Summary
Aneuploidy, chromosome stoichiometry that deviates from exact multiples of the haploid compliment of an organism, exists in eukaryotic microbes, several normal human tissues and the majority of solid tumors. Here, we review the current understanding about the cellular stress states that may result from aneuploidy. The topics of aneuploid-induced proteotoxic, metabolic, replication, and mitotic stress are assessed in the context of the gene dosage imbalance observed in aneuploid cells. We also highlight emerging findings related to the downstream effects of aneuploidy-induced cellular stress on the immune surveillance against aneuploid cells.
Introduction
Aneuploidy is a type of chromosomal aberration in which the chromosome number is abnormal (Figure 1A). For organisms whose genomes are carried on multiple chromosomes, aneuploidy encompasses thousands to billions of possible numerical combinations of chromosome numbers. As such, aneuploidy is not a single genetic state but rather a large repertoire of diverse states. The most frequent cause of aneuploidy is chromosome missegregation during meiosis or mitosis. Errors in chromosome segregation are often seen in human meiosis, which result in aneuploid gametes, leading to embryos or offspring with aneuploid cells throughout the body (constitutional aneuploidy) (Nagaoka et al., 2012). Sporadic mitotic chromosome missegregation occurs during normal human development and may lead to aneuploidy in fractions of the cells in the body (mosaic aneuploidy). Recent studies suggested that chromosomal mosaicism could be common in human preimplantation embryos (van Echten-Arends et al., 2011). Mitotic error rate is lower at later developmental stages, and therefore most of the cells in adult tissues are euploid (Mantikou et al., 2012). Still, aneuploid cells can be found in certain healthy tissues, although the prevalence of aneuploidy seems to vary with methods used, with spectral karyotyping and fluorescence in situ hybridization (FISH) giving higher estimates than single-cell sequencing. Various studies estimated ~1% to 33% of human neurons to be aneuploid (Cai et al., 2014b; Knouse et al., 2014; Rehen et al., 2001; van den Bos et al., 2016; Vitak et al., 2017), and ~4% to 50% of the normal human primary hepatocytes were aneuploid (Duncan et al., 2012; Knouse et al., 2014). The most frequent aneuploidy occurs in cancer cells. Over 90% of solid tumors and 75% of hematopoietic cancers are aneuploid (Weaver and Cleveland, 2006) as a result of chromosomal instability (CIN), which has been extensively reviewed (Funk et al., 2016; Herbert et al., 2015; Thompson et al., 2010).
Figure 1.
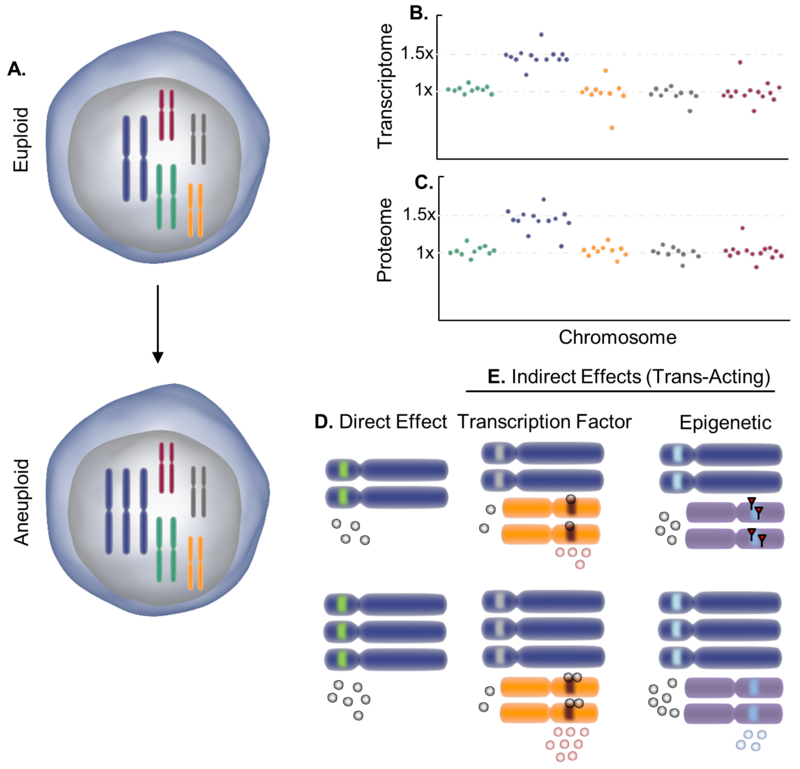
Effects of aneuploidy on the transcriptome and proteome due to the alteration of gene dosage. Changes to relative chromosome dosage (A) in aneuploidy (shown here as a euploid cell becoming trisomic for the blue chromosome) results in scaled changes to the RNA (B) and protein (C) expression level of genes carried on the aneuploid chromosome as is the case for direct effects of aneuploidy (D). However, indirect effects or trans-activating effects (E) of aneuploidy can alter the expression of genes on other chromosomes by increasing transcription factor levels to promote expression or by having an epigenetic effect such as de-silencing genes. Globular circles denote gene products that scale accordingly with chromosome number.
The past decade witnessed an increasing effort to understand the effect of aneuploidy on cellular physiology. In this review, we aim to summarize the recent progress in understanding the consequences of aneuploidy, with a focus on the various types of cellular stress likely to be elicited by aneuploidy. We first summarize chromosome-specific and genome-wide transcriptome and proteome alterations caused by aneuploidy; we then discuss the effects of these changes on cellular physiology and stress states. We will explore in depth four major types of stress caused by aneuploidy-induced gene expression changes.
Physiological consequences of aneuploidy
Because the cell is a chemical system, normal cell physiology depends on a balanced dosage of gene products. Aneuploidy alters the relative dosage of genes on the affected chromosomes. In animals, constitutional autosomal aneuploidy is highly detrimental. For example, Down syndrome is the only human autosomal aneuploidy that allows survival to adulthood. Down syndrome is caused by the gain of an extra copy of chromosome 21 (trisomy 21) and accounts for ~1/800 live births (de Graaf et al., 2015). Affected individuals display varying levels of physical and mental disability and usually have reduced life expectancy (Carfi et al., 2014; Roper and Reeves, 2006). All the other autosomal aneuploidy results in death in utero or during childhood (trisomy 13, trisomy 18) (Brewer et al., 2002). In laboratory mice, only three trisomies could survive until or beyond birth (Gropp et al., 1983). No live birth for autosomal monosomy has been reported in mammals, suggesting that missing one copy of a chromosome is even more deleterious. The fruit fly Drosophila melanogaster can tolerate the loss of a copy of chromosome 4, the smallest chromosome (Bridges, 1921), and monosomy 4 flies display decreased body size compared to diploid flies (Li, 1927). Interestingly, plants tend to be more tolerant of aneuploidy than animals. This is best exemplified by the viability of all 12 possible trisomies in jimsonweed (Datura stramonium) (Blakeslee and Belling, 1924). Each of the trisomies shows a distinctive phenotype and grows slower than the diploid plant.
Unicellular organisms exhibit even more varied levels of tolerance to aneuploidy. Extensive aneuploidy was documented in amphibian-killing chytrid fungus Batrachochytrium dendrobatidis (Rosenblum et al., 2013), the human pathogen Giardia intestinalis (Tumova et al., 2016), and the genus of parasitic protozoa Leishmania (Downing et al., 2011; Rogers et al., 2011). It appears that these micro-organisms naturally prefer the aneuploid state over a euploid one. A recent study in Leishmania donovani suggested that karyotype fluctuation could allow for selection of beneficial haplotypes under strong selection of the host environment (Prieto Barja et al., 2017). For organisms whose cells exist mostly in the euploid state, recent studies revealed that aneuploidy is generally detrimental to cellular fitness. The proliferation defect of aneuploid cells was noted over 40 years ago when the growth rate of early passage skin fibroblasts from Down syndrome patients were compared with age-matched diploid cells in several tissue culture media (Segal and McCoy, 1974). More recently, a systematic study, using a collection of disomic budding yeast (Saccharomyces cerevisiae) strains, showed reduced growth rates relative to euploid strains under standard culture conditions (Torres et al., 2007). Mouse embryonic fibroblasts (MEFs) trisomic for chromosome 1, 13, 16, or 19 were also found to have proliferation defects (Williams et al., 2008). Similar growth impairment was observed in human colorectal carcinoma HCT116 cells carrying extra copies of chromosome 3 or 5 (Stingele et al., 2012). However, specific aneuploidy can confer growth advantages under specific conditions. The spontaneously generated trisomy 8 mouse embryonic stem (ES) cells outgrew diploid ES cells during passaging in culture (Liu et al., 1997). Similarly, trisomy 12 was found to enhance the proliferation of human pluripotent stem cells (hPSCs) over diploid controls (Ben-David et al., 2014). In the colorectal adenocarcinoma cell line DLD-1, trisomy 13 cells proliferated better than the parental diploid cells in serum-free or fluorouracil-containing media (Rutledge et al., 2016).
