Abstract
Adequate gut microbiota establishment is important for lifelong health. The aim was to sequentially analyze the gut microbiota establishment in low-birth-weight preterm neonates admitted to a single neonatal intensive care unit during their first 3 weeks of life, comparing two epidemiological scenarios. Seven control infants were recruited, and another 12 during a severe S. marcescens outbreak. Meconium and feces from days 7, 14, and 21 of life were collected. Gut microbiota composition was determined by 16S rDNA massive sequencing. Cultivable isolates were genotyped by pulsed-field gel electrophoresis, with four S. marcescens submitted for whole-genome sequencing. The expected bacterial ecosystem expansion after birth is delayed, possibly related to antibiotic exposure. The Proteobacteria phylum dominates, although with marked interindividual variability. The outbreak group considerably differed from the control group, with higher densities of Escherichia coli and Serratia to the detriment of Enterococcus and other Firmicutes. Curiously, obligate predators were only detected in meconium and at very low concentrations. Genotyping of cultivable bacteria demonstrated the high bacterial horizontal transmission rate that was confirmed with whole-genome sequencing for S. marcescens. Preterm infants admitted at NICU are initially colonized by homogeneous microbial communities, most of them from the nosocomial environment, which subsequently evolve according to the individual conditions. Our results demonstrate the hospital epidemiology pressure, particularly during outbreak situations, on the gut microbiota establishing process.
Introduction
The relationship between our human cells and the microbial communities living inside us can be classified as mutualistic, commensal, or pathogenic. This consideration delimits the fine barrier that distinguishes colonization from infection, and can fluctuate over time due to influence from the host or from microbial and environmental factors. The influence of the intestinal microbiota on global human health has been confirmed [1–3], forming new research perspectives aimed to optimize the composition and the functionality of this bionetwork.
Adequate microbiota establishment in newborns is a process particularly relevant for their lifelong health [4], and its management represents a scientific challenge. Human bacterial colonization might start in utero, but the critical step begins with the exposure to maternal bacteria at birth and during the early postnatal period [5–6]. Bacterial populations fluctuate considerably during the first months of life, until a stable ecosystem is established when the infant is approximately 2–3 years of age [7–8]. Universal criteria defining a “normal” or “healthy” gut microbiota have not yet been established, which should be characterized by a high diversity, marked inter-individual variability, and conserved intraindividual stability. However, the composition of this ecosystem is influenced by numerous factors [9–10], such as the gestational age at which the neonate is born [11–12].
Preterm birth is the main cause of perinatal morbidity and mortality, as well as an important risk factor for death in the first 5 years of life [13]. A considerable increase in preterm birth rates over the past two decades has been reported worldwide, in both developed and developing countries [14]. In Spain, the preterm birth rate of all live births increased from 7.1% in 1996 to 8.2% in 2008, which is one of the highest rates in Europe [15]. This global tendency can be explained by several factors, including an early or advanced mother’s age, a small gap between pregnancies, low body mass index, multiple pregnancy, history of infectious diseases, stress, alcohol consumption, and periodontal disease [13]. Nevertheless, approximately half of the spontaneous preterm births have an unidentified cause, and it has been suggested that the composition of the maternal microbiota could play a relevant triggering role [7,16].
In low-birth-weight preterm infants (<2500 g), the gut microbiota composition and their biodiversity are aberrant, given the bacterial establishment is delayed by their prolonged hospital stay and their intense exposure to antimicrobials [12,17–19]. This fact explains, at least partially, why preterm infants have a very immature immune system and typically experience infectious complications [20–22]. In this context, Serratia marcescens is one of the most relevant emerging pathogens causing severe outbreaks in this population [23–24]. Pathogenic gut colonization during nosocomial outbreaks has frequently been reported; to our knowledge, however, the influence of an outbreak on the microbiota establishment process has not thus far been studied.
The aim of the present study was to sequentially analyze the gut microbiota establishment of low-birth-weight preterm neonates admitted to a single neonatal intensive care unit (NICU) during their first 3 weeks of life, comparing two epidemiological scenarios: a normal period and a period with a nosocomial S. marcescens outbreak.
Materials and methods
Preterm neonate inclusion criteria and sampling procedure
La Paz University Hospital (Madrid, Spain) has a 23-bed level III NICU, from which 19 low-birth-weight preterm neonates (<32 weeks gestational age) were recruited in two separate periods: (A) during an epidemiologically normal period in 2015 (control group, n = 7); and (B) during a severe S. marcescens outbreak from December 2016 to March 2017 (outbreak group, n = 12). Despite the different sampling periods, there were no significant changes in the NICU. It is important to note the data lack in the control group about antibiotic consumption, and clinical data, as a limitation of our work. From each preterm infant, four fecal samples were collected after birth: meconium, and feces from 7, 14, and 21 days of life. Although our initial aim was to extend the recruitment period, logistical limitations limited the study to the first three weeks. The samples were directly recovered from the diaper using a sterile plastic stick and immediately stored at -80°C. Although our intention was to collect fecal samples immediately after deposition, we cannot rule out the possible contact and contamination with urine. However, the contribution of the urinary microbiota should be insignificant.
