Abstract
Pharmacodynamic drug-drug interactions (DDIs) occur when the pharmacological effect of one drug is altered by that of another drug in a combination regimen. DDIs often are classified as synergistic, additive, or antagonistic in nature, albeit these terms are frequently misused. Within a complex pathophysiological system, the mechanism of interaction may occur at the same target or through alternate pathways. Quantitative evaluation of pharmacodynamic DDIs by employing modeling and simulation approaches is needed to identify and optimize safe and effective combination therapy regimens. This review investigates the opportunities and challenges in pharmacodynamic DDI studies, and highlights examples of quantitative methods for evaluating pharmacodynamic DDIs, with a particular emphasis on the use of mechanism-based modeling and simulation in DDI studies. Advancements in both experimental and computational techniques will enable the application of better, model-informed assessments of pharmacodynamic DDIs in drug discovery, development, and therapeutics.
Keywords: pharmacodynamics, drug interactions, modeling and simulation, mechanism-based modeling, quantitative systems pharmacology
Introduction
Drug-drug interactions (DDIs) occur when one drug alters the activity of another drug. Drug interactions may be both pharmacokinetic (PK) and pharmacodynamic (PD) in nature. In a recent assessment of DDIs from a de-identified electronic medical records system, prescribed pharmacotherapy regimens contained an average of 6.58 medications that had the potential to cause an average of 2.68 drug-drug interactions1. In contrast to PK interactions, which result from one drug altering the absorption, distribution, metabolism or elimination (ADME) of another, PD DDIs arise when the pharmacological effect of one drug is affected by that of another. PD DDIs are typically categorized as synergistic, additive, or antagonistic, although these terms are often used inappropriately. The definition of additivity is that the overall effect caused by a drug combination is the sum of the pharmacological effects of each individual agent in the combination. Synergy occurs when the overall effect of the drug combination is greater than additive, and antagonism occurs when the drug combination effect is less than additive. PD DDIs can be beneficial and employed deliberately, or adverse and unintended. For example, in cancer chemotherapy regimens, folinic acid is used in combination with 5-fluorouracil (5-FU) to enhance 5-FU inhibition of thymidylate synthase, resulting in synergistic cytotoxicity against cancer cells2. In a contrasting example, combining angiotensin converting enzyme (ACE) inhibitors with thiazide diuretics for hypertension may cause excessive diuresis and hypotension3.
Compared to the relatively well-defined guidance for the evaluation of PK DDIs in drug development4, PD DDIs studies lack a formal paradigm for evaluation. This situation results partly from the fact that most PD DDI investigations are limited to high-throughput in vitro screening studies. They are less commonly tested in vivo, in animal models or clinical trials, in which complex, pathophysiological, systems-level interactions can occur. The lack of definitive investigation for PD DDIs in complex systems creates knowledge gaps and residual uncertainties as to the translational impact of PD DDIs identified during drug discovery and development. Mechanism-based- and quantitative systems pharmacology (QSP) modeling can be used to predict and design novel combinatorial regimens and to assess the clinical significance of PD interactions. Despite a lack of formal guidance, there is an increasing need for PD DDI studies employing enhanced mathematical modeling strategies to assist interpretation of data, guidance of future research, and clinical and regulatory decision making.
The need for pharmacodynamic drug-drug interaction studies
Combination therapies are used widely in areas such as infectious disease, cancer, and cardiovascular diseases. One successful example is the highly active antiretroviral therapy (HAART) combination treatment that is often prescribed to patients with human immunodeficiency virus (HIV) or Acquired Immuno-Deficiency Syndrome. HAART regimens were designed to achieve substantial suppression of viral load by pharmacologically inhibiting virus entry, reverse transcription, integration, gene transcription, and replication, and these multiple objectives were achieved using different classes of drugs5. In this example, PD DDI studies can elucidate how the different drugs affect the virus-host interactions alone and in combination, demonstrate how mathematical models can be used to optimize current regimens6,7 and assist in the design of new combination therapies to decrease mortality from HIV infection. Another example of the utility of PD interaction studies is the design of sequential regimens for cell cycle-dependent anti-cancer drugs. The sequence in which exposure to multiple chemotherapy drugs occurs can be important for achieving enhanced killing of cancer cells and/or reducing drug-induced toxicity8. In one example, ex vivo studies have shown that when leukocytes obtained from patients treated with a taxane (paclitaxel or docetaxel) were subsequently incubated in vitro with a platinum agent (cisplatin), both cellular accumulation of cisplatin and the formation of platinum-DNA adducts decreased in these cells9. Moreover, clinical studies showed that patients experienced less hematopoietic toxicity when treated with paclitaxel/carboplatin compared to carboplatin alone8,10. However, tumor response rates were also lower in non-small-cell lung cancer patients receiving docetaxel before carboplatin, compared to the reverse schedule11. No differences were found in the clearance of carboplatin or docetaxel with either administration schedule11. A possible explanation of these observations is that the platinum agents induce strong S-phase arrest and cytotoxicity, whereas the taxanes induce arrest in M-phase. By reducing the intracellular concentration of cisplatin, the taxane pre-treatment would reduce platinum-DNA adduct formation, and also reduce the toxicity of the platinum-DNA adducts when the cancer cells transition out of S-phase into an M-phase block, and fail to exit mitosis in that cell cycle9. Another interesting example is that concurrent paclitaxel/carboplatin exposure, in contrast to sequential taxane/platinum exposure, was found to enhance the formation of carboplatin-DNA adducts in bladder urothelial carcinoma cells12. Mechanism-based PD DDI studies, coupled with PK/PD modeling, could provide consistent mechanistic explanations for apparently contradictory findings obtained from different temporal drug regimen designs applied in different biological systems.
Mathematical modeling and simulation in PD DDI studies provides a quantitative framework to evaluate the design of therapeutic combinations or dosing regimens. With this strategy, the contribution of each drug in a combination can be quantified, e.g., by estimating the potency of individual drugs, and then the information on single drug effects can be used to determine whether a combination or temporally-optimized regimen adds clinical value. The characteristics of a pharmacodynamic DDI can be represented quantitatively in a model by including an empirical parameter that compares the observed magnitude of effect of a combination to the effect expected if the interaction were purely additive. With appropriate mechanistic, pathophysiological system models, the sources of DDIs can be investigated at the level of biological signaling and response pathways to provide greater insight into the mechanism(s) of PD interaction.
Challenges in PD DDI studies
One of the primary challenges in the assessment of PD DDIs is a lack of knowledge of the detailed mechanism(s) of action and exposure-response relationships for each drug individually. For example, after more than a century of use, the exact mechanisms of action for aspirin are still being identified13. Traditional chemotherapeutic agents that are not molecularly targeted to specific effector pathways tend to be promiscuous; they often exhibit non-specific pharmacological effects, which further complicates the prediction of PD DDIs based on first principles. However, ‘omics technologies combined with bioinformatics hold the potential for providing new insights into mechanisms of drug action, and PD DDI studies at a detailed molecular level can reveal new targets or pathways underlying the effects of established drugs14–16.
Biological systems are highly regulated networks that are rich in redundancies and feedback loops. DDIs may result from direct action of each agent at a pharmacological target, or the effect of one drug could be altered by another drug through interaction at any node within the larger network. Therefore, a consideration of the complexity of the biological or pathophysiological system is necessary to understand PD DDIs. For example, a solid tumor consists of malignant cancer cells that are often heterogeneous in their genetic mutational burden and activity of signal transduction pathways, and the tumor cells are enveloped in the surrounding tumor microenvironment, which includes blood and lymphatic vessels, infiltrating immune cells, supporting stromal cells, and the extracellular matrix. All of these components interact with each other; the tumor-associated cellular components engage in autocrine and paracrine signaling networks, and interact with the extracellular matrix exerting its effects on tumor and stromal cell behavior17,18. In one example, a sequential regimen consisting of sustained EGFR inhibition by erlotinib, followed by the DNA-damaging agent doxorubicin, sensitized a subset of triple-negative breast cancer cells to genotoxic drugs through rewiring of oncogenic signaling pathways19. However, simultaneous co-administration did not. One reasonable strategy for designing therapeutic combinations having enhanced efficacy would combine a chemotherapeutic agent with drugs that target the tumor microenvironment. Examples already investigated include drug combinations of angiogenesis inhibitors20 or immuno-oncology agents21 combined with chemotherapy. Signaling crosstalk within the tumor microenvironment is important for tumor progression, metastasis, and response to treatments17,18. However, the operant mechanisms of tumor-microenvironment crosstalk are not well elucidated, owing, in part, to limited experimental methods for quantifying the spatiotemporal dynamics of inter- and intracellular signaling networks in each cell type, i.e., for determining how the concentrations of molecular components change as a function of time and cellular location. Single-cell technologies may provide an improved platform for exploring mechanisms of cell-cell communication and provide clues as to the existence of important signaling networks affecting concentration- and time-dependent responses in cell populations22. Improved characterization of the complex interplay between drugs and biological systems will help identify new targets of beneficial PD DDI and inspire novel combinatorial regimens.