The impact of aneuploidy on growth under diverse conditions was investigated in a comprehensive analysis of 38 aneuploid budding yeast strains derived from triploid or pentaploid meiosis (Pavelka et al., 2010). It was found that although under stress-free conditions most, though not all, aneuploids grew more poorly than euploid controls, some aneuploid strains grew significantly better than euploids under suboptimal conditions. Furthermore, different karyotypes appear to confer growth advantages under different conditions. Other studies demonstrated that unicellular organisms, such as budding yeast or the pathogenic fungus Candida albicans, although maintaining a euploid state under stress-free conditions or for sexual reproduction, use aneuploidy as a readily available source of heritable variation for evolutionary adaptation to stressful environments (Chen et al., 2012; Kaya et al., 2015; Liu et al., 2015; Pavelka et al., 2010; Ryu et al., 2016; Selmecki et al., 2006; Sunshine et al., 2015; Yona et al., 2012). Accumulating evidence also suggests that specific aneuploid chromosome patterns may be selected during the evolution of cancer (Davoli et al., 2013; Graham et al., 2017).
Effects of aneuploidy on gene expression
Effects of aneuploidy on the transcriptome
A key to understanding how aneuploidy affects cell behavior is the concept of gene dosage effects, which holds that the concentration of the primary gene product is proportional to the copy number of the gene (Epstein, 1986). Indeed, recent studies in different species indicated that aneuploid autosomes are transcribed at a level largely proportionally to their copy numbers (Figure 1B). As a result of the strong correlation between RNA expression level and chromosome copy number, transcriptome analysis revealed that approximately 8% of the yeast haploid gene deletion mutants became aneuploid for various chromosomes (Hughes et al., 2000). Subsequent studies confirmed this correlation in diverse aneuploid yeast strains without gene mutations (Pavelka et al., 2010; Torres et al., 2007). In aneuploid mouse embryonic fibroblast cell lines, the RNA expression from the gained chromosomes was also found to be overall proportional to chromosome copy numbers (Williams et al., 2008). Similar observations were also made in human monosomy 7 myeloid malignancies in a recent single-cell RNA sequencing analysis (Zhao et al., 2017).
Besides the direct dosage effect on the expression of genes located on aneuploid chromosomes, it was shown in yeast that aneuploidy also brings indirect effects to the expression of genes on other chromosomes (Figure 1D and1E) (Pavelka et al., 2010; Rancati et al., 2008). It was found that 43%-78% of these genes were downstream targets of transcription factors encoded on aneuploid chromosomes. A similar indirect effect was also seen in Arabidopsis plants with trisomy 5 (Huettel et al., 2008). In a recent study comparing the transcriptome of fetal skin primary fibroblasts from a pair of monozygotic twins discordant for trisomy 21 after normalization to chromosome stoichiometry (Letourneau et al., 2014), 182 genes were found to be significantly differentially expressed but only 6 genes were on chromosome 21. Of these differentially expressed genes not present on chromosome 21, 53 were up-regulated and 123 were down-regulated. In a recent study, 3% of random aneuploid karyotypes were found to disrupt heterochromatin assembly in yeast, leading to expression of genes that are normally silenced and consequently the destabilization of cell identity (Mulla et al., 2017). This perhaps highlights a more dramatic effect of chromosome dosage alteration on gene expression (Figure 1E).
The expression of genes on some aneuploid chromosomes can be subjected to dosage compensation, as exemplified in the triple X syndrome (47, XXX). Unlike most other aneuploidy-associated syndromes, women carrying three copies of the X chromosome usually have normal life expectancy and appear indistinguishable from the rest of the female population (Bittles et al., 2007; Tartaglia et al., 2010). This is due to the silencing of supernumerary X chromosomes mediated by the dosage compensation mechanism of X-chromosome inactivation (XCI) (Payer and Lee, 2008). XCI works through the noncoding RNA, XIST, inactivating one of the two copies of the X chromosome in diploid females. In triple X females, two of the three X chromosomes are silenced, although some X-linked genes escape the silencing and their increased expression leads to the mild phenotype of the triple X syndrome. The effect of this incomplete silencing is also seen in Klinefelter syndrome patients (47, XXY) (Brown et al., 1991). Aneuploidy of autosomes could also be dosage-compensated in some species. A recent study in Drosophila S2 cells, which contain segmental aneuploidy, found that dosage compensation not only occurs for the X chromosomes but also for aneuploid autosomes, although this compensation was imperfect and resulted in a sublinear relationship between copy number and gene expression (Zhang et al., 2010). Autosomal dosage compensation was also noted in maize and the common wheat Triticum aestivum L. (Makarevitch et al., 2008; Zhang et al., 2017). In the ten studied aneuploid wheat strains, 50-90% of the expressed genes on a given aneuploid chromosome were found be subjected to dosage compensation. Gene Ontology (GO) analysis revealed that genes showing dosage effects or dosage compensation were enriched for distinct terms, suggesting that dosage compensation is more likely to be due to functional adaptation rather than a structural mechanism. There is also a recent debate on whether dosage compensation happens in wild strains of aneuploid yeast (Gasch et al., 2016; Hose et al., 2015; Torres et al., 2016). Differences in how dosage compensation was assessed may account for the discrepancy. Gasch et al. used a gene specific approach that may account for gene-to-gene variability, while Torres et al. took a chromosome level distribution-based approach. Thus, these two studies may be focused on two sides of the same coin: while there may be no system-level dosage compensation, the expression change of specific genes and pathways may deviate from that predicted by gene dosage change either due to additional adaptive mechanisms or simply as a downstream consequence of other changes in the complex gene regulatory network.
Effects of aneuploidy on the proteome
Do the chromosome-dosage–induced transcriptomic changes also translate into the protein level? In budding yeast, changes in protein abundance of genes on aneuploid chromosomes generally scale with changes in DNA copy numbers (Figure 1C). This conclusion was derived from analysis of aneuploid strains with different chromosome stoichiometry using either the multidimensional protein identification technology (MudPIT)-based (Pavelka et al., 2010) or the stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry (Torres et al., 2010). In human cell lines, the levels of protein expression of genes on gained aneuploid chromosomes were also found to increase with chromosome copy number. However, it was less pronounced when compared with RNA expression. For instance, SILAC analysis of HCT116 cells tetrasomic for chromosome 5 in comparison with a diploid cell line found the median ratio of protein levels to be 1.6-fold for genes encoded on chromosome 5 (Stingele et al., 2012). A similar observation was made in a stable near-haploid leukemia cell line disomic for chromosome 8 (Burckstummer et al., 2013). This suggests that some genes on the aneuploid chromosome may not be translated efficiently or the protein products may be unstable. To this end, Dephoure et al. used 12 disomic yeast strains to investigate the impact of aneuploidy on the rate of translation (Dephoure et al., 2014). They found ~20% of proteins to be expressed at a lower level than expected based on copy number changes, and the majority of these proteins were components of multi-subunit complexes. Ribosomal footprint analysis did not reveal decreases in translation efficiency, whereas protein degradation seemed to be involved in the observed dosage compensation. In a recent study analyzing a group of evolutionarily conserved non-exponentially degraded (NED) proteins, it was found that NED proteins are degraded more quickly immediately after translation and are more stable later (McShane et al., 2016). Many NED proteins are core components of heteromeric protein complexes but are produced in super-stoichiometric amounts relative to their exponentially degraded (ED) counterparts. If NED proteins are encoded on a gained aneuploid chromosome, only a fraction of the extra NED proteins would be stabilized by complex formation while the rest degraded rapidly. As such, these proteins would not express as highly as predicted based on gene copy number.
