Abstract
Hepatocellular carcinoma (HCC) incidence is high in The Gambia and hepatitis B virus (HBV) infection is the main cause. People coinfected with HBV and hepatitis D virus (HDV) have an even greater risk of HCC and cirrhosis. Using a new HDV quantitative microarray antibody capture (Q-MAC) assay, we evaluated the association between HDV infection and HCC or cirrhosis among participants in The Gambia Liver Cancer Study. In this case-control study, cases had HCC (n=312) or cirrhosis (n=119). Controls (n=470) had no clinical evidence of liver disease and normal serum alpha-fetoprotein. Participants were previously tested for hepatitis B surface antigen (HBsAg); we tested HBsAg+ specimens by HDV Q-MAC, western blot, and RNA assays. We evaluated separate cutoffs of the Q-MAC assay for predicting anti-HDV and RNA positivity. Q-MAC correctly identified 29/29 subjects who were western blot positive (sensitivity=100%, specificity=99.4%) and 16/17 who were RNA positive (sensitivity=94.1%, specificity=100%). Compared to controls, cases more often had HBV monoinfection (HBsAg+/HDV RNA−; 54.1% vs. 17.0%; odds ratio [OR]= 6.28; p<0.001) or HBV-HDV coinfection (HBsAg+/HDV RNA+; 3.9% vs 0%; p<0.001). Risk estimates (for HCC or cirrhosis) based on HDV antibody status and adjusted for covariates (demographics, alcohol, smoking, body mass index, anti-HCV, and aflatoxin B1 exposure) yielded consistent results for both HBV monoinfection (adjusted OR=8.29; 95% confidence interval=5.74-11.98) and HBV-HDV coinfection (adjusted OR=30.66; 95% confidence interval=6.97-134.95). In this Gambian population, HDV Q-MAC had high sensitivity and specificity for both anti-HDV and HDV RNA. HDV infection contributed to the high risk of HCC in The Gambia.
Keywords: epidemiology, hepatitis B virus, hepatitis D virus, HDV RNA, hepatocellular carcinoma
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most common cancer and fourth leading cause of cancer-associated mortality in the world.1 Hepatitis B virus (HBV) infection is the chief cause, accounting for 33% of HCC mortality worldwide.1 HBV infection can also lead to hepatic cirrhosis, which is an important risk factor for HCC.2 Globally, ~248 million individuals have chronic HBV infection as measured by the presence of hepatitis B surface antigen (HBsAg).2
Hepatitis D virus (HDV) is a defective RNA virus that requires HBsAg for its life cycle.3 HBV-HDV coinfected individuals experience more rapid progression to cirrhosis and a higher risk of HCC than HBV monoinfected individuals.4-9 HDV testing is limited by the lack of reliable and convenient antibody assays and the lack of commercial reverse transcriptase polymerase chain reaction (RT-PCR) assays to detect HDV RNA.10 A recently developed quantitative microarray antibody capture (Q-MAC) assay demonstrated excellent performance characteristics among Mongolians11 and among injection drug users from the United States.12 However, viral antibody assays may have poorer specificity in African individuals.13-18 Therefore, it is important to evaluate the HDV Q-MAC assay in African populations.
It is also important to understand the prevalence of HDV infection in sub-Saharan Africa, where the prevalence of chronic hepatitis B is especially high (≥8%)2 and HBV infection is the major cause of HCC.1, 2 In a recent review and meta-analysis, Stockdale et al. reported that HDV prevalence in sub-Saharan African countries varies widely, with localized clusters of endemicity.19 Those authors also noted the need to define the reliability of HDV testing methods in the African setting.
The Gambia is a West African country with the third highest incidence of HCC in the world.20 About 60% of HCC-associated mortality in The Gambia has been attributed to HBV infection,1, 2, 21 but the contribution of HDV infection to HCC rates in The Gambia is unknown.19 Previous analyses of data from The Gambia Liver Cancer Study provided insights into the role of HBV, hepatitis C virus (HCV) and aflatoxin B1 exposure in the risk of HCC in this country.21, 22 Now, we have utilized data from this case-control study to: 1) evaluate the performance of the HDV Q-MAC assay; and 2) investigate the role of HDV infection in the risk of HCC or cirrhosis in The Gambia.
2. MATERIALS AND METHODS
2.1. Study population:
Details of The Gambia Liver Cancer Study are provided elsewhere.21 Briefly, subjects were recruited from the liver disease referral clinics at three tertiary care hospitals from September 1997 through January 2001: Royal Victoria Hospital, Banjul; Medical Research Council Hospital, Fajara; and Bansang Hospital, Bansang. All patients with suspected liver disease underwent a standardized ultrasound examination.