Although knowledge of beneficial PD DDIs could be used to optimize therapeutic outcomes, they are tested infrequently in animal models and clinical trials. The practical considerations in the design and evaluation of DDIs empirically include both feasibility and cost. Even in preclinical studies, such studies typically require assessment of multiple dose levels for each drug, alone and in different combination ratios23. Additionally, the evaluation of PD DDIs often relies on target- or mechanism-based PD biomarker(s) that reflect the mechanisms of action and intended activities of the drugs. Unlike diagnostic, prognostic, or predictive biomarkers, a target-based biomarker may not be indicative of clinically meaningful effects24,25. This limits the utility of some biomarkers in clinical research, and can result in a lack of reliable measurements for evaluating PD DDIs. In general, both target- and mechanism-based PD biomarkers are important for decision making, particularly for early clinical development of targeted cancer therapies. For example, in a phase II study of patients with metastatic colorectal cancer, phosphorylated EGFR (Epidermal Growth Factor Receptor) and phosphorylated ERK (Extracellular Signal-Regulated Kinase) in tumor tissues served as PD biomarkers, and the trial observed that they were reduced significantly in response to erlotinib, an EGFR tyrosine kinase inhibitor26. A typical clinical question is whether a new combination is better than the current standard of care, and studies of this nature often do not address directly the existence or nature of any underlying PD DDIs. However, the evaluation of PD DDIs ought not to be overlooked in clinical drug development, because greater insight into mechanisms of drug action could be used to design drug combinations having improved efficacy and/or reduced toxicity. Coupling mechanism-based models and appropriate quantitative translational techniques with the investigation of clinically meaningful PD response biomarkers is an approach that could reduce the numbers of animals, patients, or reduce treatment cohorts required in PD DDI drug combination studies, so that they become cost-effective for investigation.
Quantification methods for PD DDIs
A broad spectrum of mathematical modeling approaches has been used to understand PD DDIs (Figure 1). Empirical models are more common for in vitro screening, and receptor binding models can be used to determine whether interactions are synergistic, additive, or antagonistic. Such empirical assessments are used less frequently when PD evaluations transition to animal and clinical studies. At the in vitro-in vivo interface, conceptual- and physiologically-based PK/PD models play a greater role in characterizing the responses to combination regimens. Notably, quantitative systems pharmacology models can be used across all phases, scales, and biological systems, and can be used in a complimentary manner with both empirical and mechanism-based PK/PD models to provide greater insights into the mechanisms of PD DDIs.
Figure 1.
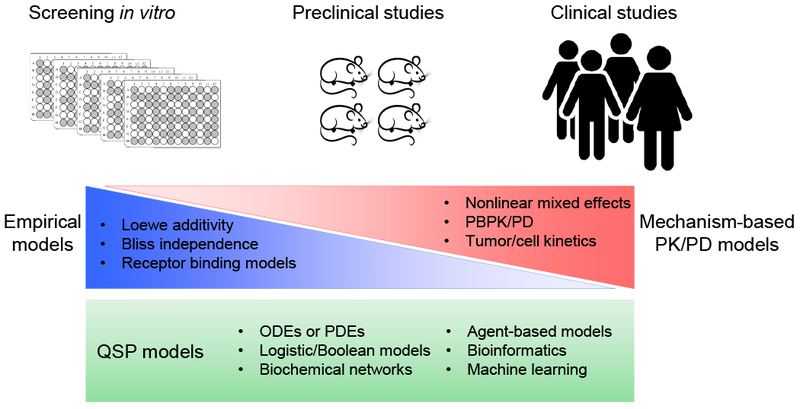
Array of mathematical modeling approaches for analyzing PD DDIs in diverse biological experimental systems. Empirical models frequently are applied to in vitro screening studies to assess the nature of potential PD DDIs. These models are used less frequently for pre-clinical animal studies and clinical studies, in which mechanism-based PK/PD models should be used to best characterize responses to drug combinations and to avoid the need for exhaustive PD DDI testing that is required for empirical assessments. Quantitative systems pharmacology (QSP) models can be constructed and calibrated across all biological systems to investigate the mechanism(s) of PD DDIs in a manner complimentary with empirical and mechanism-based models. Integration across biological systems is possible using hybrid systems models to understand and predict PD DDIs in humans. PBPK/PD: physiologically-based PK and/or PD; ODE: ordinary differential equations; PDE: partial differential equations.
Empirical evaluations
PD DDIs are more commonly studied with in vitro screens that seek to identify drug combinations having increased efficacy. For example, the NCI ALMANAC (A Large Matrix of Anti-Neoplastic Agent Combinations) study screened more than 5000 pairs of 2-drug combinations in 60 well-characterized human cancer cell lines27. This study applied a metric called the “ComboScore” to evaluate the nature of the interactions. The ComboScore was calculated as the sum of the difference between the expected versus observed cell growth fractions (Eq 1). The expected growth was assumed to conform to one of two conditions: (i) as low as the remaining cell number after cells were exposed to the more cytotoxic drug, or (ii) would equal the product of the two unaffected cell growth fractions in response to the two cytostatic agents (Eq. 3).
| Eq 1 |
| Eq 2 |
| Eq 3 |
| Eq 4 |
with representing the expected growth fraction of the ith cell line exposed to the pth concentration of drug A and qth concentration of drug B; represents the observed growth fraction under the same conditions; T0 is the time zero endpoint measurement of total cell number; represents the endpoint measurement after 2-day drug exposure; represents the endpoint measurement for the untreated control group after 2 days; and represent the observed growth fractions when exposed to drug A or drug B individually, and each is capped at 100 so that the effect of apparent drug stimulation of growth is neglected. A positive ComboScore indicates greater-than-additive activity, whereas a negative score suggests less-than-additive activity. This comprehensive screening study identified 1898 drug pairs having greater-than-additive activity in at least one cell line, and concluded that some combinations could be promising for further evaluation in tumor xenograft models and clinical trials.
In a fashion similar to the ALMANAC project, numerous in vitro screening studies are conducted to evaluate the potential of drugs for stimulation or inhibition of enzyme activities, microorganisms, or cancer cells. The first step in conducting a PD DDI study is the quantitative assessment of single-agent effects. The Hill or sigmoidal Emax function is used widely to characterize the dose- or concentration-response curve for each drug individually. For example, the expression of an inhibitory Hill equation is:
| Eq 5 |
with R as the outcome (e.g., the number of live cells remaining in response to a cytotoxic drug), R0 is the outcome in the control group, C is the drug concentration, Imax represents the inhibition capacity of the drug, which ranges from 0 to 1, IC50 is the drug concentration producing 50% of Imax, and γ is the Hill coefficient that governs the steepness of the concentration-response curve. The fraction of unaffected cells remaining after drug exposure (fu), compared to a vehicle control group, is a typical endpoint measured in cell proliferation assays. It can be derived as:
| Eq 6 |
After quantitative characterization of single-drug effects, the second step is to evaluate the drugs in combination. There exist various approaches to this assessment; the fundamental process is to compare the outcome of a reference treatment with the null hypothesis, expecting no interaction, (i.e., additive interaction). The most widely accepted functions to define “no interaction” are Loewe additivity28 and Bliss independence29. These two methods differ in their underlying assumptions, and there is no consensus as to which reference method is superior overall. Loewe additivity assumes the drugs have similar mechanisms of action, or compete for the same binding site30. As such, this method is recommended when two drugs share the same Imax and γ, and differ only in their IC50 values (i.e., have parallel concentration-response curves)31. The prediction of the unaffected cell growth fraction from Loewe additivity can be expressed as:
| Eq 7 |
with C1 and C2 representing the concentrations of drugs 1 and 2 in a combination that achieve 50% of Imax, (i.e., 50% of the inhibition of cell proliferation compared to untreated controls), and IC50,1 and IC50,2 represent the concentrations of drugs 1 and 2 as single agents that are required to achieve growth inhibition that is 50% of Imax. The combination index (CI)27, which can reflect the nature of a DDI, is defined as:
| Eq 8 |
A CI less than 1 indicates synergy, and values larger than 1 suggest antagonism. Loewe additivity can be visualized using an isobologram, as shown in Figure 2A. If the combination that achieves 50% inhibition is plotted as a point , and this point lies below the Loewe additivity line (a straight line that connects and , then the combination is Loewe synergistic. If the point lies above the Loewe additivity line, it is antagonistic. In a special case, if C1 and C2 are concentrations of the same drug, then Loewe additivity predicts that the response to the combination of C1 and C2 equals the response to this drug at the concentration of C1 + C2.