For the control group, only DNA from fecal samples was available, whereas for the outbreak group bacterial growth was obtained by culture-dependent techniques in addition to DNA. The ethics committee “Comité Ético de Investigación Clínica del Hospital Universiatrio La Paz” approved the study (reference HULP3551), and the data of all the neonates were obtained from their clinical chart. The infants were categorized according to four variables: 1) epidemiological situation (normal or S. marcescens outbreak); 2) delivery mode (vaginal or C-section); 3) gestational age (extremely preterm: <28 weeks; very preterm: 28–30 weeks; or moderately preterm: 30–32 weeks); and 4) birth weight (<1000 g; 1000–1500 g; or >1500 g).
Sample processing
Fecal samples from the S. marcescens outbreak group were slowly defrosted at -20°C for 24 h and 4°C for another 24 h, in order to avoid bacterial death. Portions between 0.3–0.5 g of each sample were inoculated into Brain Heart Infusion (BHI) broth (Difco, Detroit, Michigan) and incubated at 37°C for 24 h as a bacterial pre-enrichment. Cultivable bacteria were isolated in selective and nonselective agar media from the BHI tube, including agar plates of M-Enterococcus; De Man, Rogosa and Sharpe (MRS); mannitol salt; McConkey; and Columbia, with 5% sheep blood. The culture media were purchased from Difco, and the plates were incubated at 37°C for 24–48 h, including 5% CO2 for the blood agar plates, and anaerobic conditions for the MRS plates. Colony identification was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker, Germany), and all the isolates were conserved at -80°C in semi-skimmed milk. In parallel to bacterial cultures, total DNA was obtained from fecal aliquots of 0.3–0.5 g with the QiaAMP kit (Qiagen, Germany), determining their concentration and quality by Qubit fluorometer (Invitrogen, USA).
16S rDNA next-generation sequencing
The fecal DNA samples were sent to FISABIO (Valencia, Spain) for massive Mi-Seq 2 × 300 bp paired-end Illumina 16S rDNA sequencing (Cod. 15044223 Rev. A) from the V3 and V4 regions, which were amplified with the following primers: (Forward Primer: 5’-TCGTCGGCAG CGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG), (Reverse Primer: 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVG GGTATCTAATCC) [25]. Shannon and Chao1 indexes were used for alpha bacterial diversity estimation and were calculated, eliminating taxa with fewer than three lectures. Taxonomic affiliations were assigned using the Ribosomal Database Project (RDP) classifier, and reads with an RDP score below 0.8 were assigned to the upper taxonomic rank, leaving the last rank as unidentified. Sequence quality was measured according to the following parameters: minimum length, 250 bp; trimming quality measure type, mean; trimming quality number from 3’ extreme, 30; trimming quality window, 10 bp. Relative abundance and contingency tables included singletons and very low-represented taxons. The statistical analysis was performed using R statistical software and several open source libraries. The quantitative data of the reads were homogenized using their relative percentage from the total reads of each sample to facilitate the comparison between samples. Finally, the Galaxy Huttenhower Platform (http://huttenhower.sph.harvard.edu/galaxy) was used in order to calculate the Linear Discriminant Effect Size Analysis (LEfSe) algorithm and to obtain cladograms in which microbial taxa that explain significant differences among groups of samples were represented. A free software platform was used according to paper instructions [26]. For the statistical analysis, samples with fewer than 1,000 reads were dismissed, and all the samples from patient 7B were excluded due to a lack of some demographic and perinatal information. Fasta files are deposited in the NCBI web site under the Bioproject PRJNA510235 reference.
Pulse-field gel electrophoresis typing
Cultivable bacterial isolates were genotyped by pulse-field gel electrophoresis (PFGE), using the habitual particular settings for the PFGE protocol and also for the restriction enzymes (SmaI for staphylococci and enterococci, XbaI for Escherichia coli, and finally SpeI for Serratia and Klebsiella). The PFGE pattern analysis was made with Phoretix 5.0 software (TotalLab, Newcastle upon Tyne, UK), and the representation of the results was made based on Dice coefficients and the unweighted pair group method with arithmetic mean algorithm.
Whole genome sequencing
Four S. marcescens isolates from different infants were submitted to whole genome sequencing (WGS) by MiSeq technology (Illumina), and the genetic relationships were analyzed in the Galaxy Huttenhower Platform. The genome sequences are deposited in the European Nucleotide Archive database with the accession numbers QYRU00000000 and QYSA00000000 for Clone A, and QYRV00000000 and QYSB00000000 for Clone B.
Fungi identification
The internal transcribed spacer (ITS)-1 region was amplified from the total fecal DNA using polymerase chain reaction (PCR) in order to analyze fungi diversity, using the primers ITS1-F (5’-CGCCCGCCGCGCGCGGCGGGCGG GGCGGGGGCACGGGGGGCTTGGTCATTTAGAGGAAGTAA-3’) and ITS1-R (5’-TCCTCCGCTTATTGATATGC-3’) [27]. Afterward, the amplicons were separated with denaturing gradient gel electrophoresis (DGGE), using the D-CODE system (BioRad Laboratories, USA). The gel bands were cut, reamplified, and sequenced in an AQBI Prism 7000 apparatus.
Statistical analysis
The Kruskal-Wallis index was used for differences between two groups of samples (comparing medians) for each variable of study, and Dunn’s test was used when more than two groups were defined regarding a variable of study. The principal component analysis was applied for the multivariant analysis regarding taxonomical data in order to see differences between groups according to the variables of study. Statistical significance was adjusted to p < .005.