Cases were patients with either (1) incident HCC confirmed by liver biopsy, or ultrasound showing one or more space-occupying lesions characteristic of HCC and a serum alpha-fetoprotein of ≥20 ng/ml, or (2) cirrhosis without HCC based on ultrasound findings that were consistent with cirrhosis in the absence of space-occupying lesions. In a previous analysis of Gambia Liver Cancer data,21 defining HCC based on higher cut-off values for serum alpha-fetoprotein (100 ng/ml or 400 ng/ml) yielded similar associations between HBV infection and HCC, compared to defining HCC based on a serum alpha-fetoprotein of ≥20 ng/ml. Hence, to increase power, we used the lower cut-off value in our study. Control subjects with no clinical evidence of liver disease and normal alpha-fetoprotein levels (<5 ng/ml) were recruited from the general medical outpatient clinics of the same hospital sites and frequency matched to cases on age (10-year groupings) and gender.21 Trained study field staff collected information on demographics, socio-economic status, and lifestyle habits from study participants through a structured interview. Blood specimens were collected, processed at the Medical Research Council serology laboratory, and stored at either −20°C for serology or −70°C for nucleic acid assays. Local and international scientific and ethical review committees approved the study protocol.21 Informed consent was obtained from each participant before inclusion in the study.
2.2. Serologic analysis:
As previously described,21 samples were tested for alpha-fetoprotein and quantified by standard radiometric assay methods (DiaSorin SA, Sallugia, Italy). HBsAg, a marker of chronic HBV infection, was tested by reverse passive hemagglutination assay (Murex Diagnostics Limited, Dartford, UK) with radioimmunoassay testing of negative samples (Sorin Biomedica Diagnostics, Vercelli, Italy). HCV status was determined by detection of anti-HCV using third generation enzyme-linked immunosorbent assay (ORTHO Clinical Diagnostics, Neckargemund, Germany) with confirmation by recombinant immunoblot assay (RIBA HCV 3.0 SIA; CHIRON, Emeryville, CA). Detailed description of the procedures for testing circulating plasma DNA for the presence of Ser-249 TP53 mutation, a marker of the effect of aflatoxin exposure, is provided elsewhere.22
For the present study, we performed HDV testing on archived specimens that were collected from participants who tested HBsAg-positive (Figure 1). These specimens were tested by the Q-MAC assay as previously described.11 In brief, the assay was constructed on plasmonic gold slides with enhanced near-infrared fluorescence detection. Using a microarray printing robot, recombinant full-length HDV small antigen was placed on the slides. Slides were blocked with fetal bovine serum (FBS), washed with phosphate-buffered saline, and 1 μL of sample (diluted to 50 μL with FBS) was applied to each well. Slides were washed with phosphate-buffered saline and IRDye800-labeled donkey antihuman IgG (diluted 1:1,000 in FBS) was applied for 1 hour followed by further washing and drying. Slides were then scanned using a Licor Odyssey instrument and the fluorescent intensity measured.
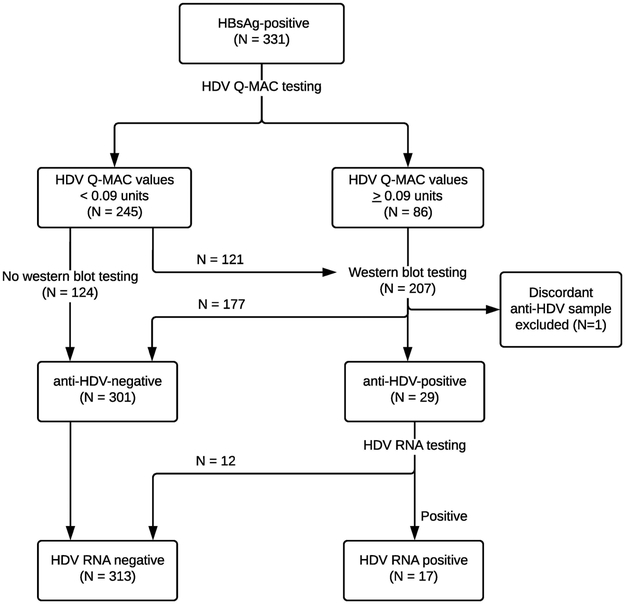
Figure 1: Flow chart to depict the HDV testing algorithm.
The flow chart shows the algorithm used to test samples for HDV in the study. All samples that were HBsAg-positive (N=331) were tested for HDV using the Q-MAC assay. Of the 245 samples that had Q-MAC values <0.09, 124 samples were not tested with western blot and were considered anti-HDV-negative, while 121 samples were tested with western blot and were all anti-HDV-negative. During replicate testing, one sample had discordant western blot result and was removed from the study, yielding 330 HBsAg-positive participants (250 cases and 80 controls) who were included in the overall analysis. Among the remaining samples that had Q-MAC values ≥0.09 units and were tested with western blot assay, 29 samples tested anti-HDV-positive, and were further tested for HDV RNA using an RT-PCR assay.
Abbreviations: HBsAg, hepatitis B surface antigen; HDV, hepatitis D virus; Q-MAC, quantitative microarray antibody capture; RNA, ribonucleic acid
Previous assessments of the performance characteristics of the Q-MAC assay among Mongolians and US injection drug users established and verified a fluorescent intensity of 0.09 units as a value that excluded a positive result by HDV western blot.11, 12 To assess that cutoff value in this Gambian population, we tested the first 121 specimens with a Q-MAC value of < 0.09 units by western blot and found that all were negative by that assay. On that basis, the remaining specimens with Q-MAC of <0.09 units (n=124) were considered negative for anti-HDV (and HDV RNA) without additional testing (Figure 1). The proportion of cases among subjects with Q-MAC values <0.09 units who were tested or not tested by western blot (76% vs. 68%; p=0.15) were similar.