Aneuploidy-associated stress
The reduced fitness of most aneuploid cells in organisms that are normally euploid led to the hypothesis that aneuploidy introduces certain stress, likely due to the unbalanced global gene expression as discussed in the above sections. While the copy-number effects predict that gene expression changes in aneuploid cells should be largely dependent on the identity of aneuploid chromosome(s), studies have looked for common gene expression signatures in multiple aneuploid models. For example, transcriptome analysis of a set of disomic budding yeast strains revealed that many, particularly disomy 4, 8, 15 and 16, exhibited gene expression signatures similar to those encompassed within the yeast environmental stress response (ESR) (Torres et al., 2007). Yeast ESR denotes the global expression programs in response to a diverse stress conditions, such as oxidative stress, heat shock and osmotic shock (Gasch et al., 2000). 615 of the ESR genes showed RNA-level changes in many disomic strains. A follow-up meta-analysis of transcriptional changes in aneuploid cells from diverse organisms, including Arabidopsis thaliana (trisomy 5), MEFs (trisomy 1, 13, 16, and 19) and human cells (trisomy 13, 18, and 21) (Sheltzer et al., 2012), found that the ESR-like signature in aneuploids could be partially attributed to the growth defect, which is consistent with a recent finding that ESR signature can largely be explained by a redistribution of cells across different cell cycle phases (O’Duibhir et al., 2014). Another study analyzed the transcriptional profiles of aneuploid human cell lines including 11 trisomic and tetrasomic cell lines and two cell lines with complex aneuploid karyotypes (Durrbaum et al., 2014). A unique aneuploidy response pattern (ARP) was identified, characterized by upregulation of genes involved in diverse functions such as the ER, Golgi and lysosome-related pathways, MHC complex and antigen processing, and metabolic pathways. These findings led to the notion that aneuploidy, regardless of specific karyotype, results in some general stress state (Figure 2 and Table 1), but the mechanism and generality of the stress identified in various studies using specific aneuploid strains or cell lines remain to be further elucidated. Furthermore, the extent of the stress could also depend on the karyotype, ploidy level, and cell type or tissue origin. Below we review the current evidence for the association of several different types of cellular stress with aneuploidy.
Figure 2.
The major categories of aneuploidy-associated stresses discussed in this review. As shown by the black arrows pointing to the four stresses. Aneuploidy leads to mitotic stress such as lagging chromosomes, replication stress via stalled and delayed replication fork progression, metabolic stress usually related to increased ROS levels in the mitochondria, and proteotoxic stress that overwhelms the proteasome. In addition to aneuploidy causing stress, some of the stresses can further perpetuate aneuploidization through causing different stress and increasing CIN (black arrows pointing between stressed and back toward aneuploidization). For example, aneuploid cells may produce more ROS that can damage DNA leading to replication stress and mitotic error.
Table 1.
List of published work supporting aneuploid-induced cellular stress, species, methods used to generate aneuploidy, and karyotype studied.
| Author | Year | Species/cell type | Aneuploidization technique | Karyotype | |
|---|---|---|---|---|---|
| Proteotoxic Stress | Torres, E. M. et al. | 2010 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 |
| Stingele, S. et al. | 2012 | Human/HCT116 and RPE-1 | Micronuclei-mediated chromosome transfer (MMCT) | Trisomies/tetrasomies | |
| Oromendia, A.B. et al. | 2012 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Dephoure, N. et al. | 2014 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Donnelly, N. et al. | 2014 | Human/HCT116 and RPE-1 | MMCT | Trisomies/tetrasomies from Stingele et al. 2012 | |
| Ohashi, A. et al. | 2015 | Human/HeLa | CENP-E inhibition in SAC-attenuated cells | Random aneuploidy | |
| Aivazidis, S. et al. | 2017 | Human/lymphoblastoid cell lines | Down syndrome patient cells | Trisomy 21 | |
| Metabolic Stress | Torres, E. M. et al. | 2007 | Budding yeast | [kar1 × wt] mediated chromosome transfer | 13 disomic strains |
| Williams, B. R. et al. | 2008 | Mouse/embryonic fibroblast | Breeding scheme, Robertsonian translocations | Trisomy 1, 13, 16, or 19 | |
| Li, M. et al. | 2010 | Mouse/embryonic fibroblast | Conditional mutation in Mad2 | Random aneuploidy | |
| Thorburn, R. R. et al. | 2013 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Dephoure, N. et al. | 2014 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Clemente-Ruiz, M. et al. | 2016 | Drosophila | RNAi-mediated knockdown of SAC genes | Changes in X chromosome number | |
| Tang, Y. et al. | 2017 | Mouse/embryonic fibroblast | Breeding scheme, Robertsonian translocations (Williams et al. 2008). | MEFs (trisomy 13 or 16) | |
| Hwang, S. et al. | 2017 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Replication Stress | Sheltzer, J. M. et al. | 2011 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 |
| Ohashi, A. et al. | 2015 | Human/HeLa | CENP-E inhibition in SAC-attenuated cells | Random aneuploidy | |
| Meena, J. K. et al. | 2015 | Human/primary fibroblasts | shRNA knockdown of GJB3, RXFP1, OSBPL3, STARD9, or MAD2L1 | Random aneuploidy | |
| Blank, H. M. et al. | 2015 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Passerini, V. et al. | 2016 | Human/HCT116 and RPE-1 | MMCT | Trisomies/tetrasomies | |
| Lamm, N. et al. | 2016 | Human/pluripotent stem cells | Acquired aneuploidy during passaging | Gain of chromosomes 17q or 12 | |
| Santaguida, S. et al. | 2017 | Human/RPE-1 | SAC kinase MPS1 inhibtion | Random aneuploidy | |
| Mitotic Stress | Reish, O. et al. | 2006 | Human/lymphocytes | Turner's syndrome patients | Monosomy X |
| Sheltzer, J. M. et al. | 2011 | Budding yeast | [kar1 × wt] mediated chromosome transfer | Disomies from Torres et al. 2007 | |
| Reish, O. et al. | 2011 | Human/lymphocytes | Patient cells | Trisomy 13, 18 or 21 | |
| Zhu, J. et al. | 2012 | Budding yeast | Triploid meiosis | Random aneuploidy | |
| Nicholson, J.M. et al. | 2015 | Human/DLD-1, amniotic fibroblasts | MMCT, patient cells | Trisomy 7 or 13 | |
| Santaguida, S. et al. | 2017 | Human/RPE-1 | SAC kinase MPS1 inhibtion | Random aneuploidy |
Proteotoxic stress
Because of unbalanced gene expression from alterations in chromosome stoichiometry in aneuploidy, it is natural to speculate that aneuploid cells may experience proteotoxic stress, broadly referring to the overburdening of cellular systems that maintain proper protein folding and homeostasis (Deshaies, 2014; Morimoto, 2008). Multi-subunit protein complexes, either structural or enzymatic, require well-defined stoichiometry to function properly, which is thought to be accomplished by tightly regulated and balanced expression of complex components (Kaizu et al., 2010). A consequence of unbalanced production of proteins could be impairment of specific cellular functions associated with the affected protein complexes. For example, gain of chromosome 6, carrying the gene encoding β-tubulin, causes lethality in yeast. However, this lethality can be rescued by additional gain of chromosome 13, carrying α-tubulin genes, thus restoring the stoichiometry of α/β-tubulin dimers (Anders et al., 2009). Another consequence of unbalanced gene expression could be a general loss of proteostasis (Dai and Sampson; Kaushik and Cuervo, 2015). It is thought that aneuploid cells, carrying extra copies of one or several chromosomes, would exhibit an overproduction of proteins relative to the chaperone systems needed to fold nascent polypeptides or the degradation systems that remove misfolded or damaged proteins (Donnelly and Storchová, 2015). This could result in accumulation of misfolded proteins as well as titration of chaperones or protein degradation machineries away from the cellular functions that depend on those activities.