Results
Characteristics of the preterm infants
The demographic and clinical characteristics of both newborn groups are shown in Table 1 and in S1 Dataset. The most relevant differences were that the control infants presented a higher weight at birth and a lower incidence of sepsis. One neonate from the control group (14.3%) suffered from early-onset sepsis, whereas late-onset sepsis was microbiologically or clinically diagnosed in five neonates from the outbreak group (41.7%), being Staphylococcus epidermidis and S. marcescens the microorganisms implicated. Data on antimicrobial therapy were only available for the outbreak group and were included the administration of prophylactic cefazolin during labor (10 infants), empiric treatment based on ampicillin, gentamicin, and clarithromycin during the first week of life (8 infants), and treatment that included vancomycin (7 infants), cefotaxime (2 infants) piperacillin/tazobactam (2 infants), meropenem (1 infant), and amikacin (3 infants) for the second and third weeks of life.
Table 1. Clinical and demographic characteristics of the preterm infants of both groups.
| Characteristic | Control Group (7 infants) |
Outbreak Group (12 infants) |
p value |
|---|---|---|---|
| Weight at birth (g) | 1462 (720–1890)a | 971 (600–1537)a | 0.009 |
| Gestational Age (weeks) | 30 (25–31)a | 28 (25–31)a | 0.26 |
| Vaginal delivery (n, %) | 5, 71.4% | 3, 25% | 0.04 |
| Male sex (n, %) | 5, 71.4% | 2, 16.6% | 0.0001 |
| Sepsis (n, %) | 1, 14.3% | 6, 50% | 0.04 |
| Length of stay (days) | 14 (5–140)a | 55 (7–89)a | 0.1 |
aValues expressed as the median value and the range (between parentheses).
Gut microbiota establishment by next-generation sequencing
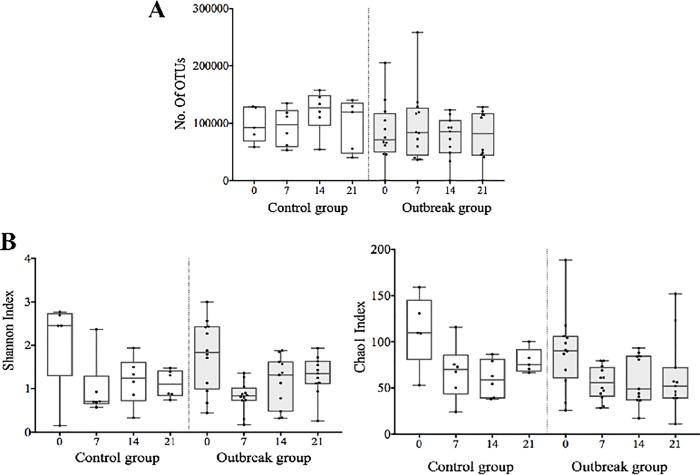
The number of operational taxonomic units (OTUs) and the alpha diversity indexes of the meconium samples were similar to the further fecal samples (Fig 1), indicating a delayed bacterial establishment process. LEfSe analysis allowed us to explore the differences in microbiota composition between the control and the outbreak groups for the four variables stated above (epidemiological situation, delivery mode, gestational age, and birth weight).
Fig 1.
Number of operational taxonomic units (OTUs) (A), and alpha diversity measured by the Chao1 index (B) in all samples studied.
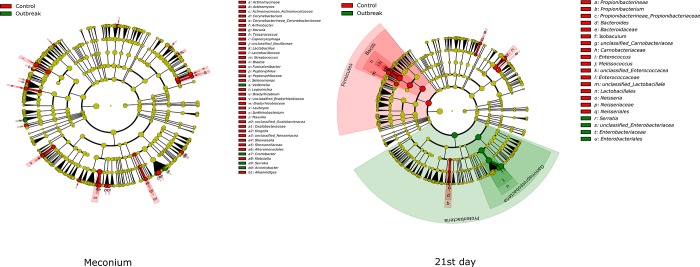
Significant differences in the gut microbiota composition of the control and outbreak groups were detected in meconium and in the day 21 samples and were more relevant in those found at day 21 (p = .0024 at the genus level and p = .073 at the phylum level) (Fig 2). The outbreak group was characterized by a higher proportion of γ-Proteobacteria, related to a higher density of Serratia, and with lower proportions of the Firmicutes and Fusobacteria phyla.
Fig 2. Cladograms showing the significant differences of gut microbiota composition in meconium and 21 days feces between control and outbreak groups.
In relation to the delivery mode, differences between vaginal delivery and C-section delivery were only significantly different at day 0 (p = .0066 at the genus level and p = .0363 at the phylum level) (Fig 3). Significant differences among bacterial communities regarding gestational age and birth weight were not detected.
Fig 3. Significant differences in the gut microbiota of meconium by the delivery mode.