Previous assessment of Q-MAC also established cut-offs of 0.164 units as positive for anti-HDV western blot and 1.659 units as positive for HDV RNA.11 To evaluate those thresholds values in the Gambians, serum samples with Q-MAC values >0.09 units (n=86) were tested with an HDV western blot assay, and those testing positive by anti-HDV western blot (n=29) were evaluated for HDV RNA levels by one-step RT-PCR (Figure 1).11 Subjects who tested negative by western blot assay were assumed to be negative for HDV RNA.
HDV RNA positive samples (n=17) were further tested for viral genotype. To determine HDV genotype, viral RNA was extracted from 140 μL of sera using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instruction. Nested RT-PCR was performed to amplify a section of the HDV genome (nt856-1275 relative to HDV reference strain JAM27), as described elsewhere.23 After purification with a QIA quick PCR Purification kit (Qiagen, Hilden, Germany), samples were sequenced using an automatic sequencer. Phylogenetic analysis of a 419 bp fragment was used to determine the HDV genotypes. All sequences obtained were subjected to an HDV BLAST search to compare them with related reference sequences in the HDV database from the Gene Bank of the National Center for Biotechnology Information.
2.3. Statistical analyses:
The performance characteristics of the Q-MAC assay were evaluated only among subjects whose samples were tested with both Q-MAC and western blot assays. Considering western blot as the gold standard, sensitivity was computed as number of subjects who tested positive by both Q-MAC (≥0.164) and western blot divided by the number of subjects who tested positive by western blot. Specificity was calculated as the number of subjects who tested negative by both Q-MAC (<0.164) and western blot divided by the number of subjects who tested negative by western blot. Similarly, we evaluated sensitivity and specificity of HDV Q-MAC for predicting an HDV RNA result among individuals tested by both Q-MAC and HDV RT-PCR (gold standard) assays, using a Q-MAC cut-off value of >1.659 units to define HDV RNA positivity.
Characteristics of cases and controls were compared using the chi-squared test. To examine disease associations, we performed separate analyses in which HDV infection status was defined based on HDV RNA positivity or anti-HDV positivity. HBsAg-negative subjects and HBsAg-positive subjects with Q-MAC of <0.09 units who did not undergo additional testing were considered negative for anti-HDV and HDV RNA. Prevalence of HBV monoinfection (HBsAg+/HDV RNA−), HBV-HDV coinfection (HBsAg+/HDV RNA+), and absence of either infection (HBsAg−/HDV RNA−) was compared among cases and controls using logistic regression. If the unadjusted odds ratios (ORs) were inestimable due to small sample size, we calculated the exact 95% confidence intervals (CIs) using Cornfield’s approximation.24 Variables that were statistically significant in univariate analyses, or otherwise clinically relevant were included in a multivariable logistic regression model and adjusted OR (aOR) estimates along with their corresponding 95% CIs were obtained. Unknown information for variables was included as a separate category to maintain the sample size. Final models were adjusted for age at enrollment, sex, study site, ethnic group, earth floor, alcohol intake, cigarette smoking, family history of cancer, body mass index (BMI), anti-HCV serostatus, and Ser-249 TP53 mutation.
We conducted some additional sensitivity analyses. To determine whether the risk of HCC or cirrhosis differs between HBV-HDV coinfected and HBV monoinfected individuals, we restricted our logistic regression analyses to people with HBV infection. We further evaluated whether HBV-HDV coinfection increases the risk of HCC or cirrhosis by fitting a polytomous logistic regression model with three possible outcomes; HCC, cirrhosis without HCC, and neither (controls: reference group). Furthermore, to determine whether the risk of HCC differed from that of cirrhosis, we conducted a case-case comparison using logistic regression. All tests were two-sided and p values < 0.05 were statistically significant.
3. RESULTS
3.1. HDV Q-MAC performance:
Of 331 HBsAg-positive participants, testing by HDV western blot was performed in all 86 with a Q-MAC result of >0.09 units plus 121 of the individuals with a Q-MAC result below that threshold (Figure 1). Replicate testing was performed on 41 of these 207 samples. One of the 41 replicated specimens yielded a discordant western blot result. That individual was excluded from further analysis, yielding 206 individuals who were included in the evaluation of the Q-MAC assay and 330 HBsAg-positive participants (250 cases and 80 controls) who were included in the overall analysis. Individuals whose samples were tested in duplicate did not differ from other subjects with regard to median Q-MAC values (0.18 vs. 0.15 units; p=0.99), case-control status (duplicate testing in 14.8% controls and 11.4% cases; p=0.20), or anti-HDV status (duplicate testing in 12.6% anti-HDV negative and 10.3% anti-HDV positive individuals; p=0.50).
All 29 individuals who tested positive by western blot, also tested positive for anti-HDV by Q-MAC assay, yielding a sensitivity of 100% (Figure 2). Among the 177 individuals who tested negative by HDV western blot, one was considered positive for anti-HDV by Q-MAC assay (Q-MAC value of 0.165), yielding a specificity of 99.4%.
