Abstract
BACKGROUND AND PURPOSE:
Differentiating pseudoprogression, a radiation-induced treatment effect, from tumor progression on imaging is a substantial challenge in glioblastoma management. Unfortunately, guidelines set by the Response Assessment in Neuro-Oncology criteria are based solely on bidirectional diametric measurements of enhancement observed on T1WI and T2WI/FLAIR scans. We hypothesized that quantitative 3D shape features of the enhancing lesion on T1WI, and T2WI/FLAIR hyperintensities (together called the lesion habitat) can more comprehensively capture pathophysiologic differences across pseudoprogression and tumor recurrence, not appreciable on diametric measurements alone.
MATERIALS AND METHODS:
A total of 105 glioblastoma studies from 2 institutions were analyzed, consisting of a training (n = 59) and an independent test (n = 46) cohort. For every study, expert delineation of the lesion habitat (T1WI enhancing lesion and T2WI/FLAIR hyperintense perilesional region) was obtained, followed by extraction of 30 shape features capturing 14 “global” contour characteristics and 16 “local” curvature measures for every habitat region. Feature selection was used to identify most discriminative features on the training cohort, which were evaluated on the test cohort using a support vector machine classifier.
RESULTS:
The top 2 most discriminative features were identified as local features capturing total curvature of the enhancing lesion and curvedness of the T2WI/FLAIR hyperintense perilesional region. Using top features from the training cohort (training accuracy = 91.5%), we obtained an accuracy of 90.2% on the test set in distinguishing pseudoprogression from tumor progression.
CONCLUSIONS:
Our preliminary results suggest that 3D shape attributes from the lesion habitat can differentially express across pseudoprogression and tumor progression and could be used to distinguish these radiographically similar pathologies.
The treatment of malignant brain tumors relies heavily on surgical resection and chemoradiation therapy, followed by at least 6 months of adjuvant temozolomide.1 This treatment regimen has been shown to improve overall prognosis in patients with brain tumors. However, a significant challenge postchemoradiation is the presence of radiation-induced side effects, such as pseudoprogression (PsP), which mimic the appearance of tumor progression on posttreatment MR imaging. PsP is an early-delayed benign treatment effect that occurs in approximately one-third of all malignant brain tumors and often stabilizes without further treatment.2 In the absence of reliable imaging tools to distinguish PsP from tumor progression,3 patients are typically kept on a “wait-and-watch” protocol, causing increased patient anxiety regarding their disease outcome and, in some cases, leading to unnecessary surgical interventions in patients with PsP with risks of morbidity. There is hence a significant clinical need to accurately differentiate PsP from true progression to improve patients' treatment management.
The Response Assessment in Neuro-Oncology (RANO) criteria4 are the currently established criteria for posttreatment response assessment in malignant glioblastomas. The most recent RANO criteria uses semiquantitative bidirectional diametric measurements of enhancing tumor on gadolinium contrast-enhanced T1WI and T2/FLAIR changes to stratify posttreatment lesions as PsP or tumor progression. However, these semiquantitative measurements result in interreader variability5 and overestimation of lesion volume,6 confusticating reliable differentiation of PsP from tumor progression. Advanced imaging modalities such as perfusion imaging,7 MR spectroscopy,8 and diffusion-weighted imaging9 have shown some promise in distinguishing tumor progression from posttreatment radiation effects. They, however, are limited in clinical applicability because they are not universally available and are often difficult to reproduce.10
Recently, “radiomics” (computational feature extraction approaches) have been used in conjunction with routinely available MR imaging sequences for survival prediction and response assessment in brain tumors.11,12 These radiomic approaches capture higher order quantitative measurements (eg, co-occurrence matrix homogeneity, neighboring gray-level dependence matrix) for modeling macro- and microscale textural and morphologic attributes within the lesion area and drawing associations of these features with clinical outcomes. While a few studies13,14 have explored distinguishing radiation necrosis (a delayed radiation-induced effect) from tumor recurrence using radiomic texture analysis, relatively little work has focused on distinguishing PsP from brain tumor recurrence, potentially on account of the poorly understood pathophysiology of PsP.3,15
Histopathologically, tumor progression is characterized by the presence of tumor cells, increased cellularity, and vascular proliferation and is known to morphologically alter white matter structure in complex ways due to infiltration, displacement, and blood-brain barrier disruption.16 Furthermore, some studies have linked the presence of tumor progression with corpus callosum involvement and subependymal spread on MR imaging.17 Evidence also suggests that a pronounced local inflammatory tissue response may develop in patients with PsP due to inherent and radiation therapy–induced capillary permeability, leading to more pronounced peritumoral brain edema.18 These changes in the pathophysiology of tumor progression and PsP are likely seen as morphometric changes in the lesion enhancement on T1-weighted MR imaging as well as perilesional T2WI/FLAIR hyperintensities but may not be appreciable using bidirectional measurements obtained from the RANO criteria alone.