Up-regulation of the ubiquitin-mediated proteasome pathway has been observed in neuronal cells from Down syndrome patients (Engidawork and Lubec, 2001), suggestive of an increased burden in protein turnover. Consistently, these cells were hypersensitive to heat or ER stressors. Unlike control euploid cells, in which expression of heat shock proteins (HSP) increased in response to heat shock stress, HSP90 and HSP70 expression showed no changes under a moderate heat shock in aneuploid cells (Aivazidis et al., 2017). This is likely due to a pre-existing stress state in the aneuploid cells that compromises the induction of HSP expression. Indeed, in other trisomic human cell lines, overexpression of the transcription factor heat shock factor 1 (HSF1) could attenuate the negative effects of the extra chromosome on protein folding (Donnelly et al., 2014). Phenotypes suggestive of proteotoxic stress were also observed in certain aneuploid yeast strains. For example, yeast strains carrying chromosome 4, 12, 13, 14 or 16 disomy were hypersensitive to the proteasome inhibitor MG132 (Torres et al., 2007). Most of yeast disomic strains also showed moderate proliferative defects in the presence of the Hsp90 inhibitor, geldanamycin, or when exposed to a temperature slightly higher than the optimal yeast growth conditions. By monitoring Hsp104-associated protein aggregates in the same series of disomic yeast strains, an increased load of protein aggregation and altered kinetics of heat adaptation were observed in 11 of 13 disomic strains.
A bottom-up approach to investigating the stress state associated with aneuploidy was to identify mutations that alter the fitness of aneuploid cells. In one study, 13 disomic strains were evolved for improved growth, and mutations associated with the adaptation were analyzed (Torres et al., 2010). A loss-of-function mutation in UBP6, encoding a deubiquitinating enzyme regulating proteasome-mediated degradation and ubiquitin recycling, rescued the growth defect in 4/11 disomic strains. Importantly, deletion of UBP6 allowed for attenuation of protein overexpression in disomy 5 and disomy 14 strains (Dephoure et al., 2014), possibly through enhancing the ubiquintin-proteosome system (UPS). In another study, Dodgson et al. performed a genome-wide screen to find synthetic interaction between single-gene deletion and chromosome disomy in yeast (Dodgson et al., 2016). This screen identified genes enriched for the GO term “vesicle-mediated transport”. Consistently, protein trafficking pathways were found to be crucial for the growth of certain disomic strains. Later, a mutation in the deubiquitinase UBP3 gene was found to impair the fitness of 6/10 disomic strains by enhancing proteotoxic stress (Oromendia et al., 2012). Taken together, indications of proteotoxic stress were observed in many but not all yeast disomic strains. In line with the observation with yeast aneuploid strains, studies in mammalian cells revealed that enhancing HSP90-dependent protein folding reduced the proliferation defects of trisomic and tetrasomic human cell lines (Donnelly et al., 2014), whereas inhibition of HSP90 exacerbated the growth defect of several trisomy mouse fibroblast cell lines (Tang et al., 2011).
Other than UPS, activation of lysosomal-mediated autophagy was also detected upon aneuploidy induction in human HCT116 and retinal pigment epithelial RPE-1 cell lines (Stingele et al., 2012). Increased p62/sequestosome expression, generally triggered by oxidative stress to sequester misfolded protein into aggregates, was found in aneuploid cells. Notably, an increase in the active autophagy marker, LC3, was observed in trisomic cell lines, suggesting that p62-dependent autophagy was activated in aneuploid cells to modulate protein homeostasis. Another study found that lysosomal stress responses were activated following chromosome missegregation and this response was important for the survival of aneuploid cells (Santaguida et al., 2015). p62 accumulation and activation of unfolded protein response were also observed in HeLa and mammary gland epithelial cell lines induced to undergo chromosome missegregation (Ohashi et al., 2015).
In sum, evidence for the presence of proteotoxic stress has been reported in studies using a variety of approaches and in model systems ranging from yeast to human cells. The results corroborate the idea that over-expression of genes associated with aneuploid chromosomes leads to disrupted proteostasis. However, there are also reasons for caution when drawing a general conclusion. Many of the studies use specific aneuploid strains or cell lines that were limited to a small number of karyotypes, compared to the vast number of possible chromosome number combinations. In fact, in many aneuploid yeast strains with simple or complex chromosome stoichiometry (Pavelka et al., 2010), a propensity for protein aggregation was not evident (our unpublished observations). Studies that involved acute induction of aneuploidy in mammalian cell lines may be complicated by the possible presence of genotoxic or metabolic stress in these cells, which are also known to induce autophagy (Balaburski et al., 2010; Santaguida et al., 2017; Santaguida et al., 2015; Soto et al., 2017; White, 2016; Xiao et al., 2015). Finally, the extent to which an aneuploid genome can tolerate extra protein expression may vary greatly from karyotype to karyotype. For example, gaining chromosomes that carry major chaperones or activators of UPS may render cells more tolerant to proteotoxic stressors (Chen et al., 2012; Kalapis et al., 2015).
Metabolic stress
The metabolic homeostasis of cells depends on a precise coordination of metabolic pathways, which depends on the stoichiometry of specified sets of enzymes and regulators. As such, it is unsurprising that metabolic alterations have been linked to aneuploid-induced stress. Disomic yeast strains exhibited changes in nucleotide and carbohydrate metabolism as well as an increased glucose uptake when compared to euploids, which may be due to upregulation of some genes encoding glucose transporters (Torres et al., 2007). However, the same study also showed that biomass production is decreased within these aneuploid cells. Amino acid levels (with the exception of aspartate and isoleucine) and many TCA cycle intermediates were found to be increased in a yeast disomy 4 (Thorburn et al., 2013). However, these changes were not observed in other disomies examined, which suggests that metabolic defects may be as difficult to generalize as the proteotoxic stress discussed above.
Altered metabolism was also observed for aneuploid mammalian cells. In MEFs trisomic for one of four specific chromosomes, each trisomy displayed alterations in glutamine use and the production of ammonium and lactate, and a subset of karyotypes exhibited increased glucose uptake (Williams et al., 2008). Trisomic and tetrasomic HCT116 cell lines generated through microcell-mediated chromosome transfer (MMCT) showed specific downregulation in proteins involved in DNA, RNA, and carbohydrate metabolism, whereas other pathways such as mitochondrial metabolism were upregulated (Stingele et al., 2012). The observed alteration of DNA metabolic pathways is consistent with DNA replication defects that have also been noted for aneuploid cells (see following section). Aneuploid cells from mice bearing a conditional knockout of Mps1, encoding a mitotic checkpoint kinase, showed a set of significantly overexpressed genes involved in cell metabolism (Foijer et al., 2014). More recently, highly aneuploid colorectal cells were found to be sensitive to an antagonist of ceramide glucosyltransferase due to overabundance of intracellular ceramide, possibly as a result of dysregulated sphingolipid metabolism (Tang et al., 2017). It was also shown that ceramide levels were increased in some aneuploid budding yeast strains and further increasing ceramide levels either genetically or pharmacologically could slow down their proliferation (Hwang et al., 2017).
There are indications that the metabolic defects associated with aneuploidy are accompanied by an increase in reactive oxygen species (ROS) levels. ROS are free radicals derived from molecular oxygen, usually as a result of metabolic output (Apel and Hirt, 2004). In yeast disomic strains, proteomic analyses revealed upregulation of genes involved in oxidative stress response pathways (such as thioredoxins and oxidoreductases) and these cells harbored higher levels of ROS (Dephoure et al., 2014). Aneuploid MEFs also contained more ROS than control diploid cells (Li et al., 2010). It remains to be understood how metabolic changes could lead to altered redox regulation in aneuploid cells. A consequence of increased ROS is oxidative DNA damage. ROS produced in aneuploid MEFs were partially responsible for activating the DNA damage checkpoint (Li et al., 2010). ROS can also lead to proteomic damage and thus potentially contributes to the proteotoxic stress discussed earlier. Interestingly, in a Drosophila model of aneuploidy, ROS play a role in activating a JNK-dependent apoptotic clearance response (Clemente-Ruiz et al., 2016). These observations point to potential interplay between different types of stress experienced by aneuploid cells and suggest that molecular signals generated by these stress states may help tissues eliminate aneuploid cells.