Figure 2.
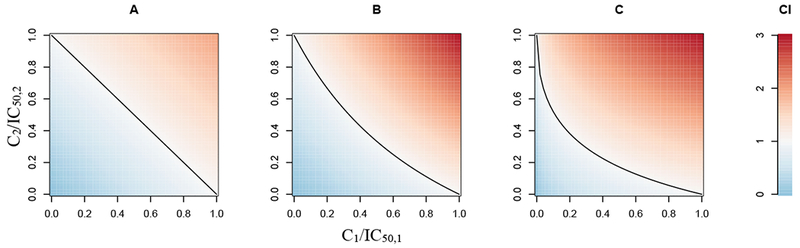
Examples of isobolograms for assessing PD drug interactions. The x- and y-axis are IC50-normalized concentrations for a drug pair. Red indicates antagonism, blue indicates synergy, and black lines represent Loewe additivity (A), Bliss independence when γ1 = γ2 = 1 (Eq. 11, B), and Bliss independence when γ1 = 0.5 and γ2 = 1 (Eq. 11, C).
Bliss independence assumes independent mechanisms of drug action, or a non-competitive interaction32, such that the combined outcome equals the product of the outcome from each drug alone. It is expressed as:
| Eq 9 |
and when Imax,1 = Imax,2 = 1 then
| Eq 10 |
Thus, the CI for Bliss independence is:
| Eq 11 |
If the observed unaffected growth fraction (e.g., the number of proliferating cells after treatment compared to untreated control) is less than the predicted unaffected fraction fu,1+2, then the interaction is considered Bliss synergistic. If the observed unaffected fraction is greater than predicted, the interaction is antagonistic. The Bliss independence method can be applied to drugs that have non-parallel concentration-response curves (e.g., different Imax and/or γ values). In contrast to the straight Loewe additivity line, when the Bliss independence line is plotted on an isobologram, it may have various shapes depending on the values of γ1 and γ233. Two examples of Bliss independent combinations on an isobologram are shown in Figure 2B (with Imax,1 = Imax,2 = 1 and γ1 = γ2 = 1) and in Figure 2C (with Imax,1 = Imax,2 = 1, γ1 = 0.5 and γ2 = 1). Paradoxically, if C1 and C2 are different concentrations of the same drug, then the Bliss independence method may predict an outcome in response to the combination of C1 and C2 that is different from the additive case of C1 + C2. The general reference model used in the ALMANAC project27 was a modified version of the Bliss independence model.
A direct comparison of experimental observations with a prediction based on the assumption of no theoretical interaction could yield a conclusion of synergy or antagonism. However, a non-interaction prediction relies solely on the single-agent concentration-response curves, and neglects all information obtained from observation of the outcomes of combination treatments. To utilize data for both single and combined agents, Greco and colleagues proposed a universal response surface approach to fit all data simultaneously33. An interaction term (e.g., α or ψ) is introduced to quantify the interaction in the combination group(s). This method is flexible and can be used to characterize Loewe additivity, Bliss independence, and other interaction models. For example, the application to Loewe additivity with parallel concentration-response curves can be expressed as:
| Eq 12 |
in which a positive α value indicates Loewe synergism and a negative value indicates antagonism. The extension of the response surface method to Bliss independence or non-competitive interaction34 can be expressed as:
| Eq 13 |
with a ψ value less than 1 indicating Bliss synergism, and a value greater than 1 indicating antagonism. An illustration of a synergistic interaction between two drugs identified by the response surface method assuming Bliss independence is shown in Figure 3. The principles of the response surface method also can be applied to indirect response models35, and additional detailed mathematical expressions have been described36. Zhu and colleagues used the response surface method in conjunction with an indirect response model of cell growth kinetics37 in a study of pancreatic cancer cells exposed to the combination of gemcitabine, a cytotoxic agent, and birinapant, a pro-apoptotic SMAC (Second Mitochondria-derived Activator of Caspases) mimetic. The analysis identified synergistic inhibition of cell growth and synergistic induction of cell death as the drug interaction mechanisms.
Figure 3.
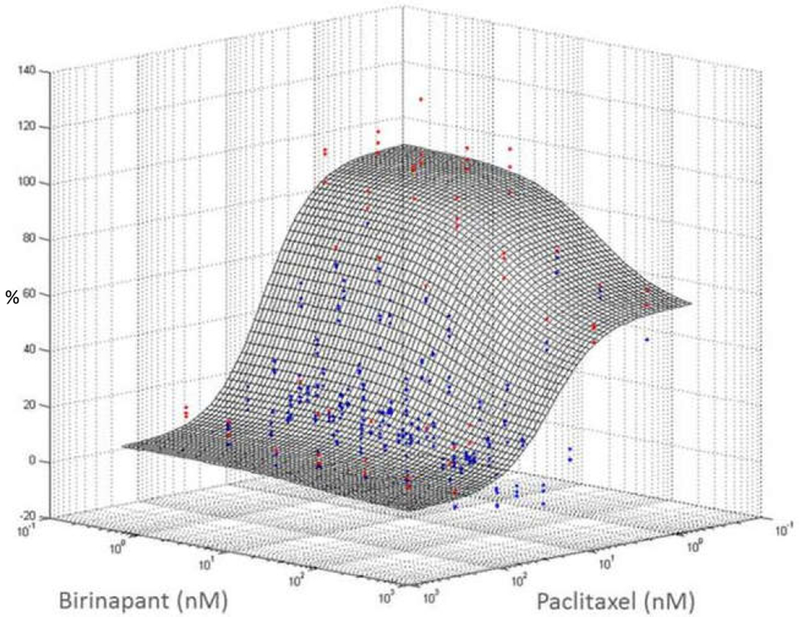
A hypothetical Bliss-independence response surface is shown for the effects of combined paclitaxel and birinapant upon the viability of PANC-1 pancreatic cancer cells. The x- and y-axes represent the concentrations of the two drugs, and the z-axis represents the magnitude of cell progression, normalized to untreated control cells, after 72h exposure to both drugs concurrently. Red dots are experimental observations above the reference additive surface (antagonism); blue dots are observations below the surface (synergism). Blue dots predominate over the red, suggesting an overall synergistic interaction that was confirmed quantitatively by model analysis. This figure is reproduced with permission of the American Society for Biochemistry and Molecular Biology, from Wang et al., 201816; permission conveyed through the Copyright Clearance Center, Inc.
Another important aspect of PD DDIs is the analysis of temporal interactions between drugs, in which the sequential exposure to drugs may elicit a greater or lesser response as compared with simultaneous treatment. The response surface method can be extended to compare drug interactions in different sequential combination regimens. For example, an additional parameter, μ, can be introduced into Eq 13 as:
| Eq 14 |
to characterize the temporal interaction in a sequential dosing regimen compared to simultaneous exposure38. A μ value less than 1 indicates a synergistic inhibition when drugs are given in a specific order compared to simultaneous exposure. This method was used to demonstrate that a sequence of bortezomib exposure for 24 hours, followed by vorinostat for an additional 24 hours, resulted in synergistic cytotoxic effects against the U266 human multiple myeloma cell line38.
There are numerous other methods available to characterize PD drug interactions in an empirical manner. For example, Jilek and colleagues defined “Degree of Independence” (DI) as the ratio of the difference between experimental observations and predicted Loewe additivity versus the difference between Bliss independence and Loewe additivity. For example, , where F represents the logarithmic measure of inhibition: log [(1 − fu) / fu], and E, B, and L represent experimental observation, Bliss independence, and Loewe additivity. This approach was applied to analyze combinations of anti-HIV drugs from different classes6. Zhao and colleagues derived expressions of “zero-interactivity” for different types of nonparallel concentration-response relationships, added the experimental uncertainties to form an “Uncertainty Envelope”, and compared the predicted range of this reference envelope to observations31. Zimmer and colleagues modified the Bliss independence technique by applying a concentration-dependent, Michaelis–Menten-like term on the effective concentration of each drug39. Some of these methods sought to provide new metrics that included statistical assessments of PD DDI studies31, and others were developed to characterize interactions in drug combinations of higher dimensionality than pairs of drugs6,39.