In this work, we attempted to evaluate the role of 3D shape features in enhancing lesion and perilesional regions on T1WI and T2WI/FLAIR hyperintensities to improve the differentiation of PsP from tumor progression on routine MR imaging. We hypothesized that uneven tumor growth due to irregular and aggressive tumor infiltration will likely induce shape and surface differences in both the enhancing lesion and the T2WI/FLAIR hyperintense perilesional components18–20 (together called the “lesion habitat”); and the radiomic shape differences in the lesion habitat will likely be different between tumor progression and PsP on routine gadolinium-enhanced T1WI (Gd-T1WI), T2WI, and FLAIR. To our knowledge, this is the first attempt at distinguishing PsP from tumor progression through jointly interrogating shape features of intratumoral and peritumoral regions from posttreatment MR imaging. We first define a tumor habitat21 by delineating 2 compartments for every posttreatment lesion: a hyperintense enhancing lesion on Gd-T1WI, and T2WI/FLAIR hyperintense perilesional components constituting both edema and the nonenhancing lesion. We then compute changes in “local” surface and “global” shape radiomic features individually from each of these delineated enhancing lesion and T2WI/FLAIR hyperintense perilesional regions to capture morphometric differences between tumor progression and PsP. We then identify the most differentiating features from each compartment obtained on the training cohort and evaluate their efficacy on the test cohort. Additionally, we also consider features combined across both compartments of the lesion habitat to distinguish PsP and tumor progression.
Materials and Methods
Study Population
This institutional review board–approved and Health Insurance Portability and Accountability Act–compliant study comprised independent training and test cohorts of patients with glioblastoma from 2 different institutions: Cleveland Clinic and Dana-Farber/Brigham and Women's Cancer Center. The 2 cohorts were identified by performing a retrospective review of all patients with brain tumors who underwent chemoradiation treatment using the Stupp protocol at the respective institutions and had an enhancing lesion within 3 months of treatment. Patients who were prescribed bevacizumab or any other treatment after receiving the standard-of-care treatment were excluded from the study. The training cohort consisted of 59 MR images obtained from the Cleveland Clinic, where 38 tumor-progression cases and 21 PsP cases were confirmed for disease presence using the criteria provided below. All 59 cases in the training cohort were IDH wild-type and were acquired from a 1.5T scanner. The testing cohort was obtained from Dana-Farber/Brigham and Women's Cancer Center and included 46 cases in which 33 tumor progression cases and 13 PsP cases were confirmed for disease presence using criteria similar to that used for the training cohort. Informed consent was obtained for all patients involved in the study. Forty-two cases in the test cohort were IDH wild-type, whereas the remaining 4 were IDH mutant. For T2WI and FLAIR scans, slice thickness was 4 mm; slice gap, 0.8 mm; and FOV, 210 mm, whereas for MPRAGE, the volumetric acquisition had a voxel size of 1 × 1 × 1 mm. The acquisition matrices for T2, FLAIR, and MPRAGE scans were 224 × 320, 168 × 256, and 256 × 256, respectively.
Table 1 summarizes the demographics for this study population.
Table 1:
Summary of study population across the training and test cohorts
| Characteristic | Training | Testing |
|---|---|---|
| No. of patients | 59 | 46 |
| Women | 20 | 16 |
| Men | 39 | 30 |
| Mean age (yr) | 60.6 | 55.6 |
| Age range (yr) | 26–74 | 25–76 |
Confirmation for Disease Presence
Our inclusion criteria consisted of the following: 1) patients treated with standard-of-care chemoradiation with no additional follow-up treatment; 2) availability of all 3 routine MR imaging sequences (Gd-T1WI, T2WI, FLAIR); 3) MR images with diagnostic image quality as determined by collaborating radiologists; and 4) patients with posttreatment enhancing lesions with >5 mm of rim enhancement and the availability of diagnostic reads of the lesion as belonging to PsP or tumor progression following disease confirmation. Confirmation for progression or pseudoprogression was obtained by either histologic analysis in some cases or follow-up imaging. Continued enhancing tumor size increase on follow-up MR imaging within the subsequent 6-month period was considered progression, while reduction in tumor size or growth within the subsequent 6-month period was considered pseudoprogression.