Replication stress
Similar to the intricate pathways involved in maintaining metabolic homeostasis, accurate eukaryotic DNA replication requires the precise choreography and proper stoichiometric balance of many proteins and protein complexes. DNA replication starts from various chromosomal sites, termed replication origins, in a two-step process involving origin licensing followed by origin firing (Masai et al., 2010). Replication licensing relies on an origin recognition complex that works with CDC6 and CDT1 to recruit the replicative helicase, MCM2-7. Several more protein complexes are then loaded to form the pre-initiation complex. Not all licensed origins will fire, and dormant origins are passively replicated or can be activated during times of replication stress to ensure faithful genome replication (Alver et al., 2014). At the start of S-phase, replication forks proceed bidirectionally from an origin forming a replication bubble. Since aneuploidy imparts changes in protein stoichiometry, one consequence of aneuploidy is impaired functionality of DNA replication complexes.
Replication stress refers to the slowing or stalling of replication fork progression (Mazouzi et al., 2014). Stalled replication forks can either be repaired following cell cycle checkpoint activation or further break down resulting in DNA double-strand breaks (DSBs). Replication stress leads to delay or complete arrest of the cell cycle. Indeed, several recent studies highlight replication stress caused by aneuploidy. For example, induction of simple aneuploidy in RPE-1 cells with an MPS1 inhibitor, which was defined by cells with genome imbalances that contained less than 5% of their genome, was shown to lead to a significant reduction in fork rate and increased replication fork stalling when compared to untreated euploid cells (Santaguida et al., 2017). Slower replication rates were also observed in trisomic and tetrasomic cells derived from both RPE-1 and HCT116 cell lines (Passerini et al., 2016). The replication defects observed were in part related to an imbalance in the production of the six subunits of the MCM2-7 helicase. This finding was consistent with a previous dataset from the same group using similar aneuploid cell lines that found a general downregulation of proteins involved in DNA replication (Stingele et al., 2012).
In addition to general replication defects such as reduced fork progression rate, some chromosome elements have also been shown to contribute to aneuploidy-associated replication stress. For example, induction of aneuploidy in fibroblasts through shRNAs knockdown of genes required for mitotic fidelity led to reduced proliferation potential and telomere-related replication stress, and this stress was rescued by expression of telomerase (Meena et al., 2015). In aneuploid cancer cells, the overall replication timing profiles appear to differ from euploid non-transformed cells. Notably, replication of usually synchronous loci became asynchronous in cells of breast cancer patients that had a higher occurrence of chromosome 17 aneuploidy (Grinberg-Rashi et al., 2010). Similarly, non-alcoholic fatty liver disease and cryptogenic cirrhosis tissues displaying high prevalence of aneuploidy showed a more asynchronous replication of two loci compared to controls where the loci replicate synchronously (Laish et al., 2016). A significant increase in centromeric replication asynchrony accompanied by a high frequency of aneuploidy in lymphocytes of hepatocellular carcinoma patients compared with those of liver cirrhosis patients and healthy control participants was also reported (Hanna et al., 2012). These studies suggest that aneuploidy can cause a disruption of the normal replication timing. However, more precise testing such as a replication timing analysis between isogenic euploids and aneuploids should be conducted to further identify differences outside of the complex cancer genome. In general, the current data has revealed various abnormalities in DNA replication associated with aneuploidy but a clear shared pattern has yet to emerge.
What are consequences of the increased levels of replication defects in aneuploid cells? Increased DNA damage, especially in the form of DSBs or increased mutational load, has been shown to accompany replication stress in aneuploid cells. DSB accumulation could be due to aberrantly exposed single-stranded DNA because of slowed or stalled replication forks. Budding yeast disomic strains contained elevated levels of DNA damage, evidenced by the accumulation of 53BP1 foci during S-phase (Blank et al., 2015). Further experiments revealed increased mutational rates for two assayed loci when compared to euploid yeast (Sheltzer et al., 2011). The mutations observed were reminiscent of mutations caused by a translesional DNA polymerase (Pol ζ). When the catalytic subunit of Pol ζ was deleted, the mutational rate of the aneuploids decreased. This study revealed that aneuploid cells may replicate some of their DNA with different polymerases than euploid cells. Consistent with results in budding yeast, DNA damage markers such as 53BP1 foci accumulated in aneuploid human RPE-1 cells (Santaguida et al., 2017). The accumulation of break point junction patterns suggestive of replication defects was observed in specific trisomic and tetrasomic human cells (Passerini et al., 2016).
In addition to increased DNA damage, aneuploid cells experiencing replication stress (including those harboring DNA damage) are also subject to several other fates, such as cell cycle delays, DNA condensation defect, inappropriate mitotic entry, senescence, and even immunological recognition and destruction (Andriani et al., 2016; Blank et al., 2015; Burrell et al., 2013; Lamm et al., 2016; Meena et al., 2015; Santaguida et al., 2017; Soto et al., 2017). Cell cycle delays associated with replication stress may be a cause of the perturbation in cell cycle/proliferative dynamics observed for aneuploid cells in the past (Segal and McCoy, 1974; Stingele et al., 2012; Williams et al., 2008).
Mitotic stress
Variation in chromosome copy number may lead to altered stoichiometry of proteins involved in the chromosome segregation machinery or spindle assembly checkpoint. Thus aneuploidy could potentially lead to mitotic stress and promote continuous generation of karyotype diversity. Multiple aneuploid budding yeast strains, generated via triploid meiosis, were found to experience repeated, nonrandom mitotic chromosome loss, suggesting aneuploidy could interfere with the fidelity of chromosome segregation (Campbell et al., 1981). This phenomenon was further explored more recently in aneuploid budding yeast. For example, using a yeast artificial chromosome (YAC) containing human DNA, 9/13 disomic strains were found to have a higher rate of YAC missegregation relative to the haploid control (Sheltzer et al., 2011). Another study systematically investigated the association of CIN with aneuploidy in diverse aneuploid progeny from triploid meiosis (Zhu et al., 2012). Evidence for genome-level and chromosome-specific determinants of CIN were revealed through tracking of karyotype changes in these aneuploid populations. For instance, aneuploid strains with equal copy of chromosomes 7 and 10 were karyotypically more stable than when the copy numbers of these two chromosomes were unequal. This effect may reflect a requirement for balanced dosage of spindle assembly checkpoint genes MAD1 (on chromosome 7) and MAD2 (on chromosome 10) for proper checkpoint function. On the genome level, the mitotic error rate appears to be positively correlated with the difference in total chromosome number of an aneuploid cell from the lower one of the closest euploid number. For example, karyotypes of 1N plus one or a few extra chromosomes tend to be more stable than those of 2N minus one or a few chromosomes. It was speculated that this was due to a growing deficit between the functional capacity of the mitotic system and an increasing chromosome segregation load.
In mammalian cells, the effect of aneuploidy on mitotic fidelity has been less clear. It was first reported that in human colon cancer cells the deviation of ploidy level from euploid was correlated with the level of CIN (Duesberg et al., 1998). When lymphocytes from Turner’s syndrome (monosomy X) patients were triggered to divide, higher levels of aneuploidy for three randomly selected chromosomes were observed relative to diploid lymphocytes (Reish et al., 2006). This was also confirmed in lymphocytes from constitutional autosomal trisomies (trisomy 13, 18 or 21) (Reish et al., 2011). A later study, however, did not report elevated rates of chromosomal mosaicism in fibroblast cells from trisomy 13, 18 and 21 patients (Valind et al., 2013). The discrepancy could be attributed to differences in cell types or in the detection methods used. A recent study directly analyzed the frequency of anaphase lagging chromosomes in aneuploid and diploid cells (Nicholson et al., 2015). In the colorectal cancer cell line DLD-1, trisomy 7 or 13 cells were found to display higher frequencies of anaphase lagging chromosomes compared to the parental diploid cells. Increased frequencies of anaphase lagging chromosomes were also observed in amniotic fibroblasts from trisomy 13 fetuses. However, another study in aneuploid cells constructed from HCT116 or RPE-1 did not find a significant increase in the frequency of lagging chromosomes (Passerini et al., 2016). Differences in cell lines or karyotype could account for the discrepancy.