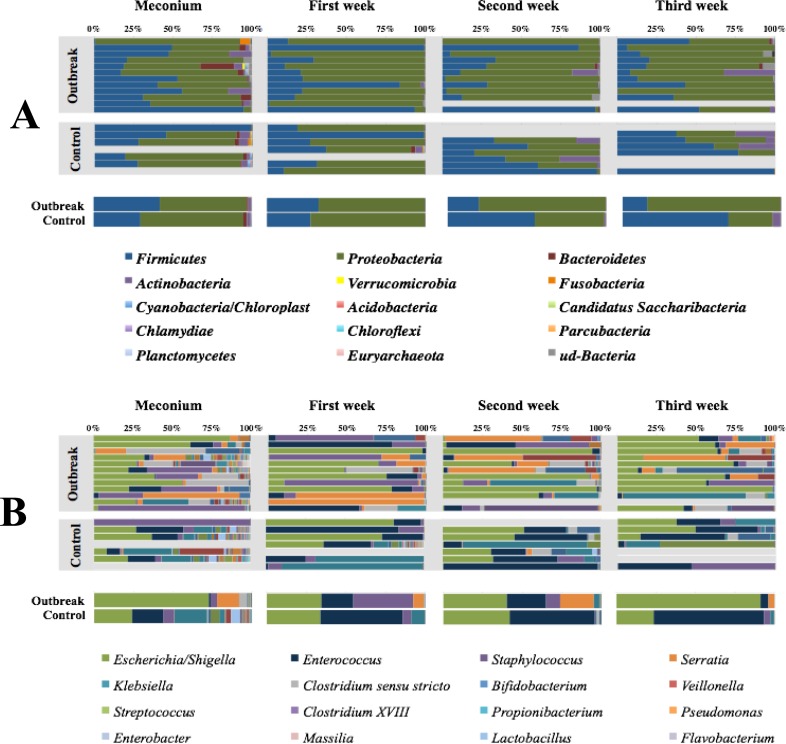
Taking into account all 55 samples, the predominant phylum during the first weeks of life of the low-weight preterm infants was Proteobacteria (median±SD 70.1% ± 26.9%, range 0.3%–99.4%), followed by Firmicutes (median±SD 22.1% ± 26.8%; range 0.05–99.4); and although up to another 23 phyla were detected, their contribution was nearly anecdotic (Fig 4A). At the genus level, the abundance of Escherichia/Shigella sp. increased over the studied period while that of Enterococcus sp. and Staphylococcus sp. decreased (Fig 4B).
Fig 4.
(A) Phyla percentage in each sample and infant, and summary of both groups expressed as the medians values. (B). Genera percentage in each sample and infant, and summary of both groups expressed as the median values. The 16 most abundant genera are highlighted in the figure, although up to 215 genera were detected in the samples analyzed in this study.
Reads accounting for the predator bacteria Bdellovibrio (2 infants), Peredibacter (1 infant), and Vampirovibrio (1 infant) were found in meconium samples from the outbreak (3 infants) and the control (1 infant) groups. The relative abundance of predatory species was extremely low (0.004%–0.11%), and none could be detected in the subsequent fecal samples.
Serratia was detected in the meconium samples from all the patients in the outbreak group (median value, 5558 OTUs), whereas this genus was considerably less abundant among the meconium samples from the control group (median value, 180 OTUs) (Table 2). Also noticeable was that the Serratia reads considerably increased just before the diagnosis of Serratia sepsis in some patients from the outbreak group, and particularly in infant O11, who finally died from S. marcescens sepsis at day 10 after birth. The gut enrichment of Serratia sequences in this infant was manifest, reaching levels of 95% of the total intestinal microbiota at day 7 (Table 2).
Table 2. Serratia abundance detected by molecular tools and distribution of the two major clones detected in the outbreak group.
The underlined isolates were submitted to whole genome sequencing. High abundance of Serratia by NGS is marked in light grey color, whereas the dark grey means a clear dominance of the Serratia genera.
| INFANT | Meconium | Day 7 | Day 14 | Day 21 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cultivable | NGS (%) | Cultivable | NGS (%) | Cultivable | NGS (%) | Cultivable | NGS (%) | ||
|
OUTBREAK GROUP |
O1 | Clone A | 5.4 | 0.002 | 12.1 | 1.0 | |||
| O2 | 12.4 | 0.003 | 0.2 | 1.7 | |||||
| O3 | 2.5 | Clone B | 17.3 | 19.0 | |||||
| O4 | 2.4 | Clone B | 2.4 | Clone B | 4.8 | 16.9 | |||
| O5 | 1.8 | 0.003 | 0.04 | Clone A | 28.8 | ||||
| O6 | 11.8 | Clone B | 5.1 | Clone B | 26.5 | Clone B | 37.4 | ||
| O7 | 2.9 | Clone B | 0.2 | 35.5 | 4.9 | ||||