Preprocessing
For every patient study, the 3 MR imaging sequences, Gd-T1WI, T2WI, and FLAIR, were coregistered to a T1-weighted brain atlas (Montreal Neurological Institute 152) using 3D Slicer (http://www.slicer.org). Registering every study to an average standard brain atlas allowed us to perform reliable cross-patient shape analyses across the 2 sites. Bias field correction was then conducted using a nonparametric nonuniform intensity normalization technique.22 Skull stripping was finally performed via the skull-stripping module in 3D Slicer.23
Segmentation of Lesion Habitat
Segments were conducted for every MR imaging slice with >5 mm of rim enhancement as recognized by an expert radiologist (V.S.). Every lesion was annotated into 2 regions: an enhancing lesion and a T2WI/FLAIR hyperintense perilesional component. T1WI scans were used to delineate the enhancing lesion, while both T2WI and FLAIR scans were used to annotate the T2WI/FLAIR hyperintense perilesional compartment. Scans were manually annotated across contiguous slices by 2 experienced board-certified neuroradiologists with >10 years of experience, V.H. and V.S. (1 for each site), via a hand-annotation tool in 3D Slicer. All segmentations of MR images were conducted on a standard desktop computer, with 24 GB RAM, and GT 730 GPU (NVIDIA, Santa Clara, California), with a Cintiq 22HD touchscreen monitor (Wacom, Saitama, Japan).
Feature Extraction
Global Features.
Fourteen global shape features were extracted from the enhancing lesion and T2WI/FLAIR hyperintense perilesional compartments for each subject. These features aim to characterize the global contour of the ROI, such as volume (number of voxels in the ROI), major and minor axes (longest and shortest diameters of the shape), and elongation (ratio between major and minor axes of the ROI). Extracted features are based on an Insight Segmentation and Registration Toolkit (ITK) implementation (www.itk.org). Descriptions of the 14 global shape features are provided in the On-line Table and Appendix.
Local Features.
The local features were mainly derived from curvature measures computed for the surface of every 3D segmented compartment (enhancing lesion or T2WI/FLAIR hyperintense perilesional regions) on a voxel basis. An isosurface is first constructed from each 3D compartment, followed by computing the first and second fundamental forms of the surface. Gaussian and mean curvatures are computed from these fundamental forms for each voxel.24 Four measures that capture the local curvature changes are then derived from Gaussian and mean curvatures; curvedness (C), sharpness (S), shape index (SI), and total curvature (KT).
For each segmented compartment, the mean, median, kurtosis, and SD of curvedness, shape index, sharpness, and total curvature were extracted (ie, a total of 16 local features were extracted per compartment per subject). Computations were implemented using in-house software implemented in Matlab R2016a platform (MathWorks, Natick, Massachusetts). A detailed description of the local features is provided in the On-line Table and the Appendix.
Feature Selection and Classification
A sequential feed-forward feature-selection algorithm25 was used so that only a subset of the top discriminative features is automatically selected for classification, and at each iteration, an additional feature is sequentially included in the feature set. The scheme starts from an empty feature vector and then gradually adds features that are most discriminative between the 2 groups until there is no improvement in the prediction. The selection algorithm was used along with a support vector machine (SVM) classifier,26 a robust machine-learning classifier that is commonly used for classification of biomedical data. In this work, we used a nonlinear radial basis function kernel within the SVM. To avoid training bias, we used a 4-fold cross-validation scheme within the training set, in which 3 folds were used for training and the fourth fold was held out for testing. The experiment was run 100 times, and the features that appeared most frequently across all runs were identified as the top discriminative features within the training cohort. Top performing features were then used within the independent test cohort for distinguishing PsP from tumor progression. Specifically, we used the following experiments:
Experiment 1: Distinguishing PsP from Tumor Progression Using the Shape Features of the Enhancing Lesion Alone.
Local and global features extracted from the lesion compartment (30 in total) were used to train an SVM classifier. The top 5 features chosen by the classifier across the 100 runs were identified as the most discriminative subset from a total of 30 features for the enhancing lesion regions and used within the independent test set to distinguish PsP from tumor progression.
Experiment 2: Distinguishing PsP from Tumor Progression Using T2WI/FLAIR Hyperintense Perilesional Shape Features Alone.
Local and global features extracted from the T2WI/FLAIR hyperintense perilesional compartment (30 in total) were used to train an SVM classifier. The top 5 features selected by the classifier across the 100 runs were identified as the most discriminative subset from a total of 30 features extracted from T2WI/FLAIR hyperintense perilesional regions and used within the independent test set for distinguishing PsP from tumor progression.