Is chromosome missegregation karyotype-specific or a general effect of aneuploidy in human cells? In the above examples, human aneuploid cells were either isolated from patients or engineered using MMCT. As such, these studies were limited to the analysis of simple karyotypes. Santaguida et al. treated RPE-1 cells with MPS1 inhibitor to generate a diverse aneuploid cell population (Santaguida et al., 2017). Followed by the washout of MPS1 inhibitor and thus in the presence of a functional SAC, 80% of the aneuploid daughter cells continued to divide and their mitotic fidelity was followed through live cell microscopy. These cells were found to exhibit a high frequency of mitotic aberrations during mitosis, lagging chromosomes during anaphase, and micronuclei in the subsequent G1. These results argue that aneuploidy could generate mitotic stress and drive chromosome instability. In addition to the two mechanisms observed in yeast, another possibility is that aneuploidy induced replication stress could impair proper mitotic chromosome segregation. For instance, unresolved replication intermediates could persists into anaphase, leading to nondisjunction of the interconnected sister chromatids (Kawabata et al., 2011). Also replication stress-associated DNA damage could cause structurally abnormal chromosomes that missegregate during mitosis, fueling a vicious cycle of genomic instability (Burrell et al., 2013).
Emerging topics
An increasing body of experimental data supports the notion that aneuploidy-associated stresses could potentiate the immunogenicity of aneuploid cells. RNA-seq analysis on fibroblasts from 12 age- and gender-matched euploid and Down syndrome individuals revealed consistent genome-wide gene expression changes in aneuploid fibroblasts that can be attributed to a transcriptional response to interferon (IFN), and this finding was confirmed in blood samples from Down syndrome individuals (Sullivan et al., 2016). Hyper-activation of the IFN pathway could be due to the fact that four of the six interferon receptors are encoded on chromosome 21 in human, and this could explain the shared phenotypes between Down syndrome and hyperactive IFN signaling disorders (“interferonopathies”) (Crow and Manel, 2015). Intriguingly, an “Interferon alpha/beta signaling” gene set was also found to be overexpressed in RPE-1 cells that became aneuploid due to chromosome missegregation. The cGAS-cGAMP-STING (stimulator of interferon genes) pathway (Cai et al., 2014a), the main mechanism for sensing cytoplasmic DNA, was also activated (Santaguida et al., 2017). In addition, these aneuploid cells also suffered from the DNA damage-induced proinflammatory senescence-associated secretory phenotype (SASP), which was also observed in aneuploid human primary fibroblasts (Andriani et al., 2016). These observations suggest that the aneuploid state could be proinflammatory.
Coincidentally, two recent studies reported that the activation of cGAS-cGAMP-STING pathway could be attributed to the formation of micronuclei after missegregation of a chromosome or chromosome fragment (Harding et al., 2017; Mackenzie et al., 2017). Micronuclei cause DNA replication defects that alter the structure of chromosomes encapsulated within them, leading to increased DNA damage and in extreme cases chromothripsis (Crasta et al., 2012; Zhang et al., 2015). Defects of the nuclear envelope around the micronuclei could expose chromosomal DNA and activate the cGAS-cGAMP-STING pathway (Harding et al., 2017; Hatch et al., 2013; Mackenzie et al., 2017). Interestingly, aneuploidy in cancer cells was found to correlate with a reduced number of infiltrating cytotoxic T cells and an increased number of M2 (immune suppressive) macrophages (Davoli et al., 2017). A new study suggested that chromosomally unstable aneuploid cancer cells, though activating STING, suppressed the downstream canonical NF-κB or IFN signaling (Bakhoum et al., 2018). Instead, these cells upregulated the non-canonical NF-κB pathway, thereby engaging immune mimicry and promoting metastasis.
Concluding remarks
In this review, we have discussed the impact of aneuploidy on the cellular transcriptome and proteome, and the resulting phenotypic consequences at both organismal and cellular levels. Several cellular stress states have been observed to be associated with aneuploidy: proteotoxic, metabolic, replication, and mitotic (Table 1). Responses to these stress states may define a set of characteristics shared by at least some, if not all, aneuploid cells of diverse karyotypes. It is important to point out that the severity of these stress states may not be equally distributed across the karyotypic spectrum, and that specific aneuploid karyotypes may even help alleviate certain stress. In unicellular organisms, karyotypic diversity constitutes the substrate for evolutionary selection. CIN, caused by replication and mitotic stress associated with an unbalanced genome may enhance this diversity. On the other hand, how an aneuploid cell population explores the fitness landscape under the constraint of the fitness loss caused by the inherent stress remains to be better understood. In animals, largely uniform diploidy is likely to be important for most tissues, though exceptions exist such as the varied estimates of aneuploidy frequencies within the brain and liver. Does aneuploidy-associated cellular stress contribute to the signal that helps tissues eliminate sporadic aneuploid cells? This could occur through cell-intrinsic mechanisms such as p53 activation or via activation of immune responses. Finally, metastatic tumor cells adopt aneuploidy as a way of life, much like microbial organisms that are constitutively aneuploid. Could there be shared mechanisms that allow these populations to alleviate or cope with stress associated with the varied gene stoichiometry? Insights into these outstanding questions would provide a deeper understanding of aneuploidy in somatic evolution and tumorigenesis. Furthermore, both karyotype-specific phenotypes and the general stress associated with aneuploidy, if it exists and can be found, may be exploited for cancer therapy.
Acknowledgements:
This work was supported by NIH grant R35-GM118172 to R. Li and Prostate Cancer Foundation Young Investigator Award (16YOUN21) to H-J. Tsai.