A common drawback of empirical methods for evaluating the nature of DDIs is that it is difficult to predict whether the interactions identified from studying 2-drug pairs are relevant to combinations of 3 or more drugs. For example, Molins and Jusko conducted a comprehensive evaluation of interactions among gemcitabine, paclitaxel, and trifluoperazine in pancreatic cancer cells, and found that the overall interaction to the 3-drug combination ranged from additive to synergistic, despite an apparent antagonism between the paclitaxel and trifluoperazine pair40. In another example, Zimmer and colleagues found a hierarchy of interactions among antibiotics, and accurately predicted the DDIs in six triplet- and two quadruplet combinations39. However, mechanisms of drug interaction(s) may differ among 2-drug pairs, and this is not considered in the empirical methods. Therefore, it may be implausible to search for a universal empirical method to predict DDIs accurately in higher dimensional combinations based solely on analysis of drug pairs.
In order to apply the empirical methods, high-throughput screening often is employed to provide rich datasets that capture concentration-response relationships for drugs as single agents and combined in different concentration ratios, and this rich data enables robust predictions as to the nature of drug interaction. However, when testing the efficacy of drug combinations in animal models or in patients, it is not feasible to obtain such rich data. More frequently, the clinical question is simply whether a combination is better than a single agent. In such situations, the limited data does not allow for robust testing of the nature of the drug interaction, nor for making projections of optimal exposures. This represents a major shortcoming of empirical modeling of DDIs, and makes the case for alternative approaches for DDI investigation that can bridge the preclinical-clinical interface, such as mechanism-based and systems pharmacology modeling.
In smaller-scale studies, such as is common in animal-based- or clinical research , the terms “additive” and “synergistic” are often misused23. Frequently they are used to describe an “enhancement” of effect suggested by a statistically greater response to a drug combination as compared to monotherapy. The misuse of “synergy” is especially common in oncology studies when tumor volume- or survival responses to a combination are compared to monotherapy or standard-of-care. The beneficial effect of a combination regimen can be the result of (i) pharmacodynamic interactions that are indeed additive or synergistic, or (ii) independent drug action without interactions, in which a patient responds to either drug as a single agent, and so the combination provides a greater probability of effective treatment41. Independent drug action and patient-to-patient variability in drug responsiveness can explain many of the combination benefits in cancer clinical trials and in xenograft models, with relatively few exceptions in which the combination therapy benefit exceeds independent drug action, and appears to exert additivity or synergy41. The usual analysis approaches for analyzing independent drug action include the highest single agent (HSA) method42 or Gaddum’s non-interaction43. By definition, these methods assume that the expected combination effect is the maximum or the best of the single-agent responses at equivalent concentrations. The HSA method is useful when a single drug dose is tested, and the same dose is used in the drug combination.
Mechanism-based PK/PD Modeling
Whereas empirical methods are useful for analyzing screening studies and identifying promising combinations for future testing, they do not provide insights into the mechanisms of action or interaction among drugs in a combination. Therefore, it is difficult to translate empirically-identified interactions directly to pre-clinical or clinical settings. In contrast, mechanism-based PK/PD models can characterize quantitatively the single-agent- and combined effects of drugs upon physiological or pathological systems. Modeling and simulations based on a mechanistic PK/PD model offer opportunities to compare different dosing regimens in patient populations and subpopulations, translate results across the stages of drug research and development, and explain non-intuitive treatment effects observed with combination therapies. For example, when ibuprofen was administered daily with aspirin, the aspirin-mediated inhibition of platelet aggregation was reported to be blocked44. The mechanism of this antagonistic interaction was characterized quantitatively using a nonlinear mixed effect (population) model that included the turnover of the drug target cyclooxygenase-1 (COX-1) according to a zero-order rate constant for COX-1 production and a first-order process for its degradation (Figure 4)45. In the model, aspirin induces a second-order, concentration-dependent, irreversible inhibition of COX-1. Ibuprofen is assumed to form a complex with COX-1 through reversible binding. A key mechanism was the assumption that the ibuprofen/COX-1 complex is protected from aspirin-induced inactivation. Platelet aggregation was modeled as being (i) proportional to total COX-1 enzyme content and (ii) regulated by ibuprofen. Simulations with the model showed that regular co-administration of aspirin and ibuprofen, or administration of ibuprofen 2 hrs before aspirin, would compromise the anti-platelet function of aspirin. This mechanism-based PK/PD model thus provided a quantitative basis for recommending only intermittent administration of ibuprofen (or other NSAIDs) in order to preserve the anti-platelet function of aspirin.
Figure 4.

Mechanism-based pharmacodynamic model of COX-1 inhibition. Aspirin (asa) induced an irreversible, 2nd-order and concentration-dependent loss of free COX-1 enzyme. Ibuprofen (ibu) inhibited the COX-1 by the formation of a reversible binding complex. C represents the drug concentration. This figure is reprinted by permission from Springer Nature; from Hong et al., 200845; permission conveyed through Springer Nature RightsLink Permissions.
The treatment of antimicrobial infections is a therapeutic area that relies on multi-drug combinations, and mechanism-based PK/PD modeling has been used to optimize combinatorial antibiotic regimens. Bulitta and colleagues developed a mechanism-based model to describe the anti-bacterial effects of tobramycin, an aminoglycoside antibiotic, on a clinical isolate of Pseudomonas aeruginosa46. The bacterial population was defined as a mixture of 3 subpopulations having different susceptibilities to the antibiotic. Two mechanisms of tobramycin-induced killing were included: (i) immediate killing as a result of bacterial outer membrane disruption, and (ii) delayed killing resulting from the synthesis of bacterial lethal protein. This model was adapted subsequently for other Gram-negative bacteria such as Acinetobacter baumannii47 and used to characterize the synergistic interaction between β-lactam antibiotics (imipenem or meropenem) and aminoglycosides (tobramycin or gentamicin)48–50. The synergy was concluded to arise from enhanced penetration of the β-lactam to its molecular target site as a result of outer membrane disruption by the aminoglycosides. A PD DDI model, based on the in vitro evaluation of bacterial killing using a hollow-fiber infection model system51, was translated in vivo to a murine thigh infection model52 and to patients53 simply by integrating the PK models of the antibiotics in those species. Simulations with the PK/PD model suggested that the continuous infusion of a high-dose β-lactam combined with an aminoglycoside would constitute a promising regimen for optimal bacterial killing without regrowth. Simulations with the model were also used to optimize the dosing regimen in patients having altered renal clearance and atypical PK profiles54.
PK/PD modeling can be applied successfully to the PD DDIs that occur when a drug interacts with both endogenous substances and complex physiological control systems that have been perturbed by pathological processes. For example, diabetes medications are given in an attempt to normalize glucose concentrations, which can vary significantly owing to food intake and the endogenous, disease-altered glucose-insulin system. Silber and colleagues developed an integrated model for the glucose-insulin system in both healthy volunteers and patients with type 2 diabetes mellitus (T2DM)55, in which plasma glucose concentrations are controlled by insulin-mediated changes in glucose production and elimination. In the model, insulin secretion is controlled by glucose concentrations in plasma and at the absorption site. Using this model along with meal tolerance test data, the primary mechanism of action of glibenclamide and its metabolites was identified as increasing insulin secretion56. The model was extended to include glucagon dynamics and to characterize the effects of the glucokinase activator LY2599506. In the model, glucagon, together with insulin and glucose, regulates hepatic glucose production, and LY2599506 both inhibits hepatic glucose production and promotes the production of insulin57. The effect of exogenous glucose and protein intake (meal ingestion) was also examined after LY2599506 administration. Although these models characterized well the short-term effects of drugs interacting with the glucose-insulin-glucagon system, the long-term progression of T2DM must be managed for effective treatment of the disease. Winter and colleagues developed a model to characterize the long-term effects of pioglitazone, metformin, and gliclazide on T2DM disease progression58 that included the homeostatic feedback relationships between fasting plasma glucose (FPG) and fasting serum insulin (FSI), as well as the physiological feed-forward relationship between FPG and glycosylated hemoglobin A1c (HbA1c). Simulations with the model showed that β-cell function is decreased gradually by long-term treatment with gliclazide, but is increased by pioglitazone. Choy and colleagues extended this model to incorporate the effect of body weight on insulin sensitivity59, and demonstrated that weight management, with diet and exercise that decreased body weight by 4.1 kg, was associated with a 30.1% increase in insulin sensitivity. These pharmacodynamic models for T2DM highlight the importance of using suitable mathematical frameworks to represent the mechanisms of drug interactions with complex physiological/pathological systems.