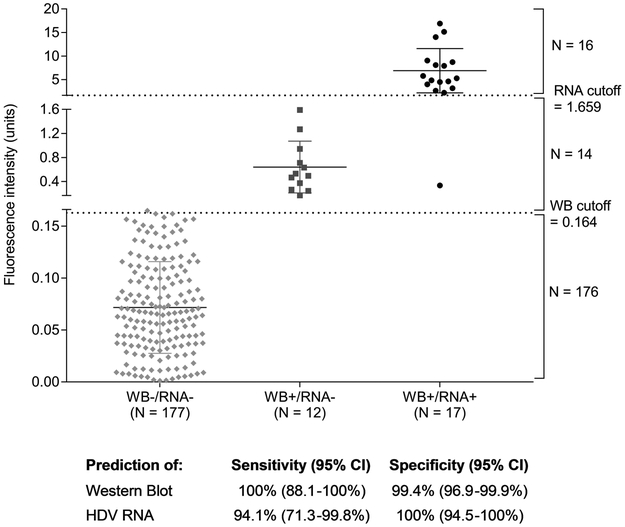
Figure 2: Performance of the HDV Q-MAC assay compared to the western blot and HDV RNA assays.
The figure plots the fluorescence intensity derived from Q-MAC assay against the findings for the same samples from western blot and HDV RNA assays. The one sample with discordant result was removed from all analyses. Of the remaining 206 samples tested, 17 were positive for both western blot and HDV RNA, 12 were positive for western blot but negative for HDV RNA, and 177 were both western blot and HDV RNA-negative. There was 100% concordance between the western blot and Q-MAC results using the proposed Q-MAC assay cutoff of 0.164 units for anti-HDV positivity 11. Of the 17 HDV RNA-positive samples, 16 exceeded 1.659 fluorescence intensity units in the Q-MAC assay, the proposed cutoff for predicting HDV RNA positivity. The sensitivity and specificity for the comparisons are provided at the bottom of the figure.
Abbreviations: HDV, hepatitis D virus; NPV, negative predictive value; PPV, positive predictive value; Q-MAC, quantitative microarray antibody capture; RNA, ribonucleic acid
Seventeen subjects tested positive for HDV RNA by RT-PCR (Figure 2); based on the Q-MAC threshold of 1.659 units for HDV RNA positivity,16 of the 17 were classified as HDV RNA positive by Q-MAC, yielding a sensitivity of 94.1%. All 12 individuals who were classified as negative for HDV RNA by Q-MAC also tested negative for HDV RNA by RT-PCR, thus yielding a specificity of 100%. Twelve subjects who tested positive for anti-HDV by western blot and negative for HDV RNA by the RT-PCR assay were correctly classified as being anti-HDV positive and HDV RNA negative by HDV Q-MAC (Figure 2).
3.2. HDV genotype:
Twelve participants had genotype 5 infection (70.6%) and 5 had genotype 1 infection (29.4%). The participant who was excluded due to discordant western blot results also had genotype 1 infection.
3.3. Study population:
We included 431 cases (HCC, n=312; cirrhosis without HCC, n=119), and 470 controls in our analysis, after exclusion of the discordant sample. The comparison of characteristics between cases and controls and prevalence of HBsAg-positivity among controls are presented in Table 1. Despite the attempt to frequency-match by age and sex, cases and controls differed in the distributions of these variables, hence, these variables were included in the multivariable logistic regression analyses. Cases compared to controls were more likely to be older (age ≥60 years, 23.9% vs. 19.6%), men (76.3% vs. 70.0%), recruited from the Royal Victoria Hospital (42.2% vs. 27.7%), and belonging to Fula or Wollof ethnic groups (48.8% vs. 35.8%). Cases more often belonged to a lower socio-economic status than controls, as evidenced by the higher proportion who never attended school (21.1% vs. 11.0%) or had earth floor houses (58.2% vs. 49.6%). More cases than controls were former smokers (33.0% vs. 22.8%), had a family history of cancer (8.8% vs. 3.6%), and had a BMI <18 kg/m2 (30.4% vs. 22.4%). There was no significant difference between cases and controls with respect to alcohol consumption. The prevalence of HBsAg-positivity (58.0% vs. 17.0%), anti-HCV positivity (12.3% vs. 3.0%) and Ser-249 TP53 mutation (18.8% vs. 3.6%) was significantly higher among cases than controls. Comparison of characteristics of people with cirrhosis and HCC are presented in Table 2.
Table 1:
Characteristics of the study population.