| O8 | 1.9 | 0.7 | 1.3 | 0.02 | |||||
| O9 | 2.0 | 0.004 | 0.004 | 0.006 | |||||
| O10 | Clone A | 59.5 | Clone A | 78.5 | Clone A | 0.8 | Clone A | 0.4 | |
| O11 | 3.1 | Clone B | 94.5 | ||||||
| O12 | 0.3 | Clone A | 1.3 | 0.1 | 1.2 | ||||
| CONTROL GROUP | C1 | 0.004 | 0.005 | 0.008 | |||||
| C2 | 0.006 | 0.001 | 0.001 | 0.006 | |||||
| C3 | 0.1 | 0.0007 | 0 | 0 | |||||
| C4 | 0.06 | 0.2 | 0.01 | ||||||
| C5 | 0.8 | 3.1 | |||||||
| C6 | 0.06 | 0 | 0 | ||||||
| C7 | 0.0008 | 0 | 0.002 | ||||||
Characterization of cultivable isolates from the outbreak group
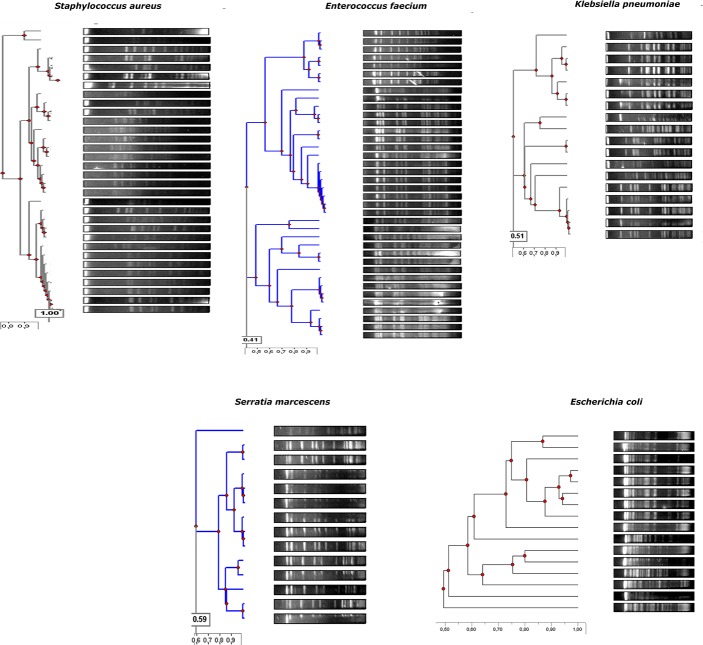
A total of 16 E. coli (8 infants), 35 Enterococcus faecalis (11 infants), 2 Enterococcus faecium (1 infant), 12 Klebsiella oxytoca (7 infants), 6 Klebsiella pneumoniae (5 infants), 32 S. epidermidis (all 12 infants), 1 Serratia liquefaciens and 14 S. marcescens (8 infants) isolates were recovered. Regarding the meconium samples, the cultivable microorganisms were S. epidermidis (7 infants), E. faecalis (3 infants), and S. marcescens (2 infants). Only four meconium samples (33.3%) did not yield viable microorganisms. PFGE analysis showed that isolates recovered from different samples from the same infant were identical or closely related (Fig 5).
Fig 5. Dendrograme showing the genetic relationship among the cultivable isolates based on the Dice’s coefficient.
Similarly, a single E. faecalis pulsotype was detected, colonizing three infants and two major clones of K. pneumoniae in two infants. Among the Serratia isolates, two genetically unrelated clones were detected, affecting four and five infants each. The relative abundance of Serratia in the gut microbiota of each infant is presented in Table 2, which compares both next-generation sequencing (NGS) and microbiological culture techniques. It shows the high counts of Serratia in all samples from the outbreak group, confirming the results previously observed in the NGS analysis.
WGS of Serratia clones
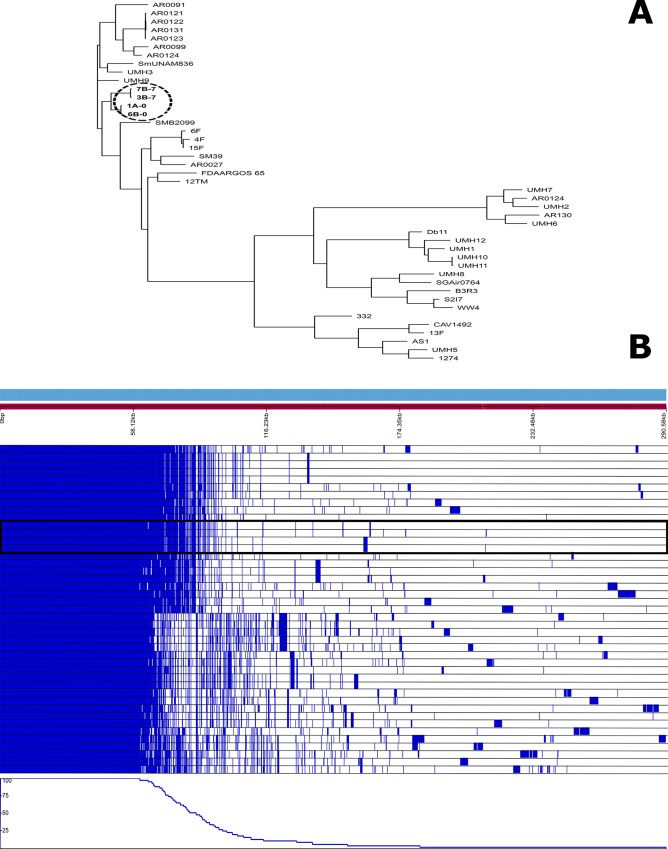
Four Serratia isolates representing the two dominant PFGE clones associated with sepsis in the outbreak group were submitted to WGS (Fig 6). The genome analysis confirmed two phylogenic linages unrelated to others previously published. It is important to note that the isolate causing the sepsis and death of infant O11 was identical to the isolate obtained from infant O7, who had a satisfactory clinical evolution. WGS allowed the characterization of the resistome (aac(6’)-Ic-1, blaSRT-2-1, blaSRT-1-1, and qnrE) and the virulome (cheB, cheR, cheW, cheY, flgB, flgC, flgG, flgI, flhA, flhC, flhD, fliA, fliG, fliI, fliM, fliN, fliP, fliQ, and fliZ), which was uniform in all four isolates. Nevertheless, our isolates remained susceptible to most antibiotics, given the resistant genes detected were located on the chromosome and not transferable.