References:
- Aivazidis S, Coughlan CM, Rauniyar AK, Jiang H, Liggett LA, Maclean KN, and Roede JR (2017). The burden of trisomy 21 disrupts the proteostasis network in Down syndrome. PloS one 12, e0176307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver RC, Chadha GS, and Blow JJ (2014). The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA repair 19, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders KR, Kudrna JR, Keller KE, Kinghorn B, Miller EM, Pauw D, Peck AT, Shellooe CE, and Strong IJT (2009). A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genetics 10, 36–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriani GA, Almeida VP, Faggioli F, Mauro M, Tsai WL, Santambrogio L, Maslov A, Gadina M, Campisi J, Vijg J, et al. (2016). Whole Chromosome Instability induces senescence and promotes SASP. Scientific reports 6, 35218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, and Hirt H (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual review of plant biology 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaburski GM, Hontz RD, and Murphy ME (2010). p53 and ARF: unexpected players in autophagy. Trends in cell biology 20, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, Narwani K, Clark AT, Andrews PW, Benvenisty N, et al. (2014). Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nature communications 5, 4825. [DOI] [PubMed] [Google Scholar]
- Bittles AH, Bower C, Hussain R, and Glasson EJ (2007). The four ages of Down syndrome. European journal of public health 17, 221–225. [DOI] [PubMed] [Google Scholar]
- Blakeslee AF, and Belling J (1924). CHROMOSOMAL MUTATIONS IN THE JIMSON WEED, DATURA STRAMONIUM. Journal of Heredity 15, 195–206. [Google Scholar]
- Blank HM, Sheltzer JM, Meehl CM, and Amon A (2015). Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Molecular biology of the cell 26, 1440–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CM, Holloway SH, Stone DH, Carothers AD, and FitzPatrick DR (2002). Survival in trisomy 13 and trisomy 18 cases ascertained from population based registers. Journal of medical genetics 39, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB (1921). Genetical and Cytological Proof of Non-disjunction of the Fourth Chromosome of Drosophila Melanogaster. Proceedings of the National Academy of Sciences of the United States of America 7, 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, and Willard HF (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Banning C, Hainzl P, Schobesberger R, Kerzendorfer C, Pauler FM, Chen D, Them N, Schischlik F, Rebsamen M, et al. (2013). A reversible gene trap collection empowers haploid genetics in human cells. Nature methods 10, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. (2013). Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, and Chen ZJ (2014a). The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell 54, 289–296. [DOI] [PubMed] [Google Scholar]
- Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, and Walsh CA (2014b). Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell reports 8, 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Doctor JS, Feuersanger JH, and Doolittle MM (1981). Differential mitotic stability of yeast disomes derived from triploid meiosis. Genetics 98, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfi A, Antocicco M, Brandi V, Cipriani C, Fiore F, Mascia D, Settanni S, Vetrano DL, Bernabei R, and Onder G (2014). Characteristics of adults with down syndrome: prevalence of age-related conditions. Frontiers in medicine 1, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, and Li R (2012). Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, Murillo-Maldonado JM, Benhra N, Barrio L, Perez L, Quiroga G, Nebreda AR, and Milan M (2016). Gene Dosage Imbalance Contributes to Chromosomal Instability-Induced Tumorigenesis. Developmental cell 36, 290–302. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, and Pellman D (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, and Manel N (2015). Aicardi-Goutieres syndrome and the type I interferonopathies. Nature reviews Immunology 15, 429–440. [DOI] [PubMed] [Google Scholar]
- Dai C, and Sampson SB HSF1: Guardian of Proteostasis in Cancer. Trends in cell biology 26, 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, and Elledge SJ (2017). Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science (New York, NY) 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, and Elledge SJ (2013). Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf G, Buckley F, and Skotko BG (2015). Estimates of the live births, natural losses, and elective terminations with Down syndrome in the United States. American journal of medical genetics Part A 167a, 756–767. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, and Torres EM (2014). Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ (2014). Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC biology 12, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson SE, Kim S, Costanzo M, Baryshnikova A, Morse DL, Kaiser CA, Boone C, and Amon A (2016). Chromosome-Specific and Global Effects of Aneuploidy in Saccharomyces cerevisiae. Genetics 202, 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Durrbaum M, Stingele S, and Storchova Z (2014). HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. The EMBO journal 33, 2374–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, and Storchová Z (2015). Aneuploidy and proteotoxic stress in cancer. Molecular & Cellular Oncology 2, e976491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, et al. (2011). Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome research 21, 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P, Rausch C, Rasnick D, and Hehlmann R (1998). Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proceedings of the National Academy of Sciences of the United States of America 95, 13692–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, and Grompe M (2012). Frequent aneuploidy among normal human hepatocytes. Gastroenterology 142, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, and Storchova Z (2014). Unique features of the transcriptional response to model aneuploidy in human cells. BMC genomics 15, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engidawork E, and Lubec G (2001). Protein expression in Down syndrome brain. Amino acids 21, 331–361. [DOI] [PubMed] [Google Scholar]
- Epstein CJ (1986). The consequences of chromosome imbalance Principles, mechanisms and models (London: Cambridge University Press; ). [Google Scholar]
- Foijer F, Xie SZ, Simon JE, Bakker PL, Conte N, Davis SH, Kregel E, Jonkers J, Bradley A, and Sorger PK (2014). Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proceedings of the National Academy of Sciences of the United States of America 111, 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk LC, Zasadil LM, and Weaver BA (2016). Living in CIN: Mitotic Infidelity and Its Consequences for Tumor Promotion and Suppression. Developmental cell 39, 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Hose J, Newton MA, Sardi M, Yong M, and Wang Z (2016). Further support for aneuploidy tolerance in wild yeast and effects of dosage compensation on gene copy-number evolution. eLife 5, e14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, and Brown PO (2000). Genomic expression programs in the response of yeast cells to environmental changes. Molecular biology of the cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham NA, Minasyan A, Lomova A, Cass A, Balanis NG, Friedman M, Chan S, Zhao S, Delgado A, Go J, et al. (2017). Recurrent patterns of DNA copy number alterations in tumors reflect metabolic selection pressures. Molecular systems biology 13, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg-Rashi H, Cytron S, Gelman-Kohan Z, Litmanovitch T, and Avivi L (2010). Replication Timing Aberrations and Aneuploidy in Peripheral Blood Lymphocytes of Breast Cancer Patients. Neoplasia (New York, NY) 12, 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp A, Winking H, Herbst EW, and Claussen CP (1983). Murine trisomy: developmental profiles of the embryo, and isolation of trisomic cellular systems. The Journal of experimental zoology 228, 253–269. [DOI] [PubMed] [Google Scholar]
- Hanna MO, Zayed NA, Darwish H, and Girgis SI (2012). Asynchronous DNA replication and aneuploidy in lymphocytes of hepatocellular carcinoma patients. Cancer genetics 205, 636–643. [DOI] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, and Greenberg RA (2017). Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, and Hetzer MW (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert M, Kalleas D, Cooney D, Lamb M, and Lister L (2015). Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harbor perspectives in biology 7, a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hose J, Yong CM, Sardi M, Wang Z, Newton MA, and Gasch AP (2015). Dosage compensation can buffer copy-number variation in wild yeast. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kreil DP, Matzke M, and Matzke AJ (2008). Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS genetics 4, e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, et al. (2000). Widespread aneuploidy revealed by DNA microarray expression profiling. Nature genetics 25, 333–337. [DOI] [PubMed] [Google Scholar]
- Hwang S, Gustafsson HT, O’Sullivan C, Bisceglia G, Huang X, Klose C, Schevchenko A, Dickson RC, Cavaliere P, Dephoure N, et al. (2017). Serine-Dependent Sphingolipid Synthesis Is a Metabolic Liability of Aneuploid Cells. Cell reports 21, 3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu K, Moriya H, and Kitano H (2010). Fragilities Caused by Dosage Imbalance in Regulation of the Budding Yeast Cell Cycle. PLoS genetics 6, e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapis D, Bezerra AR, Farkas Z, Horvath P, Bódi Z, Daraba A, Szamecz B, Gut I, Bayes M, Santos MAS, et al. (2015). Evolution of Robustness to Protein Mistranslation by Accelerated Protein Turnover. PLoS biology 13, e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2015). Proteostasis and aging. Nature Medicine 21, 1406. [DOI] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, and Shima N (2011). Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Molecular cell 41, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Gerashchenko MV, Seim I, Labarre J, Toledano MB, and Gladyshev VN (2015). Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proceedings of the National Academy of Sciences of the United States of America 112, 10685–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, and Amon A (2014). Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proceedings of the National Academy of Sciences of the United States of America 111, 13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laish I, Mannasse-Green B, Hadary R, Konikoff FM, Amiel A, and Kitay-Cohen Y (2016). Aneuploidy and asynchronous replication in non-alcholic fatty liver disease and cryptogenic cirrhosis. Gene 593, 162–166. [DOI] [PubMed] [Google Scholar]
- Lamm N, Ben-David U, Golan-Lev T, Storchova Z, Benvenisty N, and Kerem B (2016). Genomic Instability in Human Pluripotent Stem Cells Arises from Replicative Stress and Chromosome Condensation Defects. Cell stem cell 18, 253–261. [DOI] [PubMed] [Google Scholar]
- Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, et al. (2014). Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508, 345–350. [DOI] [PubMed] [Google Scholar]