| Characteristics | Cases (N=431) N (%) |
Controls (N=470) N (%) |
p value | HBsAg prevalence among controls (%) |
|---|---|---|---|---|
| Age, years | ||||
| < 35 | 101 (23.4) | 151 (32.1) | 0.01 | 23.8 |
| 35 - 49 | 145 (33.6) | 131 (27.9) | 16.0 | |
| 50 - 59 | 82 (19.1) | 96 (20.4) | 14.6 | |
| ≥ 60 | 103 (23.9) | 92 (19.6) | 9.8 | |
| Sex | ||||
| Males | 329 (76.3) | 329 (70.0) | 0.03 | 19.8 |
| Females | 102 (23.7) | 141 (20.0) | 10.6 | |
| Study site | ||||
| RVH | 182 (42.2) | 130 (27.7) | < 0.001 | 18.5 |
| MRC | 124 (28.8) | 126 (26.8) | 23.8 | |
| BSG | 125 (29.0) | 214 (45.5) | 12.1 | |
| Ethnic group | ||||
| Madinka | 111 (26.2) | 156 (33.2) | 0.003 | 16.7 |
| Fula | 114 (27.0) | 93 (19.8) | 12.9 | |
| Wollof | 92 (21.8) | 75 (16.0) | 17.3 | |
| Others/unknown | 114 (26.5) | 146 (31.0) | 19.9 | |
| Attended school | ||||
| Never | 91 (21.1) | 52 (11.0) | < 0.001 | 13.5 |
| Ever | 332 (77.0) | 412 (87.7) | 16.8 | |
| Unknown | 8 (1.9) | 6 (1.3) | ||
| Earth floor house | ||||
| No | 169 (39.2) | 231 (49.2) | 0.004 | 16.7 |
| Yes | 251 (58.2) | 233 (49.6) | 16.0 | |
| Unknown | 11 (2.6) | 6 (1.3) | ||
| Alcohol intake | ||||
| Never | 364 (84.5) | 418 (88.9) | 0.14 | 16.5 |
| Ever | 50 (11.6) | 39 (8.3) | 18.0 | |
| Unknown | 17 (3.9) | 13 (2.8) | ||
| Cigarette smoking | ||||
| Never smoker | 205 (47.6) | 277 (58.9) | 0.001 | 14.4 |
| Former smoker | 142 (33.0) | 107 (22.8) | 23.4 | |
| Current smoker | 70 (16.2) | 77 (16.4) | 14.3 | |
| Unknown | 14 (3.2) | 9 (1.9) | ||
| Family history of cancer | ||||
| No | 385 (89.3) | 444 (94.5) | 0.005 | 16.0 |
| Yes | 38 (8.8) | 17 (3.6) | 23.5 | |
| Unknown | 8 (1.9) | 9 (1.9) | ||
| Marital status | ||||
| Never married | 52 (12.1) | 62 (13.2) | 0.81 | 27.4 |
| Ever married | 371 (86.1) | 401 (85.3) | 14.2 | |
| Unknown | 8 (1.7) | 7 (1.5) | ||
| Body mass index, kg/m2 | ||||
| < 18.5 | 131 (30.4) | 105 (22.4) | < 0.001 | 17.1 |
| ≥ 18.5 | 144 (33.4) | 285 (60.6) | 16.1 | |
| Unknown | 156 (36.2) | 80 (17.0) | ||
| anti-HCV | ||||
| Negative | 329 (76.3) | 429 (91.3) | < 0.001 | 17.3 |
| Positive | 53 (12.3) | 14 (3.0) | 7.1 | |
| Unknown | 49 (11.4) | 27 (5.7) | ||
| Hepatitis B surface antigen | ||||
| Negative | 181 (42.0) | 390 (83.0) | < 0.001 | … |
| Positive | 250 (58.0) | 80 (17.0) | … | |
| Ser-249 TP53 mutation | ||||
| Absent | 182 (42.2) | 332 (70.6) | < 0.001 | 14.2 |
| Present | 81 (18.8) | 17 (3.6) | 11.8 | |
| Unknown | 168 (39.0) | 121 (25.7) |
Abbreviations: HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; RVH, Royal Victoria Hospital, Banjul; MRC, Medical Research Council Hospital, Fajara; BSG, Bansang Hospital, Bansang
Table 2:
Characteristics of people with cirrhosis or hepatocellular carcinoma.
| Characteristics | HCC cases (N=312) N (%) |
Cirrhosis cases (N=119) N (%) |
p value |
|---|---|---|---|
| Age, years | 0.05 | ||
| < 35 | 65 (20.8) | 36 (30.2) | |
| 35 - 49 | 101 (32.4) | 44 (37.0) | |
| 50 - 59 | 65 (20.8) | 17 (14.3) | |
| ≥ 60 | 81 (26.0) | 22 (18.5) | |
| Sex | 0.001 | ||
| Males | 251 (80.5) | 78 (65.6) | |
| Females | 61 (19.5) | 41 (34.4) | |
| Study site | 0.11 | ||
| RVH | 124 (39.7) | 58 (48.7) | |
| MRC | 98 (31.4) | 26 (21.9) | |
| BSG | 90 (28.9) | 26 (29.4) | |
| Ethnic group | 0.60 | ||
| Madinka | 83 (26.6) | 28 (23.5) | |
| Fula | 78 (25.0) | 36 (30.3) | |
| Wollof | 65 (20.8) | 27 (22.7) | |
| Others/unknown | 86 (27.6) | 28 (23.5) | |
| Attended school | 0.98 | ||
| Never | 66 (21.2) | 25 (21.0) | |
| Ever | 240 (76.9) | 92 (77.3) | |
| Unknown | 6 (1.9) | 2 (1.7) | |
| Earth floor house | 0.55 | ||
| No | 126 (40.4) | 43 (36.1) | |
| Yes | 177 (56.7) | 74 (62.2) | |
| Unknown | 9 (2.9) | 2 (1.7) | |
| Alcohol intake | 0.48 | ||
| Never | 260 (83.3) | 104 (87.4) | |
| Ever | 40 (12.8) | 10 (8.4) | |
| Unknown | 12 (3.9) | 5 (4.2) | |
| Cigarette smoking | 0.06 | ||
| Never smoker | 136 (43.6) | 69 (58.0) | |
| Former smoker | 110 (35.3) | 32 (26.9) | |