Experiment 3: Distinguishing PsP from Tumor Progression Using Integrated Tumor Habitat Shape Features.
All 60 features extracted from both the enhancing lesion and T2WI/FLAIR hyperintense perilesional compartments were used in conjunction to train an SVM classifier. The top 5 features selected by the classifier from both enhancing lesion and T2WI/FLAIR hyperintense perilesional features across 100 runs of cross-validation were identified as the most discriminative from the 60 features extracted across the lesion habitat and used within the independent test set to distinguish PsP from tumor progression.
Statistical Analysis
Statistical analysis was performed using a nonparametric Wilcoxon signed ranked test to further evaluate whether there are significant differences in PsP and tumor progression, across the top performing features that were selected by the classifier.
Evaluating Variability of Top Shape Radiomic Features across the 2 Sites
To test the variability of the top features selected by the classifier across the 2 sites, we computed their correlation coefficients between the training and testing sets across true progression and PsP. The correlation coefficients were computed for every feature, separately for every class (PsP or tumor progression) across the 2 sites, with high correlation values reflecting less variability across features across the 2 sites and low values reflecting more variability.
Results
Experiment 1: Distinguishing PsP from Tumor Progression Using Enhancing Lesion Shape Features Alone
Classification in the Training Cohort.
Following SVM classification using shape features of the enhancing lesion alone, the following 5 features were identified as the top discriminative features between tumor progression (TP) and PsP cases: 3 global features constituting roundness (0.5 ± 0.16 TP, 0.33 ± 0.15 PsP), eccentricity (0.8 ± 0.1 TP, 0.912 ± 0.08 PsP), and compactness (0.3 ± 0.2 TP, 0.1 ± 0.24 PsP) and 2 local features including the mean of KT (0.11 ± 0.01 TP, 0.09 ± 0.01 PsP), and the SD of S (4 ± 2 TP, 1 ± 0.9 PsP). Adding additional features to this subset did not improve the classification accuracy. Three of the top 5 features, compactness, roundness, and the SD of S, also were statistically significantly different between PsP and tumor progression (Table 2). Figure 1 demonstrates color maps of the enhancing lesion subcompartment of a tumor progression case and a PsP case, reflecting 3 of the top discriminative local and global features. Using the shape features of the enhancing lesion within the SVM classifier, we correctly classified 50 of 59 subjects (accuracy = 84.75%). Five of the misclassified cases were tumor progression, while the other 4 were PsP.
Table 2:
Statistically significant features using the Wilcoxon rank sum test for the training cohort
| Compartment | Feature | P Value |
|---|---|---|
| Enhancing lesion | SD of S | .05 |
| Enhancing lesion | Roundness | .0057 |
| Enhancing lesion | Compactness | .00027 |
| T2WI/FLAIR hyperintense perilesion | Minor axis length | .02 |
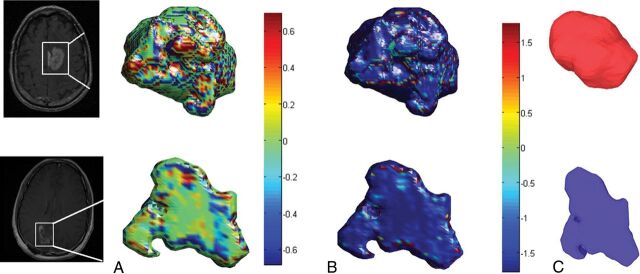
Fig 1.
Color maps visualizing the significant local features on an enhancing lesion region for a TP case (upper row) and a PsP case (lower row). The KT measure is shown in A, whereas the S measure is shown in B. C, Surface-rendering for the 2 cases shows the compactness global feature that was discriminative by the classifier. The PsP mass is more elliptic, whereas the tumor appears to be more compact.
Classification in the Test Cohort.
When we applied the top 5 discriminative features obtained on the training set for enhancing lesions to the test set (n = 41, 29 tumor progression cases and 12 PsP cases), the classifier missed 3 tumor progression cases and 4 PsP cases (accuracy = 83%).
Experiment 2: Distinguishing PsP from Tumor Progression Using the T2WI/FLAIR Hyperintense Perilesional Shape Features Alone
Classification in the Training Cohort.