Similar to diabetes, cancer is a complex system pathophysiology consisting of malignant cells, the tumor microenvironment (including the vasculature and lymphatic systems), the immune system, supporting stromal cells, the extracellular matrix, and complex communications among these components17. Targeting multiple elements in the tumor microenvironment is a strategy for combination cancer therapy, as well as an emerging field for DDI studies. Several mechanism-based PK/PD models have been developed to investigate combination treatment strategies in such pathophysiological systems. For example, Hahnfeldt and colleagues proposed a mathematical model to describe the dynamics of tumor growth under angiogenic control60, using a lung cancer xenograft model system treated with the angiogenic inhibitors endostatin, angiostatin, and TNP-470. In the model, the maximal tumor volume (V) is limited by the vascular carrying capacity (K):
| Eq 15 |
where λ1 is the first-order tumor growth rate constant, and K is determined by the tumor angiogenic effect, the existing vasculature, and exogenous angiogenesis inhibitors. When tumor volume V is less than capacity K, is greater than 0, the tumor grows. Conversely, when V is greater than K, the tumor shrinks. Hutchinson and colleagues modified this model to investigate the temporal characteristics of vessel normalization after bevacizumab or vanucizumab treatment in a breast cancer xenograft model61 . Simulations showed that the effects of chemotherapy were enhanced greatly if chemotherapy was administered during a discrete time window of vessel normalization. Imbs and colleagues used this model to predict successfully that a regimen having a 3-day gap between bevacizumab treatment and pemetrexed-cisplatin administration would result in better tumor control compared to a regimen having an 8-day-gap62. These proof-of-concept cases demonstrate the utility of quantitative PK/PD DDI models of tumor-microenvironment interaction to optimize dosing regimens.
Tumor-microenvironment crosstalk also plays a role in drug resistance63. Picco and colleagues investigated intrinsic- and environment-mediated drug resistance (EMDR) in BRAF-mutated melanoma xenografts, and proposed a mathematical model that characterized the DDI between a stroma-targeted FAK (focal adhesion kinase) inhibitor and a tumor-targeted BRAF inhibitor (BRAFi)64. The model features two subpopulations of tumor cells that are either sensitive or resistant to the BRAFi, and two subpopulations of stromal cells that are either normal or activated. In the model, the activated stroma induces EMDR by stimulating the proliferation of resistant tumor cells. The BRAFi inhibits the proliferation of drug-sensitive tumor cells, and also stimulates stromal activation. The FAKi counters these effects by inhibiting the EMDR effect. Tumor growth data from both cell culture- and xenograft studies were obtained to identify the contribution of intrinsic- vs. environment-mediated drug resistance pathways. Simulations based on the model suggested that the optimal drug regimen would be to administer the BRAFi with a 1-day-on/2-day-off schedule, whereas the FAKi should be administered continuously. A binary function was used in this example to model the effect of both the BRAF and FAK inhibitors. However, this function could be replaced by one representing the duration over which drug concentrations remained above effective concentrations, which could be identified experimentally from dose-response relationships and PK information.
Tumor-immune system interactions have garnered increasing attention because immuno-oncology therapies, such as adoptive T cell transfer and immune checkpoint inhibitors, show promising anti-cancer treatment effects65. Kirschner and Panett applied a mathematical model to characterize the dynamic interactions between tumor cells (T) and immune-effector cells (E), along with the influence of the cytokine interleukin-2 (IL-2, IL)66. The effector cells in the model represent activated immune cells, such as cytotoxic T-cells and natural killer cells that are recruited by tumor antigenicity (represented by c, below, which is proportional to the tumor mass) and are stimulated to proliferate by IL-2. The stimulation by IL-2 was described by a saturable process having a maximum proliferation rate constant of p1 and a Michaelis constant g1. Tumor cells are eliminated by the immune effector cells according to Michaelis–Menten kinetics, and IL-2 is secreted when the effector cells are stimulated by the tumor, also with Michaelis–Menten kinetics.
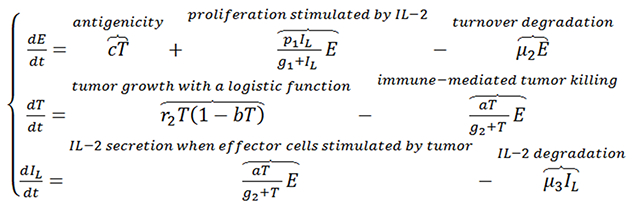 |
Eq 16 |
In this model, tumor proliferation is described by the logistic function, and the degradation of both immune effector cells and IL-2 follow first-order processes. This model establishes a quantitative foundation for the assessment of IL-2 treatment influence upon adoptive cell transfer. Several other models have been developed to investigate the combination of chemotherapy and/or radiotherapy with different types of immunotherapies67–69, and the use of mathematical models to explore approaches to overcome the challenges of cancer immunotherapy has been reviewed recently70. Owing to the complexity of the immune system, many of the models incorporate various immune cell types, along with inter- and intracellular signaling pathways in both tumor and immune cells. QSP approaches (below) are particularly useful in handling these types of complex dynamic interplay. For example, Kosinsky and colleagues developed a QSP model to characterize the synergy between radiation and anti-PD-1/PD-L1 therapy, and to optimize the administration sequences and schedules of the agents in this combination71.
Quantitative systems pharmacology models
QSP modeling is a relatively new tool in drug discovery and development, and in regulatory science72. Through mathematical modeling, QSP models can integrate and recapitulate the fundamental interactions in biological systems, simulate drug activity as perturbations of those systems, and evaluate drug effects within the context of the properties of the system, such as steady states, stability, and limits73. QSP can connect network-based system modeling with basic PK/PD principles74, and is enabled by “big data” technologies such as genomics, proteomics, and other ‘omics platforms75. QSP approaches can facilitate PD DDI studies by mapping the mechanisms of interaction of a combination onto the relevant physiological system(s), generating hypothetical combinations to achieve defined outcomes through simulations of different target perturbations, and integrating knowledge from different scales or informational sources for translational purposes.
Various methods are used by in QSP modeling, ranging from bioinformatics and data mining to logic-based- and differential equation-based models for characterizing system-specific spatiotemporal dynamics. QSP models for studying PD DDIs often are fit for the purpose of designing or optimizing a new drug combination to achieve a desired outcome, or to identify the source(s) of an observed PD interaction. For example, a logic-based Boolean network model was developed to represent a key signaling network in multiple myeloma cells76 and minimal intervention analysis was conducted to identify potential 2- and 3-drug combinations to achieve apoptosis77. This is a “bottom-up” strategy to build models based on knowledge of intracellular signal transduction pathways. In an integrated example, Zhu and colleagues discovered a synergistic interaction between the SMAC mimetic birinapant and the cytotoxic gemcitabine in pancreatic cancer cells37, used bioinformatic analyses of quantitative proteomic data78 to identify a drug interaction at the level of DNA damage response and repair pathways, and developed a multiscale model to integrate the protein- and pathway dynamics with cell cycle progression and cell growth kinetics (Figure 5)15. The interaction of birinapant and gemcitabine was attributed to birinapant blocking the gemcitabine-induced protein ATM (Ataxia-Telangiectasia Mutated), PP5 (Protein Phosphatases 5) and PP2CB (Protein Phosphatases 2A Beta), and also to mediating the degradation of cIAP1/2 (Cellular Inhibitor of Apoptosis Protein 1/2) and gemcitabine-induced FasL (Fas Ligand). The protein expression changes propagated temporally through the signaling transduction network so as to perturb the cell cycle and result in apparent synergy15,37. Sensitivity analysis suggested that a key protein to inhibit cell growth was RFC1 (Replication Factor C Subunit 1), and the model predicted that a sequential regimen of 24h pre-treatment with gemcitabine first, followed by birinapant exposure, would produce considerably greater tumor cell growth inhibition compared to simultaneous drug exposure15. This hypothesis was then experimentally validated37.
Figure 5.