| Current smoker | 54 (17.3) | 16 (13.5) | |
| Unknown | 12 (3.9) | 2 (1.7) | |
| Family history of cancer | 0.66 | ||
| No | 276 (88.5) | 109 (91.6) | |
| Yes | 30 (9.6) | 8 (6.7) | |
| Unknown | 6 (1.9) | 2 (1.7) | |
| Marital status | 0.45 | ||
| Never married | 34 (10.9) | 18 (15.1) | |
| Ever married | 272 (87.2) | 99 (83.2) | |
| Unknown | 6 (1.9) | 2 (1.7) | |
| Body mass index, kg/m2 | < 0.001 | ||
| < 18.5 | 111 (35.6) | 20 (16.8) | |
| ≥ 18.5 | 82 (26.3) | 62 (52.1) | |
| Unknown | 119 (38.1) | 37 (31.1) | |
| anti-HCV | |||
| Negative | 230 (73.7) | 99 (83.2) | 0.04 |
| Positive | 46 (14.7) | 7 (5.9) | |
| Unknown | 36 (11.5) | 13 (10.9) | |
| Ser-249 TP53 mutation | < 0.001 | ||
| Absent | 98 (31.4) | 84 (70.6) | |
| Present | 66 (21.2) | 15 (12.6) | |
| Unknown | 148 (47.4) | 20 (16.8) | |
| HDV status (based on HDV RNA positivity) | 0.65 | ||
| HBsAg− / HDV RNA− | 129 (41.4) | 52 (43.7) | |
| HBsAg+ / HDV RNA− | 172 (55.1) | 61 (51.3) | |
| HBsAg+ / HDV RNA+ | 11 (3.5) | 6 (5.0) | |
| HDV status (based on anti-HDV positivity) | 0.66 | ||
| HBsAg− / anti-HDV− | 129 (41.4) | 52 (43.7) | |
| HBsAg+ / anti-HDV− | 165 (52.9) | 58 (48.7) | |
| HBsAg+ / anti-HDV+ | 18 (5.8) | 9 (7.6) |
Abbreviations: HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; HDV, hepatitis D virus; RVH, Royal Victoria Hospital, Banjul; MRC, Medical Research Council Hospital, Fajara; BSG, Bansang Hospital, Bansang
Among controls, a higher prevalence of HBsAg-positivity was observed for young participants, males, those recruited at the Medical Research Council hospital, ever alcohol users, former smokers, people with a family history of cancer, those who were never married, and participants who were anti-HCV seronegative (Table 1).
3.4. Association between HDV infection and HCC or cirrhosis:
More cases than controls had evidence of HBV monoinfection (54.1% vs. 17.0%; p<0.001) and HBV-HDV coinfection (HDV RNA positivity: 3.9% vs. 0.0%; p<0.001; Table 3). None of the control participants who tested positive for HBsAg (n=80) tested positive for HDV RNA. In unadjusted logistic regression analysis, participants with HBV monoinfection were 6.28 times more, and those with HBV-HDV coinfection were infinitely more likely to have HCC or cirrhosis (without HCC) than those without HBV infection (Table 3). These effect sizes persisted after adjusting for potential confounding variables in analyses of HBV monoinfection (aOR=8.35; 95%CI=5.80-12.03) or HBV-HDV coinfection (aOR could not be determined). Similar results were obtained when HDV infection status was determined by anti-HDV positivity (Table 3).
Table 3:
Association between HBV and HDV infection with hepatocellular carcinoma or cirrhosis.
| HBV and HDV status |
Cases N (%) |
Controls N (%) |
Unadjusted OR (95% CI) |
Adjusted OR (95% CI) † |
|---|---|---|---|---|
| HDV status determined by HDV RNA positivity | ||||
| HBsAg− / HDV RNA− | 181 (42.0) | 390 (83.0) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / HDV RNA− | 233 (54.1) | 80 (17.0) | 6.28 (4.61, 8.55) | 8.35 (5.80, 12.03) |
| HBsAg+ / HDV RNA+ | 17 (3.9) | 0 (0.0) | − (9.49, −) ‡ | − |
| HDV status determined by anti-HDV positivity | ||||
| HBsAg− / anti-HDV− | 181 (42.0) | 390 (83.0) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / anti-HDV− | 223 (51.7) | 78 (16.6) | 6.16 (4.51, 8.42) | 8.29 (5.74, 11.98) |
| HBsAg+ / anti-HDV+ | 27 (6.3) | 2 (0.4) | 29.09 (6.84, 123.65) | 30.66 (6.97, 134.95) |
Abbreviations: CI, confidence intervals; HBsAg, hepatitis B surface antigen; HDV, hepatitis D virus; OR, odds ratio
Cannot be estimated
Adjusted for age, sex, study site, ethnic group, earth floor, alcohol intake, cigarette smoking, family history of cancer, body mass index, anti-HCV, and Ser-249 TP53 mutation
Exact 95% confidence intervals were calculated for the unadjusted OR and calculation of the confidence intervals for the adjusted OR was not possible.