For the T2WI/FLAIR hyperintense perilesional regions, the top 5 discriminative shape features as identified by the SVM classifier were the following: 2 global features constituting the elongation shape factor (0.29 ± 0.2 TP, 0.6 ± 0.43 PsP) and minor axis length (0.5 ± 0.23 TP, 0.4 ± 0.24 PsP) and 3 local features including a median of S (0.1 ± 0.03 TP, 0.09 ± 0.01 PsP), a median of C (0.05 ± 0.03 TP, 0.03 ± 0.03 PsP), and a median of SI (0.05 ± 0.01 TP, 0.03 ± 0.01 PsP). These features correctly classified 52 of 59 patients (accuracy = 88.14%), with 4 PsP cases and 3 tumor progression cases misclassified. Minor axis length was statistically significant as well (Table 2). Figure 2 demonstrates the SI and the elongation shape factor features for T2WI/FLAIR hyperintense perilesional areas of both a TP and a PsP case.
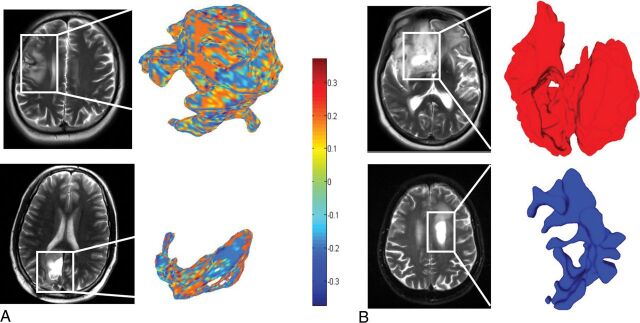
Fig 2.
A, Color maps visualizing a significant local feature (shape index) on a peritumoral region for a tumor progression case (upper row) and a PsP case (lower row). B, Surface-rendering for 2 other cases that visualize the elongation shape factor feature as a discriminative feature between tumor progression and PsP. The higher elongation shown in the lower row for the PsP case is aligned with the previous findings reporting a rather elongated and elliptic shape of benign masses.31
Classification in the Test Cohort.
Of the 46 test studies that included the T2WI/FLAIR hyperintense perilesional compartment, the top 5 features obtained by the classifier from the T2WI/FLAIR hyperintense perilesional areas as identified on the training set correctly classified 27 of the 33 subjects with tumor progression, as well as 11 of the 13 PsP cases (accuracy = 82.6%).
Experiment 3: Distinguishing PsP from Tumor Progression Using Integrated Lesion Habitat Features
Classification in the Training Cohort.
When integrating shape and surface features from the enhancing lesion as well as the T2WI/FLAIR hyperintense perilesional regions within an SVM classifier, we selected a set of top 5 features from the training set. The top 5 features included the T2WI/FLAIR elongation shape factor of the perilesion, and the median of C as well as the mean of KT, roundness, and eccentricity of the enhancing lesion. These top features correctly identified 54 of 59 subjects (accuracy = 91.5%), with 2 tumor progression and 3 PsP cases misclassified. A summary of the top discriminative features for all the 3 experiments is provided in Table 3.
Table 3:
Experiments conducted on the training and testing cohorts and the corresponding classifier performance
| Compartment | Features | Training | Testing |
|---|---|---|---|
| Enhancing lesion | Mean of KT, SD of S, roundness, eccentricity, compactness | 38 TP, 21 PsP: Accuracy = 84.75% | 29 TP, 12 PsP: Accuracy = 83% |
| T2WI/FLAIR hyperintense perilesion | Median of C, median of S, median of SI, minor axis length, elongation shape factor | 38 TP, 21 PsP: Accuracy = 88.14% | 33 TP, 13 PsP: Accuracy = 82.6% |
| Lesion habitat | Lesion: Mean of KT roundness, eccentricity T2WI/FLAIR hyperintense Perilesion: median of C, elongation shape factor |
38 TP, 21 PsP: Accuracy = 91.5% | 29 TP, 12 PsP: Accuracy = 90.2% |
Classification in the Test Cohort.
Using integrated top features from both enhancing lesion and T2WI/FLAIR hyperintense perilesional areas (5 in total across both compartments) on the test cases resulted in 37 of the 41 cases being correctly classified (accuracy = 90.2%). All 29 tumor-progression cases were correctly classified, and 8 of 12 PsP cases were correctly classified, suggesting that including T2WI/FLAIR hyperintense perilesional shape features along with the enhancing lesion shape features improved the performance of the classifier.