Fig 6.
Phylogenic representation of the 4 S.marcescens genomes sequenced in this study and comparison with other 32 public S. marcescens genomes (A). The second part of the figure (B) represents the common core of all 36 genomes versus the isolate-specific genes. Our 4 isolates grouped together in both analysis and are marked.
Fungi detection
PCR-DGGE yielded positive amplifications in infant 6 (days 21 and 28) and infant 8 (days 14 and 21), and the nucleotide sequences of these amplicons corresponded to Candida albicans.
Discussion
In the present study, we have described the succession of the gut microbiota composition from meconium to the first 3 weeks of life of low-weight preterm infants, comparing two epidemiological scenarios from the same NICU. The primary result of our work is related to the S. marcensens outbreak influence on infant gut microbiota patterns, including meconium, which might have been modified even before birth in relation to the hospital admission of mothers to prevent premature birth. Although there is marked inter-individual variability, the gut microbiota of low-weight premature infants is dominated by Proteobacteria phylum, particularly E. coli. Moreover, the expected bacterial ecosystem expansion after birth appears to be delayed, probably in relation to antibiotic exposition. In that sense it is important to remark the data lack about antibiotic prescription on the control group, that represent an important limitation of our work.
Regarding to the outbreak group, all but two infants received empiric antibiotics immediately after birth, and most required culture-guided antibiotic treatments during their admittance, including up to three different antimicrobial families. However, the high incidence of infectious complications among preterm neonates is the main argument used to justify the wide empiric and culture-guided use of antimicrobials in preterm infants, particularly in those with low birth weight, as in our case. Early biomarkers of preterm sepsis, together with the development of microbiome-based approaches, are urgently required to reduce antibiotic use in NICUs [28–29].
Prematurity is the main cause of neonatal morbidity and mortality, and the establishment of an adequate gut microbiota appears to be one of the most promising strategies to improve preterm infants’ health and to reduce the impact of sequelae later in life [30–31]. Preterm infants admitted to an NICU have a high risk of infection, and S. marcescens is one of the most relevant nosocomial pathogens;23 their intestinal carriage has been identified as a potential reservoir [32].
Our results demonstrated considerable Proteobacteria enrichment in both preterm infant groups, although the enrichment was significantly higher in the outbreak group from the second week of life, in concordance with other authors [33–36]. Previous studies found that Proteobacteria (mostly Enterobacter and Photorhabdus), Firmicutes (mostly Enterococcus and Lactobacillus), and Actinobacteria (Bifidobacterium) dominated the microbial composition of meconium [37]. E. coli, Staphylococcus sp., Klebsiella sp., and a high rate of facultative anaerobes also commonly appear in the meconium of preterm neonates [38–39]. Our study demonstrated significant differences between the control group dominated by E. coli and Enterococcus and the outbreak group with higher densities of E. coli and Serratia. Recent data have shown that current 16S rDNA technology is not applicable for the gut ecosystem of premature infants [40]; this is an important limitation of our work, probably showing lower detection of the Bifidobacterium population. A recent work also demonstrated that Bifidobacterium density is related to the gestational age [12].
Curiously, the meconium samples from both groups were characterized by high interindividual variability and similar alpha diversity as the subsequent fecal samples, pointing to a marked delay in the establishment of the ecosystem. Similar results have been reported in very-low-birth-weight preterm infants [17]. Other authors have described an increase of fecal microbiota complexity during the NICU stay [41].
Although intrauterine fetal gut colonization is still a controversial issue [6–8], the detection of the Serratia microorganism in meconium samples suggests the possibility that its preterm colonization could start before birth. Most of the mothers in our study (7 of 12) were admitted to prevent a premature birth by various causes for a median period of 9 days before birth, with a range from 1 to 49 days. In contrast, the mother whose infant died from S. marcescens sepsis was admitted only on the day before delivery. Whereas this microorganism was scarcely represented in the meconium, it was very dominant at day 7, a fact that probably preceded the blood translocation and the sepsis episode that occurred at day 10. Therefore, a systematic routine exploration for potential enrichment of specific gut bacterial populations could possibly contribute to prevention of bacteremia in susceptible groups of patients, such as preterm infants. Recently, a novel functional methodology using volatile organic compounds as biomarkers for early detection of gut bacterial enrichment was reported [42]. Our results also demonstrated that some epidemic microorganisms, such as S. marcescens, are able to colonize and eventually infect preterm neonates even when state-of-the-art preventive measures have been applied. The PFGE analysis grouped isolates colonizing the 12 infants in the outbreak group into two major clones, whereas the WGS revealed a close relationship between them, suggesting the existence of a common ancestor. These molecular techniques also revealed that the virulome of the strain causing bacteremia and death was identical to other strains with clinical successful evolution, reinforcing the hypothesis that the unclear barrier delimiting colonization from infection is influenced by numerous factors.