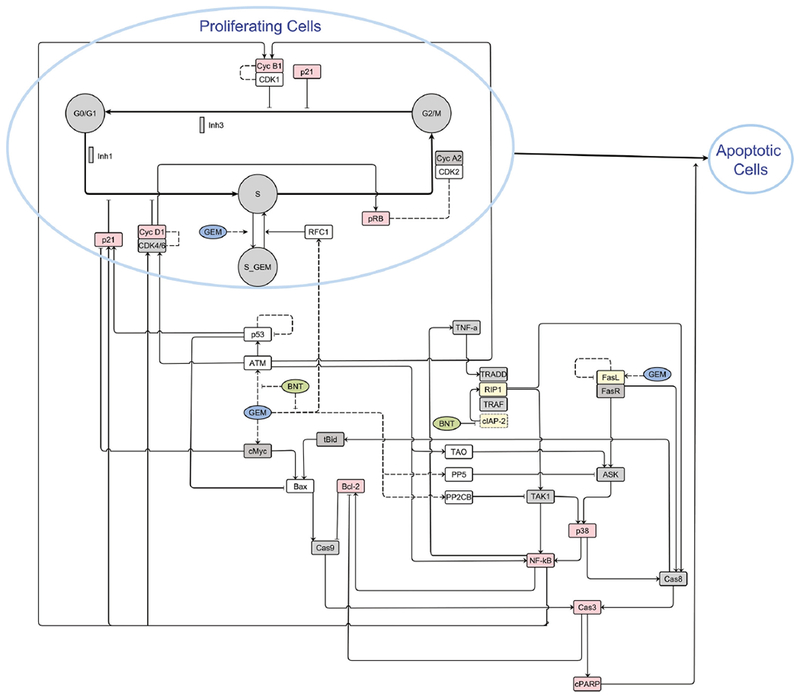
Multi-scale network model integrating systems information and gemcitabine- and birinapant-mediated effects on signaling pathways, cell cycle distributions (G0/G1-, S-, and G2/M-phases), and apoptosis in pancreatic cancer cells. Blue open circles represent proliferating and apoptotic cells. Grey circles represent cell cycle stages, and arrow-head lines indicate activation or transitions. Bar-head lines indicates inhibition. Squares represent the specific intracellular proteins as indicted. Box color denotes the source of the data for the figure (red: western blot analysis; yellow: published literature; clear: LC/MS proteomics; grey: quantitative data not available). Dashed lines represent hypothesized interactions. This figure is reprinted with the permission from John Wiley and Sons; from Zhu et al., 201878.
QSP models have made considerable progress in understanding and predicting PD DDIs. However, standards and best practices are critical for model development and validation owing to the complexity and flexibility in building QSP models79–81. Currently, there is no consensus on systematic approaches for exploring parameter space and increasing confidence in model predictions.
Prospectus
Drug discovery and development is evolving from the ‘one-drug/one-target/one-disease process’ paradigm toward identifying “sweet spots” or the key nexuses of interaction at which to intervene in order to restore pathophysiological systems. By integrating QSP analysis with physiologically-based pharmacokinetic (PBPK) models, multiscale modeling could predict potential PD DDIs, and, via clinical trial simulations, create testable hypotheses as to their potential clinical significance. PD DDI studies can leverage “big data” to identify novel targets and/or covariates for drug interactions, evaluate the strength of DDIs in patients of different genetic/environment backgrounds, and help select the “right” patients for clinical trials. In one example, machine-learning algorithms have been used to predict DDIs for adverse reactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties82. In a second example, the use of machine-learning, QSP, and PK/PD modeling techniques to integrate genomic and transcriptomic data with experimental studies of patient-derived, induced pluripotent stem cells (iPSC), which can recapitulate the inter-individual variabilities in clinical drug responses of their donors, resulted in the identification of genomic predictors of adverse drug reactions83. This new paradigm can be applied to expand PD DDI studies by utilizing rich information sources such as electronic health records, and in this way, patients potentially could be selected based on their genomic/transcriptomic/proteomic susceptibility to DDIs before combination therapies are administered.
A PD DDI study can be implemented in clinical settings with proper design and assessment methods, and this would address the problem that clinical studies are too costly to be designed based upon empirical discovery of potentially beneficial PD DDIs. Adequate evidence for clinically-significant PD DDIs should be obtained before a clinical trial. First, the hypothesis generation step for candidate combinations would be performed and hypotheses tested in silico or with in vitro screening studies. Subsequently, a detailed investigation of the mechanism(s) of suspect PD DDIs in the combination would be conducted, and PD DDI mechanisms would be represented by a QSP model that has been properly calibrated and qualified. PBPK modeling that predicts the physiological concentrations of drugs at target site(s) would be integrated with the QSP model to predict in vivo exposure-response relationships for each drug individually vs. combined. Regimens then would be optimized toward the desired outcome(s) in terms of the dose and schedule design, and “proof-of-concept” animal studies could be used for parameter calibration and model validation purposes. Translational PK/PD models, based on either PBPK or allometric relationships, could be used for projections in humans84. Eventually, the derived model could be used to generate clinical simulations that include considerations of model- and clinical uncertainties. Employing this paradigm, only one treatment combination (or a small number of combinations) that was predicted to result in significantly altered clinical endpoints compared to single-drug treatments would require clinical testing. Virtual patient populations, generated by simulation using identified covariates and proper parameter ranges, could be used to assess inter-patient variability in such QSP models applied to clinical studies85.
Summary
Pharmacodynamic DDIs are common in drug combination therapies, and mechanistic understanding of PD DDIs can be useful for the rational design of treatment regimens. There is a broad spectrum of approaches, as well as a lack of quantitative standards, for the evaluation of PD-based DDIs. Empirical methods are suitable for high-throughput screening of drug combinations, and useful for generating testable hypotheses as to combinations that may produce desired outcomes. Mechanism-based PK/PD modeling and simulation are useful for comparing hypothesized combination regimens, and have multi-scale, translational potential. QSP models that characterize complex pathophysiological systems in considerable detail have applications along the entire spectrum of drug development activities, from the prediction of possible beneficial combinations in silico, to the rational design and evaluation of preclinical animal experiments and clinical studies. Despite the infrequent evaluation of PD DDIs in clinical settings, advances in experimental and computational techniques provide precedents and opportunities for PD DDI modeling and simulation to guide the development and analysis of clinical trials, and result in improved patient response to drug therapies.
Acknowledgements
This work includes descriptions of research that was funded in part by NIH Grant CA198096. We regret that space limitations preclude the inclusion of many highly meritorious applications and examples.
Funding
This work includes descriptions of research that was funded, in part, by NIH Grant # CA198096.
Footnotes
Conflicts of Interest
The authors declared no competing interests for this work.