Evaluating Variability of Top Shape Radiomic Features across the 2 Sites
The On-line Figure shows boxplots for the top 3 features (SD of S of the enhancing lesion and T2WI/FLAIR median of C and median of S of the hyperintense perilesion) for both true progression and PsP classes across the 2 cohorts. The correlation coefficients for the lesion SD of S for both true progression and PsP across the 2 sites were 0.88 and 0.77, respectively, whereas they were 0.75 and 0.81 for T2WI/FLAIR median of C of the hyperintense perilesion and 0.9 and 0.775 for T2WI/FLAIR median of S of the hyperintense perilesion. These relatively high values of correlation coefficients between the 2 sites for both groups seem to suggest low variability across the 2 sites (Cleveland Clinic and Dana-Farber/Brigham and Women's Cancer Center) for the best performing features.
Discussion
Distinguishing tumor progression from PsP is currently one of the greatest clinical challenges in neuro-oncology.2 We present the first approach at assessing the effectiveness of 3D shape and surface radiomic features extracted from the tumor habitat (enhancing lesion and T2WI/FLAIR hyperintense perilesional regions) to differentiate tumor progression from PsP on conventional MR images (Gd-T1WI, T2WI, FLAIR). Our work was based on the rationale that there are observable 3D shape and surface irregularities encountered on the enhancing lesion as well as the perilesional boundaries in patients with tumor recurrence (potentially due to aggressive tumor infiltration and disruption) compared with those with benign pseudoprogression, which will likely have more regular boundaries.16,19,20
Multiple experiments were conducted, including using shape features from the enhancing lesion alone and from T2WI/FLAIR hyperintense perilesional areas alone and finally using the feature set of both compartments for distinguishing tumor progression from PsP. Results summarized in Table 3 suggest that using the integrated set of features from the lesion habitat provided the best classification accuracies in distinguishing PsP from tumor progression. When we used integrated shape features from the lesion habitat, classification accuracies were improved for both training and testing cohorts, with significant improvement in the test cohort (90.2% accuracy versus 83% for the lesion alone and 82.6% for T2WI/FLAIR hyperintense perilesion). One hundred percent sensitivity was obtained for identifying tumor progression cases for the test cohort, and only 2 cases of 38 in the training cohort were misclassified. A study has previously suggested the presence of more defined peritumoral edema in patients with PsP due to local inflammatory tissue response caused by inherent and radiation therapy-induced capillary permeability.18 These changes are likely seen as changes in shape features in the perilesional compartment contributing to improved distinction of PsP from tumor progression.
For the global radiomic features, minor axis length and the elongation shape factor of T2WI/FLAIR hyperintense perilesional areas were the most discriminative between tumor progression and PsP. The elongation shape factor had higher values in PsP, which agree with previous findings that benign masses tend to be more elongated and elliptic than tumor-progression cases.20,27 The discriminative features of the enhancing lesion compartment (eccentricity, roundness, and compactness) also agree with previous findings that emphasized the anisotropic, irregular structure of lesion regions in tumor progression compared with a rather elliptic shape of benign brain masses.20 This finding was supported by the higher eccentricity values of PsP cases (ie, more elliptic and elongated) than tumor progression cases shown in our results. Our reported findings in global shape differences between tumor progression and PsP are also supported by findings in breast tumors that aimed at quantifying tumor boundaries using various shape descriptors (eg, compactness, moments) for tumor classification.27,28 Other studies showed that measures of circularity (ie, roundness), size, and irregularity could distinguish primary glioblastomas and metastases.16,29
Surface features measuring local curvatures showed higher dominance in the enhancing lesion and in T2WI/FLAIR hyperintense perilesional regions of tumor progression cases (Fig 1). This could be attributed to tumors tending to alter the structure of white matter through infiltration and disruption, which eventually causes many shape irregularities leading to notable tumor surface changes.16 A study on local curvature analysis for classifying breast tumors similarly demonstrated that malignant tumor masses had more variations in their local curvature measures than benign masses.30
The stability analysis of the top features showed relatively high values of correlation coefficients of the top features across the 2 sites for tumor progression and PsP. This finding suggests that the most discriminative shape features of PsP versus tumor progression are also likely stable across the 2 sites.
This study, however, has its limitations. The results reported are preliminary because our training and testing cohorts were limited to a relatively small sample size. A large independent multisite validation should be performed to further validate our findings. While the training and test data within the respective sites were largely consistent, the potential variability introduced due to differences in imaging sequence parameters (ie, slice thickness, scanner) across the training and test sets was not explicitly studied in this work and will be a part of a future study. Additionally, the cohorts included in this study did not have information regarding the administration of steroids posttreatment; hence, this could not be studied. The ground truth for most of the studies was obtained from follow-up imaging scans and lacked histologic confirmation.