Some OTUs assigned to the mandatory predator bacteria Bdellovibrio, Vampirovibrio, and Peredibacter were detected in the three meconium samples analyzed in this study. Such bacteria need to predate other bacteria to grow and reproduce and are considered to be important ecological balancers of the microbial communities [43]. Few studies have focused on these bacteria in human ecosystems; however, their presence in meconium samples suggests that they might not be infrequent in the gut microbiota. A predator’s inoculation could represent an ecological tool to modulate bacterial communities, taking into account predator-prey specificity [44].
The preterm nutrition policy of the hospital specifies neonates be fed maternal milk, although this is typically combined with human milk from donors and with preterm-adapted formulas. All the participating preterm infants received all three types of milk during the study, and although such data are not detailed, we are aware that this factor also influences the gut microbiota establishment. Maternal milk can reshape the infant gut microbiota [9], contributing its own site-specific microbiota [45–47], but also promoting the increase of a precise population by its prebiotic action [48]. Thus, it would be suitable to include in the microbiota profiling scheme the differentiation between living and dead bacteria in order to identify real colonizers from casual bacterial passengers associated with food intake [49].
Globally, our results indicate that, regardless of their perinatal settings, preterm neonates admitted to the same NICU are initially colonized by similar microbial communities that later evolve according to individual conditions. A Serratia outbreak influence on the establishment of the gut microbiota appears to be universal from the first days of admission; however, our results might also be applied to outbreaks caused by other microorganisms. This highlights the importance of the environment regarding the pattern of gut colonization of hospitalized preterm infants.
Supporting information
(XLSX)
Acknowledgments
The author would like to thank Dr. Manuel Ponce-Alonso for his helpful with genomes analysis.
Data Availability
The clinical data have been uploaded as a Supporting Information file. Additionally, the genome sequences are deposited in the European Nucleotide Archive database with the accession numbers QYRU00000000 and QYSA00000000 for Clone A (submission numbers SUB4493714 and SUB4514092); and QYRV00000000 and QYSB00000000 for Clone B (submission numbers SUB4510090 and SUB4525283). Fasta files are deposited in the NCBI web site under the Bioproject PRJNA510235 reference.
Funding Statement
Claudio Alba was supported by “Consejería de Educación, Juventud y Deporte de la Comunidad de Madrid” and European Social Funds. Claudia Saralegui was supported by “Fundación Mutua Madrileña” grant to Rosa del Campo achieved in 2017 call with reference number AP165902017.
References
- 1.Sommer F, Bäckhed F. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol. 2013,11:227–38. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 2.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016, 65:330–9. 10.1136/gutjnl-2015-309990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016, 375:2369–79. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 4.Walker WA. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res. 2017, 82:387–95. 10.1038/pr.2017.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rautava S, Luoto R, Salminen S, Isolauri E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 2012, 9:565–76. 10.1038/nrgastro.2012.144 [DOI] [PubMed] [Google Scholar]
- 6.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the "sterile womb" and "in utero colonization" hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017, 5:48 10.1186/s40168-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014, 6(237). 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017, 81(4). 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015, 17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 10.Madan JC, Farzan SF, Hibberd PL, Karagas MR. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr Opin Pediatr. 2012, 24:753–9. 10.1097/MOP.0b013e32835a1ac8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A. 2014, 111:12522–7. 10.1073/pnas.1409497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chernikova DA, Madan JC, Housman ML, Zain-Ul-Abideen M, Lundgren SN, Morrison HG, et al. The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr Res. 2018, 84:71–9. 10.1038/s41390-018-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blencowe H, Cousens S, Chou D, et al. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013, 10:Supp1:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blencowe H, Cousens S, Oestergaard MZ, Oestergaard M, Say L, Moller AB, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012, 379:2162–72. 10.1016/S0140-6736(12)60820-4 [DOI] [PubMed] [Google Scholar]
- 15.Zeitlin J, Szamotulska K, Drewniak N, Mohangoo AD, Chalmers J, Sakkeus L, et al. Preterm birth time trends in Europe: a study of 19 countries. BJOG. 2013, 120:1356–65. 10.1111/1471-0528.12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015, 112:11060–5. 10.1073/pnas.1502875112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel AL, Mutlu EA, Sun Y, Koenig L, Green S, Jakubowicz A, et al. Longitudinal survey of microbiota in hospitalized preterm very low birth weight infants. J Pediatr Gastroenterol Nutr. 2016, 62:292–303. 10.1097/MPG.0000000000000913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unger S, Stintzi A, Shah P, Mack D, O'Connor DL. Gut microbiota of the very-low-birth-weight infant. Pediatr Res. 2015, 77:205–13. 10.1038/pr.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moles L, Gómez M, Jiménez E, Fernández L, Bustos G, Chaves F, et al. Preterm infant gut colonization in the neonatal ICU and complete restoration 2 years later. Clin Microbiol Infect. 2015, 21:936.e1–10. 10.1016/j.cmi.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 20.Elgin TG, Kern SL, McElroy SJ. Development of the neonatal intestinal microbiome and its association with necrotizing enterocolitis. Clin Ther. 2016, 38:706–15. 10.1016/j.clinthera.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 21.Groer MW, Gregory KE, Louis-Jacques A, Thibeau S, Walker WA. The very low birth weight infant microbiome and childhood health. Birth Defects Res C Embryo Today. 2015, 105:252–64. 10.1002/bdrc.21115 [DOI] [PubMed] [Google Scholar]