References
- 1.Butkiewicz M, Restrepo NA, Haines JL & Crawford DC Drug-Drug Interaction Profiles of Medication Regimens Extracted from a De-Identified Electronic Medical Records System. AMIA Jt Summits Transl Sci Proc 2016, 33–40 (2016). [PMC free article] [PubMed] [Google Scholar]
- 2.Keyomarsi K & Moran RG Folinic acid augmentation of the effects of fluoropyrimidines on murine and human leukemic cells. Cancer Res 46, 5229–5235 (1986). [PubMed] [Google Scholar]
- 3.Mignat C & Unger T ACE inhibitors. Drug interactions of clinical significance. Drug Saf 12, 334–347, doi: 10.2165/00002018-199512050-00005 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Huang SM, Temple R, Throckmorton DC & Lesko LJ Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther 81, 298–304, doi: 10.1038/sj.clpt.6100054 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Barbaro G, Scozzafava A, Mastrolorenzo A & Supuran CT Highly active antiretroviral therapy: current state of the art, new agents and their pharmacological interactions useful for improving therapeutic outcome. Curr Pharm Des 11, 1805–1843 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Jilek BL et al. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med 18, 446–451, doi: 10.1038/nm.2649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castiglione F, Pappalardo F, Bernaschi M & Motta S Optimization of HAART with genetic algorithms and agent-based models of HIV infection. Bioinformatics 23, 3350–3355, doi: 10.1093/bioinformatics/btm408 (2007). [DOI] [PubMed] [Google Scholar]
- 8.McLeod HL Clinically relevant drug-drug interactions in oncology. Br J Clin Pharmacol 45, 539–544 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J et al. Docetaxel and paclitaxel inhibit DNA-adduct formation and intracellular accumulation of cisplatin in human leukocytes. Cancer Chemother Pharmacol 37, 382–384, doi: 10.1007/s002800050401 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Kearns CM & Egorin MJ Considerations regarding the less-than-expected thrombocytopenia encountered with combination paclitaxel/carboplatin chemotherapy. Semin Oncol 24, S2-91–S92-96 (1997). [PubMed] [Google Scholar]
- 11.Ando M et al. Sequence effect of docetaxel and carboplatin on toxicity, tumor response and pharmacokinetics in non-small-cell lung cancer patients: a phase I study of two sequences. Cancer Chemother Pharmacol 55, 552–558, doi: 10.1007/s00280-004-0921-z (2005). [DOI] [PubMed] [Google Scholar]
- 12.Jiang S et al. Paclitaxel Enhances Carboplatin-DNA Adduct Formation and Cytotoxicity. Chem Res Toxicol 28, 2250–2252, doi: 10.1021/acs.chemrestox.5b00422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadavid AP Aspirin: The Mechanism of Action Revisited in the Context of Pregnancy Complications. Front Immunol 8, 261, doi: 10.3389/fimmu.2017.00261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelkonen O et al. Omics and its potential impact on R&D and regulation of complex herbal products. J Ethnopharmacol 140, 587–593, doi: 10.1016/j.jep.2012.01.035 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Shen X, Qu J, Straubinger RM & Jusko WJ Multi-Scale Network Model Supported by Proteomics for Analysis of Combined Gemcitabine and Birinapant Effects in Pancreatic Cancer Cells. CPT Pharmacometrics Syst Pharmacol 7, 549–561, doi: 10.1002/psp4.12320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X et al. Temporal Effects of Combined Birinapant and Paclitaxel on Pancreatic Cancer Cells Investigated via Large-Scale, Ion-Current-Based Quantitative Proteomics (IonStar). Mol Cell Proteomics 17, 655–671, doi: 10.1074/mcp.RA117.000519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quail DF & Joyce JA Microenvironmental regulation of tumor progression and metastasis. Nat Med 19, 1423–1437, doi: 10.1038/nm.3394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binnewies M et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550, doi: 10.1038/s41591-018-0014-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MJ et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794, doi: 10.1016/j.cell.2012.03.031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma J & Waxman DJ Combination of antiangiogenesis with chemotherapy for more effective cancer treatment. Mol Cancer Ther 7, 3670–3684, doi: 10.1158/1535-7163.MCT-08-0715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antonia SJ, Larkin J & Ascierto PA Immuno-oncology combinations: a review of clinical experience and future prospects. Clin Cancer Res 20, 6258–6268, doi: 10.1158/1078-0432.CCR-14-1457 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Proserpio V & Lonnberg T Single-cell technologies are revolutionizing the approach to rare cells. Immunol Cell Biol 94, 225–229, doi: 10.1038/icb.2015.106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou TC Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res 70, 440–446, doi: 10.1158/0008-5472.Can-09-1947 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Amur S, LaVange L, Zineh I, Buckman-Garner S & Woodcock J Biomarker Qualification: Toward a Multiple Stakeholder Framework for Biomarker Development, Regulatory Acceptance, and Utilization. Clin Pharmacol Ther 98, 34–46, doi: 10.1002/cpt.136 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Goossens N, Nakagawa S, Sun X & Hoshida Y Cancer biomarker discovery and validation. Transl Cancer Res 4, 256–269, doi: 10.3978/j.issn.2218-676X.2015.06.04 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsley CA et al. Phase II study of erlotinib (OSI-774) in patients with metastatic colorectal cancer. Br J Cancer 94, 1136–1143, doi: 10.1038/sj.bjc.6603055 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holbeck SL et al. The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity. Cancer Res 77, 3564–3576, doi: 10.1158/0008-5472.CAN-17-0489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loewe SM, H. Effect of combinations: Mathematical basis of problem. Arch. Exp. Pathol. Pharmacol 114, 313–326 (1926). [Google Scholar]
- 29.Bliss CI The Toxicity of Poisons Applied Jointly 1. Ann Appl Biol 26, 585–615, doi:doi: 10.1111/j.1744-7348.1939.tb06990.x (1939). [DOI] [Google Scholar]
- 30.Ariëns EJ & Simonis AM A molecular basis for drug action. J Pharm Pharmacol 16, 137–157, doi:doi: 10.1111/j.2042-7158.1964.tb07437.x (1964). [DOI] [PubMed] [Google Scholar]
- 31.Zhao L, Au JL & Wientjes MG Method to Assess Interactivity of Drugs with Nonparallel Concentration Effect Relationships. Curr Cancer Drug Targets 17, 735–755, doi: 10.2174/1568009617666170330154054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ariëns EJ & Simonis AM A molecular basis for drug action: The interaction of one or more drugs with different receptors. J Pharm Pharmacol 16, 289–312, doi:doi: 10.1111/j.2042-7158.1964.tb07461.x (1964). [DOI] [PubMed] [Google Scholar]
- 33.Greco WR, Bravo G & Parsons JC The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47, 331–385 (1995). [PubMed] [Google Scholar]
- 34.Chakraborty A & Jusko WJ Pharmacodynamic interaction of recombinant human interleukin-10 and prednisolone using in vitro whole blood lymphocyte proliferation. J Pharm Sci 91, 1334–1342, doi: 10.1002/jps.3000 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Earp J, Krzyzanski W, Chakraborty A, Zamacona MK & Jusko WJ Assessment of drug interactions relevant to pharmacodynamic indirect response models. J Pharmacokinet Pharmacodyn 31, 345–380 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Koch G, Schropp J & Jusko WJ Assessment of non-linear combination effect terms for drug-drug interactions. J Pharmacokinet Pharmacodyn 43, 461–479, doi: 10.1007/s10928-016-9490-0 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X, Straubinger RM & Jusko WJ Mechanism-based mathematical modeling of combined gemcitabine and birinapant in pancreatic cancer cells. J Pharmacokinet Pharmacodyn 42, 477–496, doi: 10.1007/s10928-015-9429-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanavati C & Mager DE Sequential Exposure of Bortezomib and Vorinostat is Synergistic in Multiple Myeloma Cells. Pharm Res 34, 668–679, doi: 10.1007/s11095-017-2095-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmer A, Katzir I, Dekel E, Mayo AE & Alon U Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc Nat Acad Sci 113, 10442–10447, doi: 10.1073/pnas.1606301113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molins EAG & Jusko WJ Assessment of three-drug combination pharmacodynamic interactions in pancreatic cancer cells. AAPS J. 20, 80, doi: 10.1208/s12248-018-0235-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer AC & Sorger PK Combination Cancer Therapy Can Confer Benefit via Patient-to-Patient Variability without Drug Additivity or Synergy. Cell 171, 1678–1691 e1613, doi: 10.1016/j.cell.2017.11.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehar J et al. Chemical combination effects predict connectivity in biological systems. Mol Syst Biol 3, 80, doi: 10.1038/msb4100116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berenbaum MC What is synergy? Pharmacol Rev 41, 93 (1989). [PubMed] [Google Scholar]