While this study is preliminary, the top features identified in the training cohort (an overall accuracy of 91.5%) seemed to perform well in the independent cohort, with an accuracy of 90.2%. Future studies will involve a large-scale validation of these findings to create a “lockdown” classifier by identifying the most discriminative and stable features using data cohorts from multiple institutions.
Future work will also focus on integrating the most discriminative shape features with radiomic texture features13 as well as other clinical parameters (age, sex, Karnofsky performance score) and investigating their performance in distinguishing PsP from tumor progression.
Conclusions
The results presented suggest that a combination of local surface and global shape attributes from the enhancing lesion and T2WI/FLAIR hyperintense perilesional areas (lesion habitat) from routinely acquired MR images could improve the distinction of PsP from tumor progression over using features from enhancing lesions or T2WI/FLAIR hyperintense perilesional regions alone.
Supplementary Material
ABBREVIATIONS:
- C
curvedness
- Gd
gadolinium
- KT
measure of the total curvature
- PsP
pseudoprogression
- RANO
Response Assessment in Neuro-Oncology
- S
sharpness
- SI
shape index
- SVM
support vector machine
- TP
tumor progression
Footnotes
Disclosures: Virginia Hill—UNRELATED: Consultancy: Google, Comments: Google asked me radiology questions for 30 minutes, and I will be paid $75; OTHER RELATIONSHIPS: Some of my coworkers and I are in discussions with Enlitic to embark on a joint research project automating detection of head CT emergencies. This has nothing to do with this article. Manmeet Aahluwalia—UNRELATED: Consultancy: Monteris Medical, AstraZeneca, Bristol-Myers Squibb, Abbvie, Varian Medical Systems, Flatiron, CBT Pharmaceuticals, Incyte; Grants/Grants Pending: Novartis, Bristol-Myers Squibb, AstraZeneca, NovoCure, Pharmacyclics, Incyte, Abbvie, Merck; Stock/Stock Options: MimiVax, Doctible.
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award Nos. 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, R01 CA220581-01A1; the National Center for Research Resources under award No. 1 C06 RR12463-01; the Department of Defense Prostate Cancer Idea Development Award; the Department of Defense Lung Cancer Idea Development Award; the Dana Foundation David Mahoney Neuroimaging Program; the Ohio Third Frontier Technology Validation Fund; the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering; and the Clinical and Translational Science Award Program at Case Western Reserve University.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Wick W, Chinot OL, Bendszus M, et al. . Evaluation of pseudoprogression rates and tumor progression patterns in a phase III trial of bevacizumab plus radiotherapy/temozolomide for newly diagnosed glioblastoma. Neuro Oncol 2016;18:1434–41 10.1093/neuonc/now091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clarke JL, Chang S. Pseudoprogression and pseudoresponse: challenges in brain tumor imaging. Curr Neurol Neurosci Rep 2009;9:241–46 10.1007/s11910-009-0035-4 [DOI] [PubMed] [Google Scholar]
- 3. Parvez K, Parvez A, Zadeh G. The diagnosis and treatment of pseudoprogression, radiation necrosis and brain tumor recurrence. Int J Mol Sci 2014;15:11832–46 10.3390/ijms150711832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen PY, Macdonald DR, Reardon DA, et al. . Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol 2010;28:1963–72 10.1200/JCO.2009.26.3541 [DOI] [PubMed] [Google Scholar]
- 5. Provenzale JM, Ison C, DeLong D. Bidimensional measurements in brain tumors: assessment of interobserver variability. AJR Am J Roentgenol 2009;193:W515–22 10.2214/AJR.09.2615 [DOI] [PubMed] [Google Scholar]
- 6. Dempsey MF, Condon BR, Hadley DM. Measurement of tumor “size” in recurrent malignant glioma: 1D, 2D, or 3D? AJNR Am J Neuroradiol 2005;26:770–76 [PMC free article] [PubMed] [Google Scholar]
- 7. Mitsuya K, Nakasu Y, Horiguchi S, et al. . Perfusion weighted magnetic resonance imaging to distinguish the recurrence of metastatic brain tumors from radiation necrosis after stereotactic radiosurgery. J Neurooncol 2010;99:81–88 10.1007/s11060-009-0106-z [DOI] [PubMed] [Google Scholar]
- 8. Elias AE, Carlos RC, Smith EA, et al. . MR spectroscopy using normalized and non-normalized metabolite ratios for differentiating recurrent brain tumor from radiation injury. Acad Radiol 2011;18:1101–08 10.1016/j.acra.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 9. Asao C, Korogi Y, Kitajima M, et al. . Diffusion-weighted imaging of radiation-induced brain injury for differentiation from tumor recurrence. AJNR Am J Neuroradiol 2005;26:1455–60 [PMC free article] [PubMed] [Google Scholar]