- 22.Yusef D, Shalakhti T, Awad S, Algharaibeh H, Khasawneh W. Clinical characteristics and epidemiology of sepsis in the neonatal intensive care unit in the era of multi-drug resistant organisms: A retrospective review. Pediatr Neonatol. 2017, 59:35–41. 10.1016/j.pedneo.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 23.Martineau C, Li X, Lalancette C, Perreault T, Fournier E, Tremblay J, et al. Serratia marcescens outbreak in a neonatal intensive care unit (NICU): new insights from next-generation sequencing applications. J Clin Microbiol. 2018, 27;56(9). pii: e00235–18. 10.1128/JCM.00235-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J, Quach C. Outbreaks in the neonatal ICU: a review of the literature. Curr Opin Infect Dis. 2017, 30:395–403. 10.1097/QCO.0000000000000383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41:1–11. 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Yu Y, Cai Z, Bartlam M, Wang Y. Comparison of ITS and 18S rDNA for estimating fungal diversity using PCR-DGGE. World J Microbiol Biotechnol. 2015, 31:1387–95. 10.1007/s11274-015-1890-6 [DOI] [PubMed] [Google Scholar]
- 28.Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: a research priority. Microbiome. 2014, 2:38 10.1186/2049-2618-2-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma D, Farahbakhsh N, Shastri S, Sharma P. Biomarkers for diagnosis of neonatal sepsis: a literature review. J Matern Fetal Neonatal Med. 2018, 31:1646–59. 10.1080/14767058.2017.1322060 [DOI] [PubMed] [Google Scholar]
- 30.Ruiz L, Moles L, Gueimonde M, Rodriguez JM. Perinatal microbiomes' influence on preterm birth and preterms' health: influencing factors and modulation strategies. J Pediatr Gastroenterol Nutr. 2016, 63:e193–e20. 10.1097/MPG.0000000000001196 [DOI] [PubMed] [Google Scholar]
- 31.Stinson LF, Payne MS, Keelan JA. Planting the seed: origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit Rev Microbiol. 2017, 43:352–69. 10.1080/1040841X.2016.1211088 [DOI] [PubMed] [Google Scholar]
- 32.Montagnani C, Cocchi P, Lega L, Campana S, Biermann KP, Braggion C, et al. Serratia marcescens outbreak in a neonatal intensive care unit: crucial role of implementing hand hygiene among external consultants. BMC Infect Dis. 2015, 15:11 10.1186/s12879-014-0734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, et al. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One. 2016, 11:e0152751 10.1371/journal.pone.0152751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017, 5:4 10.1186/s40168-016-0213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underwood MA, Sohn K. The microbiota of the extremely preterm infant. Clin Perinatol. 2017, 44:407–27. 10.1016/j.clp.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogacka A, Salazar N, Suárez M, Milani C, Arboleya S, Solís G, et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome. 2017, 5:93 10.1186/s40168-017-0313-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One. 2015, 10:e0133320 10.1371/journal.pone.0133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PloS One. 2014, 9:e90784 10.1371/journal.pone.0090784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moles L, Gómez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One. 2013, 8:e66986 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcon-Giner C, Caim S, Mitra S, Ketskemety J, Wegmann U, Wain J, et al. Optimisation of 16S rRNA gut microbiota profiling of extremely low birth weight infants. BMC Genomics. 2017, 18:841 10.1186/s12864-017-4229-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, et al. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. 2011, 158:390–6. 10.1016/j.jpeds.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 42.Berkhout DJC, van Keulen BJ, Niemarkt HJ, Bessem JR, de Boode WP, Cossey V, et al. Late-onset sepsis in preterm infants can be detected preclinically by fecal volatile organic compound analysis: a prospective, multicenter cohort study. Clin Infect Dis. 2018, June 21, In press 10.1093/cid/ciy383 [DOI] [PubMed] [Google Scholar]
- 43.Iebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A, De Biase RV, et al. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One. 2013, 8:e61608 10.1371/journal.pone.0061608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadouri DE, To K, Shanks RMQ, Doi Y. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PloS One. 2013, 8:e63397 10.1371/journal.pone.0063397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013, 69:1–10. 10.1016/j.phrs.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 46.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171:647–54. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macpherson AJ, de Agüero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol. 2017, 17:508–17. 10.1038/nri.2017.58 [DOI] [PubMed] [Google Scholar]
- 48.Moukarzel S, Bode L. Human milk oligosaccharides and the preterm infant: a journey in sickness and in health. Clin Perinatol. 2017, 44:193–207. 10.1016/j.clp.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 49.Young GR, Smith DL, Embleton ND, Berrington JE, Schwalbe EC, Cummings SP, et al. Reducing viability bias in analysis of gut microbiota in preterm infants at risk of NEC and sepsis. Front Cell Infect Microbiol. 2017, 7:237 10.3389/fcimb.2017.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
The clinical data have been uploaded as a Supporting Information file. Additionally, the genome sequences are deposited in the European Nucleotide Archive database with the accession numbers QYRU00000000 and QYSA00000000 for Clone A (submission numbers SUB4493714 and SUB4514092); and QYRV00000000 and QYSB00000000 for Clone B (submission numbers SUB4510090 and SUB4525283). Fasta files are deposited in the NCBI web site under the Bioproject PRJNA510235 reference.