Participants who had HBV-HDV coinfection had a higher risk of HCC or cirrhosis compared to those who had HBV monoinfection (unadjusted OR= -; 95%CI=1.51, -). When HDV infection status was determined by anti-HDV positivity, risk of HCC or cirrhosis was significant in unadjusted (OR=4.72; 95% CI=1.10-20.32) and borderline significant in the adjusted analyses (aOR=4.00; 95% CI=0.90-17.76) (Data not shown in Tables).
Results of analyses to evaluate whether the risk of HCC differs from cirrhosis without HCC are presented in Table 4. In unadjusted models, HBV monoinfected individuals had a higher risk of cirrhosis (without HCC) (OR=5.72; 95%CI=3.68-8.89), and HCC (OR=6.50; 95%CI, 4.67-9.06) compared to HBV uninfected individuals. HBV-HDV coinfected individuals also had a higher risk of cirrhosis (OR= -; 95%CI=11.49, -) and HCC (OR= -; 95%CI=8.59, -) compared to HBV uninfected people. These risk estimates remained significant after adjusting for potential confounders, although the aORs for HBV-HDV coinfection were not estimable because no control subject had HBV-HDV coinfection. In the case-case comparison, there was no significant difference between the risk of HCC and cirrhosis (without HCC) for both HBV monoinfected and HBV-HDV coinfected individuals. Comparable OR estimates were obtained when anti-HDV positivity was used to ascertain HDV infection status.
Table 4:
Association between HBV and HDV infection with hepatocellular carcinoma or cirrhosis.
| HBV and HDV status |
HCC vs. Controls† | Cirrhosis without HCC vs. Controls† |
Cirrhosis without HCC vs. HCC‡ |
|---|---|---|---|
| OR (95% CI) | OR (95% CI) | OR (95% CI) | |
| HDV status determined by HDV RNA positivity | |||
| Unadjusted models | |||
| HBsAg− / HDV RNA− | 1.0 (Reference) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / HDV RNA− | 6.50 (4.67, 9.06) | 5.72 (3.68, 8.89) | 0.88 (0.57, 1.36) |
| HBsAg+ / HDV RNA+ | − (8.59, −) | − (11.49, −) | 1.35 (0.48, 3.85) |
| Adjusted models¶ | |||
| HBsAg− / HDV RNA− | 1.0 (Reference) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / HDV RNA− | 9.11 (6.09, 13.61) | 7.36 (4.49, 12.07) | 0.64 (0.37, 1.11) |
| HBsAg+ / HDV RNA+ | − | − | 0.70 (0.22, 2.24) |
| HDV status determined by anti-HDV positivity | |||
| Unadjusted models | |||
| HBsAg− / anti-HDV − | 1.0 (Reference) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / anti-HDV − | 6.40 (4.58, 8.94) | 5.58 (3.57, 8.71) | 0.87 (0.56, 1.35) |
| HBsAg+ / anti-HDV+ | 27.21 (6.23, 118.86) | 33.75 (7.10, 160.49) | 1.24 (0.52, 2.94) |
| Adjusted models¶ | |||
| HBsAg− / anti-HDV− | 1.0 (Reference) | 1.0 (Reference) | 1.0 (Reference) |
| HBsAg+ / anti-HDV− | 9.09 (6.06, 13.63) | 7.22 (4.38, 11.90) | 0.63 (0.36, 1.09) |
| HBsAg+ / anti-HDV+ | 31.25 (6.81, 143.36) | 31.31 (6.32, 155.10) | 0.80 (0.30, 2.14) |
Abbreviations: CI, confidence intervals; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HDV, hepatitis D virus; OR, odds ratio
Cannot be estimated
OR estimates derived from polytomous logistic regression model comparing HCC cases and cirrhosis without HCC cases to controls.
OR estimates derived from logistic regression models comparing cirrhosis without HCC cases and HCC cases
Adjusted for age, sex, ethnicity, study site, earth floor, alcohol abuse, cigarette smoking, family history of cancer, HCV antibody, Ser-249 TP53 mutation, and BMI
4. DISCUSSION
HBV infection is endemic in The Gambia and ~60% of HCC cases have been attributed to that infection.21 We now show that HDV infection also contributes to the high risk of cirrhosis or HCC in this population. To our knowledge, this is the first report on the role of HDV in cirrhosis or HCC risk in The Gambia, a country that ranks among the top three in the world for liver cancer incidence.20 HDV prevalence was low among the 80 HBsAg-positive controls, as none were positive for HDV RNA and only two (2.5%) were positive for anti-HDV. Our controls were selected from hospital clinics and, therefore, may not be representative of the general population. However, the 2.5% prevalence of HDV in our HBsAg-positive controls is quite close to the 2.0% prevalence among 394 HBsAg-positive Gambians during community testing in 2011-2014.25
HDV has been reported to increase the risk of HCC compared to HBV monoinfected individuals,4-6, 8 but the evidence for a causal role of HDV in HCC development has not been considered sufficient to define this virus as a human carcinogen.26 Consistent with previous reports, HBV-HDV coinfected Gambians had a highly increased risk of HCC or cirrhosis (without HCC) compared to uninfected or HBV monoinfected people. We found no evidence that HBV-HDV coinfection had a greater effect on the risk of HCC compared to that for cirrhosis alone, suggesting that chronic HDV infection leads to HCC by enhancing hepatic inflammation and injury, and thereby accelerating the progression of fibrosis.