- Li J. c. (1927). DEVELOPMENT IN DROSOPHILA MELANOGASTER. Genetics 12, 1–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, and Zhang P (2010). The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 107, 14188–14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yong MY, Yurieva M, Srinivasan KG, Liu J, Lim JS, Poidinger M, Wright GD, Zolezzi F, Choi H, et al. (2015). Gene Essentiality Is a Quantitative Property Linked to Cellular Evolvability. Cell 163, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu H, Loring J, Hormuzdi S, Disteche CM, Bornstein P, and Jaenisch R (1997). Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Developmental dynamics : an official publication of the American Association of Anatomists 209, 85–91. [DOI] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Phillips RL, and Springer NM (2008). Profiling expression changes caused by a segmental aneuploid in maize. BMC genomics 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S, and Mastenbroek S (2012). Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochimica et biophysica acta 1822, 1921–1930. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, and Oda M (2010). Eukaryotic chromosome DNA replication: where, when, and how? Annual review of biochemistry 79, 89–130. [DOI] [PubMed] [Google Scholar]
- Mazouzi A, Velimezi G, and Loizou JI (2014). DNA replication stress: causes, resolution and disease. Experimental cell research 329, 85–93. [DOI] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchova Z, Marsh JA, et al. (2016). Kinetic Analysis of Protein Stability Reveals Age-Dependent Degradation. Cell 167, 803–815.e821. [DOI] [PubMed] [Google Scholar]
- Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang Z-Q, Kestler HA, et al. (2015). Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. The EMBO journal 34, 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulla WA, Seidel CW, Zhu J, Tsai HJ, Smith SE, Singh P, Bradford WD, McCroskey S, Nelliat AR, Conkright J, et al. (2017). Aneuploidy as a cause of impaired chromatin silencing and mating-type specification in budding yeast. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, and Hunt PA (2012). Human aneuploidy: mechanisms and new insights into an age-old problem. Nature reviews Genetics 13, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JM, Macedo JC, Mattingly AJ, Wangsa D, Camps J, Lima V, Gomes AM, Doria S, Ried T, Logarinho E, et al. (2015). Chromosome mis-segregation and cytokinesis failure in trisomic human cells. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Duibhir E, Lijnzaad P, Benschop JJ, Lenstra TL, van Leenen D, Groot Koerkamp MJ, Margaritis T, Brok MO, Kemmeren P, and Holstege FC (2014). Cell cycle population effects in perturbation studies. Molecular systems biology 10, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A, Ohori M, Iwai K, Nakayama Y, Nambu T, Morishita D, Kawamoto T, Miyamoto M, Hirayama T, Okaniwa M, et al. (2015). Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nature communications 6, 7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, and Amon A (2012). Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26, 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, and Storchova Z (2016). The presence of extra chromosomes leads to genomic instability. Nature communications 7, 10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, and Li R (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, and Lee JT (2008). X chromosome dosage compensation: how mammals keep the balance. Annual review of genetics 42, 733–772. [DOI] [PubMed] [Google Scholar]
- Prieto Barja P, Pescher P, Bussotti G, Dumetz F, Imamura H, Kedra D, Domagalska M, Chaumeau V, Himmelbauer H, Pages M, et al. (2017). Haplotype selection as an adaptive mechanism in the protozoan pathogen Leishmania donovani. Nature ecology & evolution 1, 1961–1969. [DOI] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, and Li R (2008). Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, and Chun J (2001). Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences of the United States of America 98, 13361–13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reish O, Brosh N, Gobazov R, Rosenblat M, Libman V, and Mashevich M (2006). Sporadic aneuploidy in PHA-stimulated lymphocytes of Turner’s syndrome patients. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 14, 527–534. [DOI] [PubMed] [Google Scholar]
- Reish O, Regev M, Kanesky A, Girafi S, and Mashevich M (2011). Sporadic aneuploidy in PHA-stimulated lymphocytes of trisomies 21, 18, and 13. Cytogenetic and genome research 133, 184–189. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H, et al. (2011). Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome research 21, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, and Reeves RH (2006). Understanding the basis for Down syndrome phenotypes. PLoS genetics 2, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, Rodriguez D, Eastman JM, Richards-Hrdlicka K, Joneson S, Jenkinson TS, et al. (2013). Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proceedings of the National Academy of Sciences 110, 9385–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, and Cimini D (2016). Selective advantage of trisomic human cells cultured in non-standard conditions. Scientific reports 6, 22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HY, Wilson NR, Mehta S, Hwang SS, and Hochstrasser M (2016). Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes Dev 30, 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, Iyer DR, M’Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A, and Amon A (2017). Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Developmental cell 41, 638–651.e635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, and Amon A (2015). Aneuploidy-induced cellular stresses limit autophagic degradation. Genes & Development 29, 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, and McCoy EE (1974). Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. Journal of cellular physiology 83, 85–90. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, and Berman J (2006). Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science (New York, NY) 313, 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, and Amon A (2011). Aneuploidy drives genomic instability in yeast. Science (New York, NY) 333, 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, and Amon A (2012). Transcriptional consequences of aneuploidy. Proceedings of the National Academy of Sciences of the United States of America 109, 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M, Raaijmakers JA, Bakker B, Spierings DCJ, Lansdorp PM, Foijer F, and Medema RH (2017). p53 Prohibits Propagation of Chromosome Segregation Errors that Produce Structural Aneuploidies. Cell reports 19, 2423–2431. [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, and Storchova Z (2012). Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Molecular systems biology 8, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Lewis HC, Hill AA, Pandey A, Jackson LP, Cabral JM, Smith KP, Liggett LA, Gomez EB, Galbraith MD, et al. (2016). Trisomy 21 consistently activates the interferon response. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine AB, Payen C, Ong GT, Liachko I, Tan KM, and Dunham MJ (2015). The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS biology 13, e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, and Amon A (2011). Identification of aneuploidy-selective antiproliferation compounds. Cell 144, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Yuwen H, Wang K, Bruno PM, Bullock K, Deik A, Santaguida S, Trakala M, Pfau SJ, Zhong N, et al. (2017). Aneuploid Cell Survival Relies upon Sphingolipid Homeostasis. Cancer research 77, 5272–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia NR, Howell S, Sutherland A, Wilson R, and Wilson L (2010). A review of trisomy X (47,XXX). Orphanet journal of rare diseases 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, and Compton DA (2010). Mechanisms of chromosomal instability. Current biology : CB 20, R285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, and Amon A (2013). Aneuploid yeast strains exhibit defects in cell growth and passage through START. Molecular biology of the cell 24, 1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, and Amon A (2010). Identification of aneuploidy-tolerating mutations. Cell 143, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, and Amon A (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science (New York, NY) 317, 916–924. [DOI] [PubMed] [Google Scholar]
- Torres EM, Springer M, and Amon A (2016). No current evidence for widespread dosage compensation in S. cerevisiae. eLife 5, e10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumova P, Uzlikova M, Jurczyk T, and Nohynkova E (2016). Constitutive aneuploidy and genomic instability in the single-celled eukaryote Giardia intestinalis. MicrobiologyOpen 5, 560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valind A, Jin Y, Baldetorp B, and Gisselsson D (2013). Whole chromosome gain does not in itself confer cancer-like chromosomal instability. Proceedings of the National Academy of Sciences 110, 21119–21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bos H, Spierings DC, Taudt AS, Bakker B, Porubsky D, Falconer E, Novoa C, Halsema N, Kazemier HG, Hoekstra-Wakker K, et al. (2016). Single-cell whole genome sequencing reveals no evidence for common aneuploidy in normal and Alzheimer’s disease neurons. Genome Biol 17, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, and Repping S (2011). Chromosomal mosaicism in human preimplantation embryos: a systematic review. Human reproduction update 17, 620–627. [DOI] [PubMed] [Google Scholar]
- Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, and Adey A (2017). Sequencing thousands of single-cell genomes with combinatorial indexing. Nature methods 14, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, and Cleveland DW (2006). Does aneuploidy cause cancer? Current opinion in cell biology 18, 658–667. [DOI] [PubMed] [Google Scholar]
- White E (2016). Autophagy and p53. Cold Spring Harbor perspectives in medicine 6, a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, and Amon A (2008). Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science (New York, NY) 322, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T, Liu M, Jin M, Wu K, and Yuan J (2015). FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes & Development 29, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, and Dahan O (2012). Chromosomal duplication is a transient evolutionary solution to stress. Proceedings of the National Academy of Sciences of the United States of America 109, 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Li N, Gong L, Gou X, Wang B, Deng X, Li C, Dong Q, Zhang H, and Liu B (2017). Global Analysis of Gene Expression in Response to Whole-Chromosome Aneuploidy in Hexaploid Wheat. Plant Physiol 175, 828–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, and Pellman D (2015). Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, and Oliver B (2010). Expression in aneuploid Drosophila S2 cells. PLoS biology 8, e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Gao S, Wu Z, Kajigaya S, Feng X, Liu Q, Townsley DM, Cooper J, Chen J, Keyvanfar K, et al. (2017). Single-cell RNA-seq reveals a distinct transcriptome signature of aneuploid hematopoietic cells. Blood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, and Li R (2012). Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS genetics 8, e1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]