- 44.Catella-Lawson F et al. Cyclooxygenase inhibitors and the antiplatelet effects of aspirin. N Engl J Med 345, 1809–1817, doi: 10.1056/NEJMoa003199 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Hong Y, Gengo FM, Rainka MM, Bates VE & Mager DE Population pharmacodynamic modelling of aspirin- and Ibuprofen-induced inhibition of platelet aggregation in healthy subjects. Clin Pharmacokinet 47, 129–137, doi: 10.2165/00003088-200847020-00006 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Bulitta JB et al. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 59, 2315–2327, doi: 10.1128/AAC.04099-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yadav R, Landersdorfer CB, Nation RL, Boyce JD & Bulitta JB Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59, 2286–2298, doi: 10.1128/AAC.04379-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yadav R, Bulitta JB, Nation RL & Landersdorfer CB Optimization of Synergistic Combination Regimens against Carbapenem- and Aminoglycoside-Resistant Clinical Pseudomonas aeruginosa Isolates via Mechanism-Based Pharmacokinetic/Pharmacodynamic Modeling. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.01011-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landersdorfer CB et al. Optimization of a Meropenem-Tobramycin Combination Dosage Regimen against Hypermutable and Nonhypermutable Pseudomonas aeruginosa via Mechanism-Based Modeling and the Hollow-Fiber Infection Model. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.02055-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadav R et al. Aminoglycoside Concentrations Required for Synergy with Carbapenems against Pseudomonas aeruginosa Determined via Mechanistic Studies and Modeling. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.00722-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landersdorfer CB et al. Combating Carbapenem-Resistant Acinetobacter baumannii by an Optimized Imipenem-plus-Tobramycin Dosage Regimen: Prospective Validation via Hollow-Fiber Infection and Mathematical Modeling. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.02053-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yadav R, Bulitta JB, Wang J, Nation RL & Landersdorfer CB Evaluation of Pharmacokinetic/Pharmacodynamic Model-Based Optimized Combination Regimens against Multidrug-Resistant Pseudomonas aeruginosa in a Murine Thigh Infection Model by Using Humanized Dosing Schemes. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.01268-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rees VE et al. Meropenem Combined with Ciprofloxacin Combats Hypermutable Pseudomonas aeruginosa from Respiratory Infections of Cystic Fibrosis Patients. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.01150-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yadav R et al. Optimization and Evaluation of Piperacillin-Tobramycin Combination Dosage Regimens against Pseudomonas aeruginosa for Patients with Altered Pharmacokinetics via the Hollow-Fiber Infection Model and Mechanism-Based Modeling. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.00078-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silber HE, Jauslin PM, Frey N & Karlsson MO An integrated model for the glucose-insulin system. Basic Clin Pharmacol Toxicol 106, 189–194, doi: 10.1111/j.1742-7843.2009.00510.x (2010). [DOI] [PubMed] [Google Scholar]
- 56.Choy S, Henin E, van der Walt JS, Kjellsson MC & Karlsson MO Identification of the primary mechanism of action of an insulin secretagogue from meal test data in healthy volunteers based on an integrated glucose-insulin model. J Pharmacokinet Pharmacodyn 40, 1–10, doi: 10.1007/s10928-012-9281-1 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Schneck KB, Zhang X, Bauer R, Karlsson MO & Sinha VP Assessment of glycemic response to an oral glucokinase activator in a proof of concept study: application of a semi-mechanistic, integrated glucose-insulin-glucagon model. J Pharmacokinet Pharmacodyn 40, 67–80, doi: 10.1007/s10928-012-9287-8 (2013). [DOI] [PubMed] [Google Scholar]
- 58.de Winter W et al. A mechanism-based disease progression model for comparison of long-term effects of pioglitazone, metformin and gliclazide on disease processes underlying type 2 diabetes mellitus. J Pharmacokinet Pharmacodyn 33, 313–343, doi: 10.1007/s10928-006-9008-2 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Choy S, Kjellsson MC, Karlsson MO & de Winter W Weight-HbA1c-insulin-glucose model for describing disease progression of type 2 diabetes. CPT Pharmacometrics Syst Pharmacol 5, 11–19, doi: 10.1002/psp4.12051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hahnfeldt P, Panigrahy D, Folkman J & Hlatky L Tumor development under angiogenic signaling: a dynamical theory of tumor growth, treatment response, and postvascular dormancy. Cancer Res 59, 4770–4775 (1999). [PubMed] [Google Scholar]
- 61.Hutchinson LG et al. Modeling Longitudinal Preclinical Tumor Size Data to Identify Transient Dynamics in Tumor Response to Antiangiogenic Drugs. CPT Pharmacometrics Syst Pharmacol 5, 636–645, doi: 10.1002/psp4.12142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imbs DC et al. Revisiting Bevacizumab + Cytotoxics Scheduling Using Mathematical Modeling: Proof of Concept Study in Experimental Non-Small Cell Lung Carcinoma. CPT Pharmacometrics Syst Pharmacol 7, 42–50, doi: 10.1002/psp4.12265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meads MB, Gatenby RA & Dalton WS Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer 9, 665–674, doi: 10.1038/nrc2714 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Picco N, Sahai E, Maini PK & Anderson ARA Integrating Models to Quantify Environment-Mediated Drug Resistance. Cancer Res 77, 5409–5418, doi: 10.1158/0008-5472.Can-17-0835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Houot R, Schultz LM, Marabelle A & Kohrt H T-cell-based Immunotherapy: Adoptive Cell Transfer and Checkpoint Inhibition. Cancer Immunol Res 3, 1115–1122, doi: 10.1158/2326-6066.CIR-15-0190 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Kirschner D & Panetta JC Modeling immunotherapy of the tumor-immune interaction. J Math Biol 37, 235–252 (1998). [DOI] [PubMed] [Google Scholar]
- 67.de Pillis LG, Gu W & Radunskaya AE Mixed immunotherapy and chemotherapy of tumors: modeling, applications and biological interpretations. J Theor Biol 238, 841–862, doi: 10.1016/j.jtbi.2005.06.037 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Serre R et al. Mathematical Modeling of Cancer Immunotherapy and Its Synergy with Radiotherapy. Cancer Res 76, 4931–4940, doi: 10.1158/0008-5472.CAN-15-3567 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Wilson S & Levy D A mathematical model of the enhancement of tumor vaccine efficacy by immunotherapy. Bull Math Biol 74, 1485–1500, doi: 10.1007/s11538-012-9722-4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konstorum A, Vella AT, Adler AJ & Laubenbacher RC Addressing current challenges in cancer immunotherapy with mathematical and computational modelling. J R Soc Interface 14, doi: 10.1098/rsif.2017.0150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kosinsky Y et al. Radiation and PD-(L)1 treatment combinations: immune response and dose optimization via a predictive systems model. J Immunother Cancer 6, 17, doi: 10.1186/s40425-018-0327-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson MC & Riggs MM FDA Advisory Meeting Clinical Pharmacology Review Utilizes a Quantitative Systems Pharmacology (QSP) Model: A Watershed Moment? CPT Pharmacometrics Syst Pharmacol 4, e00020, doi: 10.1002/psp4.20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knight-Schrijver VR, Chelliah V, Cucurull-Sanchez L & Le Novere N The promises of quantitative systems pharmacology modelling for drug development. Comput Struct Biotechnol J 14, 363–370, doi: 10.1016/j.csbj.2016.09.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mager DE & Kimko HHC Systems Pharmacology and Pharmacodynamics: An Introduction. AAPS Adv Pharm Sci 23, 3–14, doi: 10.1007/978-3-319-44534-2_1 (2016). [DOI] [Google Scholar]
- 75.Hart T & Xie L Providing data science support for systems pharmacology and its implications to drug discovery. Expert Opin Drug Discov 11, 241–256, doi: 10.1517/17460441.2016.1135126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chudasama VL, Ovacik MA, Abernethy DR & Mager DE Logic-Based and Cellular Pharmacodynamic Modeling of Bortezomib Responses in U266 Human Myeloma Cells. J Pharmacol Exp Ther 354, 448–458, doi: 10.1124/jpet.115.224766 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bloomingdale P, Nguyen VA, Niu J & Mager DE Boolean network modeling in systems pharmacology. J Pharmacokinet Pharmacodyn 45, 159–180, doi: 10.1007/s10928-017-9567-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu X, Shen X, Qu J, Straubinger RM & Jusko WJ Proteomic Analysis of Combined Gemcitabine and Birinapant in Pancreatic Cancer Cells. Front Pharmacol 9, 84, doi: 10.3389/fphar.2018.00084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirouac DC How Do We “Validate” a QSP Model? CPT Pharmacometrics Syst Pharmacol 7, 547–548, doi: 10.1002/psp4.12310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Timmis J et al. Building confidence in quantitative systems pharmacology models: An engineer’s guide to exploring the rationale in model design and development. CPT Pharmacometrics Syst Pharmacol 6, 156–167, doi: 10.1002/psp4.12157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Friedrich CM A model qualification method for mechanistic physiological QSP models to support model-informed drug development. CPT Pharmacometrics Syst Pharmacol 5, 43–53, doi: 10.1002/psp4.12056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ryu JY, Kim HU & Lee SY Deep learning improves prediction of drug-drug and drug-food interactions. Proc Natl Acad Sci 115, E4304–E4311, doi: 10.1073/pnas.1803294115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Hassselt JGC & Iyengar R Systems pharmacology-based identification of pharmacogenomic determinants of adverse drug reactions using human iPSC-derived cell lines. Curr Opin Syst Biol 4, 9–15, doi: 10.1016/j.coisb.2017.05.006 (2017). [DOI] [Google Scholar]
- 84.Mager DE & Jusko WJ Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther 83, 909–912, doi:clpt200852 [pii] 10.1038/clpt.2008.52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rieger TR et al. Improving the generation and selection of virtual populations in quantitative systems pharmacology models. Prog Biophys Mol Biol, doi: 10.1016/j.pbiomolbio.2018.06.002 (2018). [DOI] [PubMed] [Google Scholar]