- 10. Brandsma D, Stalpers L, Taal W, et al. . Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol 2008;9:453–61 10.1016/S1470-2045(08)70125-6 [DOI] [PubMed] [Google Scholar]
- 11. Kickingereder P, Burth S, Wick A, et al. . Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016;280:880–89 10.1148/radiol.2016160845 [DOI] [PubMed] [Google Scholar]
- 12. Yang D, Rao G, Martinez J, et al. . Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys 2015;42:6725–35 10.1118/1.4934373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tiwari P, Prasanna P, Wolansky L, et al. . Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: a feasibility study. AJNR Am J Neuroradiol 2016;37:2231–36 10.3174/ajnr.A4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Yang J, Ho A, et al. . A predictive model for distinguishing radiation necrosis from tumor progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol 2018;28:2255–63 10.1007/s00330-017-5154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellingson BM, Chung C, Pope WB, et al. . Pseudoprogression, radionecrosis, inflammation or true tumor progression? Challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. J Neurooncol 2017;134:495–504 10.1007/s11060-017-2375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldberg-Zimring D, Talos IF, Bhagwat JG, et al. . Statistical validation of brain tumor shape approximation via spherical harmonics for image-guided neurosurgery. Acad Radiol 2005;12:459–66 10.1016/j.acra.2004.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar AJ, Leeds NE, Fuller GN, et al. . Malignant gliomas: MR imaging spectrum of radiation therapy-and chemotherapy-induced necrosis of the brain after treatment. Radiology 2000;217:377–84 10.1148/radiology.217.2.r00nv36377 [DOI] [PubMed] [Google Scholar]
- 18. Brandes AA, Tosoni A, Spagnolli F, et al. . Disease progression or pseudoprogression after concomitant radiochemotherapy treatment: pitfalls in neurooncology. Neuro Oncol 2008;10:361–67 10.1215/15228517-2008-008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bharath K, Kurtek S, Rao A, et al. . Radiologic image-based statistical shape analysis of brain tumors. Journal of the Royal Statistical Society: Series C (Applied Statistics). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu P, Xie K, Zheng Y, et al. . Brain tumors classification based on 3D shape. In: Jin D, Lin S, eds. Advances in Future Computer and Control Systems. Berlin: Springer; 2012;160:277–83 [Google Scholar]
- 21. Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology 2013;269:8–15 10.1148/radiol.13122697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tustison NJ, Avants BB, Cook PA, et al. . N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310–20 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tao X, Chang MC. A skull stripping method using deformable surface and tissue classification. In: Medical Imaging: Image Processing. Ohio: International Society for Optics and Photonics; 2010;7623:76233L [Google Scholar]
- 24. Pienaar R, Fischl B, Caviness V, et al. . A methodology for analyzing curvature in the developing brain from preterm to adult. Int J Imaging Syst Technol 2008;18:42–68 10.1002/ima.20138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcano-Cedeno A, Quintanilla-Domínguez J, Cortina-Januchs MG, et al. . Feature selection using sequential forward selection and classification applying artificial metaplasticity neural network. In: Proceedings of the Annual Conference of the Institute of Electrical and Electronics Engineers Industrial Electronics Society Glendale, Arizona; November 7–10, 2010;2845–50 [Google Scholar]
- 26. Guyon I, Weston J, Barnhill S, et al. . Gene selection for cancer classification using support vector machines. Machine Learning 2002;46:389–422 10.1023/A:1012487302797 [DOI] [Google Scholar]
- 27. Chen Y, Gunawan E, Low KS, et al. . Effect of lesion morphology on microwave signature in 2-D ultra-wideband breast imaging. IEEE Trans Biomed Eng 2008;55:2011–21 10.1109/TBME.2008.921136 [DOI] [PubMed] [Google Scholar]
- 28. Tsui PH, Liao YY, Chang CC, et al. . Classification of benign and malignant breast tumors by 2-D analysis based on contour description and scatterer characterization. IEEE Trans Med Imaging 2010;29:513–22 10.1109/TMI.2009.2037147 [DOI] [PubMed] [Google Scholar]
- 29. Zacharaki EI, Wang S, Chawla S, et al. . Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009;62:1609–18 10.1002/mrm.22147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J, Nishikawa RM, Reiser I, et al. . Local curvature analysis for classifying breast tumors: preliminary analysis in dedicated breast CT. Med Phys 2015;42:5479–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.