2017;50:e12349 10.1111/cpr.12349
Yang Yu and Jian Yang are Equal contributors.
Funding Information
This study was supported by the: Project of Standard Diagnosis and Treatment of Key Disease of Jiangsu Province (BE2015722); Project of the peak of the six talents of Jiangsu Province (WSN‐018) and Scientific Research Foundation for Health of Jiangsu Province (H201408).
References
- 1. Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eddy SR. Non‐coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919‐929. [DOI] [PubMed] [Google Scholar]
- 3. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide‐coding capacity and parallels with 3′ UTRs. RNA. 2012;18:825‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao J, Song X, Wang K. lncScore: alignment‐free identification of long noncoding RNA from assembled novel transcripts. Sci Rep. 2016;6:34838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia H, Osak M, Bogu GK, et al. Genome‐wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478‐1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saha A, Bhattacharya S, Bhattacharya A. Serum stress responsive gene EhslncRNA of Entamoeba histolytica is a novel long noncoding RNA. Sci Rep. 2016;6:27476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu X, Feng Y, Zhang D, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun M, Nie F, Wang Y, et al. LncRNA HOXA11‐AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299‐6310. [DOI] [PubMed] [Google Scholar]
- 10. Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA‐EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guttman M, Rinn JL. Modular regulatory principles of large non‐coding RNAs. Nature. 2012;482:339‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang H, Liu P, Zhang J, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR‐193b. Oncogene. 2016;35:3647‐3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi X, Sun M, Wu Y, et al. Post‐transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36:503‐513. [DOI] [PubMed] [Google Scholar]
- 15. Li Z, Yu X, Shen J. ANRIL: a pivotal tumor suppressor long non‐coding RNA in human cancers. Tumour Biol. 2016;37:5657‐5661. [DOI] [PubMed] [Google Scholar]
- 16. Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR‐99a/miR‐449a. Oncotarget. 2014;5:2276‐2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45‐R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma C, Shi X, Zhu Q, et al. The growth arrest‐specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437‐1444. [DOI] [PubMed] [Google Scholar]
- 20. Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neumann O, Kesselmeier M, Geffers R, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56:1817‐1827. [DOI] [PubMed] [Google Scholar]
- 22. Chen WM, Huang MD, Sun DP, et al. Long intergenic non‐coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773‐9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yue B, Cai D, Liu C, et al. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016; 24:2064‐2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao J, Liu Y, Zhang W, et al. Long non‐coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112‐3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiao S, Wang H, Shi Z, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166‐180. [DOI] [PubMed] [Google Scholar]
- 26. Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non‐coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441‐5447. [DOI] [PubMed] [Google Scholar]
- 27. Li Q, Shao YF, Zhang XJ, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015;36:2007‐2012. [DOI] [PubMed] [Google Scholar]
- 28. Yang T, Zeng H, Chen W, et al. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147‐153. [DOI] [PubMed] [Google Scholar]
- 29. Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia T, Liao Q, Jiang XM, et al. Long noncoding RNA associated‐competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eggert T, Wolter K, Ji J, et al. Distinct functions of senescence‐associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30:533‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245‐1255. [DOI] [PubMed] [Google Scholar]
- 33. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. [DOI] [PubMed] [Google Scholar]
- 34. Giannelli G, Koudelkova P, Dituri F, et al. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65:798‐808. [DOI] [PubMed] [Google Scholar]
- 35. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 36. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF‐beta promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666‐681. [DOI] [PubMed] [Google Scholar]
- 37. Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501‐1512e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813‐42824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets. 2009;9:237‐249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsukamoto S, Ishikawa T, Iida S, et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17:2444‐2450. [DOI] [PubMed] [Google Scholar]
- 41. Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9‐29. [DOI] [PubMed] [Google Scholar]
- 42. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104‐117. [DOI] [PubMed] [Google Scholar]
- 43. Koblansky AA, Truax AD, Liu R, et al. The innate immune receptor NLRX1 functions as a tumor suppressor by reducing colon tumorigenesis and key tumor‐promoting signals. Cell Rep. 2016;14:2562‐2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fung KY, Nice E, Priebe I, et al. Colorectal cancer biomarkers: to be or not to be? Cautionary tales from a road well travelled. World J Gastroenterol. 2014;20:888‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fang L, Lu W, Choi HH, et al. ERK2‐dependent phosphorylation of CSN6 is critical in colorectal cancer development. Cancer Cell. 2015;28:183‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu AX, Hong TS, Hezel AF, et al. Current management of gallbladder carcinoma. Oncologist. 2010;15:168‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scanu T, Spaapen RM, Bakker JM, et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe. 2015;17:763‐774. [DOI] [PubMed] [Google Scholar]
- 48. Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang SH, Ma F, Tang ZH, et al. Long non‐coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR‐342‐3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai Q, Wang ZQ, Wang SH, et al. Upregulation of long non‐coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8:4068‐4081. [PMC free article] [PubMed] [Google Scholar]
- 51. He X, Sun G, Guo F, et al. Knockdown of long non‐coding RNA FTX inhibits proliferation, migration, and invasion in renal cell carcinoma cells. Oncol Res. 2016;5:157‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119‐1132. [DOI] [PubMed] [Google Scholar]
- 53. Rini BI, Atkins MB. Resistance to targeted therapy in renal‐cell carcinoma. Lancet Oncol. 2009;10:992‐1000. [DOI] [PubMed] [Google Scholar]
- 54. LaGory EL, Wu C, Taniguchi CM, et al. Suppression of PGC‐1alpha is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. 2015;12:116‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seliger B, Dressler SP, Lichtenfels R, et al. Candidate biomarkers in renal cell carcinoma. Proteomics. 2007;7:4601‐4612. [DOI] [PubMed] [Google Scholar]
- 56. Xu Z, Yang F, Wei D, et al. Long noncoding RNA‐SRLR elicits intrinsic sorafenib resistance via evoking IL‐6/STAT3 axis in renal cell carcinoma. Oncogene. 2016. [DOI] [PubMed] [Google Scholar]
- 57. Wu Y, Tan C, Weng WW, et al. Long non‐coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6:285‐299. [PMC free article] [PubMed] [Google Scholar]
- 58. Ferreira LB, Palumbo A, de Mello KD, et al. PCA3 noncoding RNA is involved in the control of prostate‐cancer cell survival and modulates androgen receptor signaling. BMC Cancer. 2012;12:507. [DOI] [PMC free article] [PubMed] [Google Scholar]