It is possible to estimate the proportion of cases of HCC or cirrhosis attributable to HDV infection in The Gambia (i.e., the population attributable fraction) based on the formula Pd*(OR-1/OR), where ‘ Pd’ is the proportion of cases with HDV infection.27 Because the observed odds ratios are very high, the population attributable fraction for HCC or cirrhosis effectively equals the prevalence of HDV among the cases. Therefore, based on our results, we estimate that ~4-6% of HCC or cirrhosis cases in The Gambia can be attributed to HDV infection.
In this study, we evaluated the HDV Q-MAC assay for predicting positivity for both anti-HDV and HDV RNA, with separate cut-off values for each parameter.11 Previous studies have shown that some assays for viral antibodies have poor specificity in African populations.13-15 However, compared to HDV western blot, Q-MAC had 99.4% specificity (and 100% sensitivity) in this West African population. An unusual feature of HDV Q-MAC is that, by applying a higher cutoff value than for antibody testing, the assay also may predict HDV RNA positivity.11, 12 The present results support that contention, as test characteristics for HDV RNA (sensitivity=94.1%; specificity=100%) were very good. Results from the three studies of HDV Q-MAC to date suggest it may be possible to employ an HDV testing algorithm in which Q-MAC values <0.164 are considered negative for both anti-HDV and HDV RNA; values >1.659 are considered positive for anti-HDV and HDV RNA; and values of 0.164-1.659 are considered positive for anti-HDV, but indeterminate for HDV RNA. This approach could improve efficiency and reduce costs by limiting PCR testing to fewer individuals than an algorithm in which all individuals with antibodies to HDV are tested for HDV RNA.
In the present study, HDV RNA was not detected in plasma of 12/29 (41.4%) subjects who tested anti-HDV positive by Q-MAC and western blot, which is a higher proportion than we have observed in other populations.11, 12 Several possible explanations for this result should be considered. Samples that are anti-HDV antibody positive, but negative for HDV RNA could represent false-positive antibody results, although that seems unlikely given the consistent results seen for testing by Q-MAC and western blot. HBV-HDV coinfection may be followed by HDV RNA clearance,28 and perhaps that occurred more frequently in this Gambian population than in the Mongolian and US populations that we examined previously. Finally, the samples for this study were collected under field conditions in a developing country, transported to the United States and stored for up to 20 years. Although samples were promptly processed and maintained at −70° C, HDV RNA may have degraded in some specimens.
Estimating the prevalence of HDV infection by traditional antibody-based assays may be complicated by high false-positive rates that have been observed in the past. Due to high costs associated with RNA testing, prevalence estimates for infections such as HCV and HDV have often been limited to enzyme-linked immunosorbent assay-based antibody tests, and confirmation of positive results with RNA testing by RT-PCR. Due to high prevalence of other infectious diseases such as malaria, syphilis, or HIV in sub-Saharan African countries, it has been hypothesized that cross-reacting antibodies contribute to a high frequency of false-positive anti-HCV results.14, 15 Such cross-reactivity would inflate prevalence estimates. Studies in various sub-Saharan African and other developing countries are needed to determine the performance of the Q-MAC and other anti-HDV tests.
In the future, antiviral therapy for HDV could help reduce the risk of chronic liver disease and HCC. Current treatment for HDV infection depends on interferon-based therapies with a viral response rate of less than 30%.29 The recent development of Myrcludex-B, which inhibits HDV entry into hepatocytes by blocking the binding of HBsAg to the entry receptor,30 and lonafarnib, which inhibits prenylation of HDV antigen,31 are encouraging. The possibility of more effective treatment highlights the need to identify HDV-infected individuals so that they receive appropriate medical care.
Because HDV requires HBV for its life cycle, an increase in HBV vaccination coverage should decrease the prevalence of HDV infection. The Gambia Hepatitis Intervention Study is a collaboration of the International Agency for Research in Cancer, the Government of the Republic of The Gambia, and the Medical Research Council of the United Kingdom.32 This effort was launched in 1986 to evaluate the effectiveness of childhood HBV vaccination on HCC incidence in The Gambia; since 1990 HBV vaccine has been offered to all newborns in The Gambia.32 Childhood HBV vaccination in that era did not affect our findings, as our study was conducted among adults from 1997 to 2001. However, given that HCC incidence among Gambians begins to peak at around 25-30 years of age,32 HBV vaccination coverage initiated in 1990 might begin to impact HCC incidence by 2015. Despite that important public health advance, many older Gambians with chronic hepatitis B remain at risk for HCC, particularly if they are coinfected with HDV. Early identification of such individuals and initiation of therapy to control HBV replication could further reduce the high burden of HCC in this country.
Our study had some limitations. The Gambia Liver Cancer Study was conducted during 1997-2001 and, therefore, may not reflect the current contribution of HDV infection to HCC in The Gambia. Previous analyses revealed that horizontal transmission from close intrafamilial and household contacts at a young age may be responsible for a majority of chronic HBV infection cases in The Gambia,21 but the prevalence of HDV infection among our controls (representatives of the general population) was too low for a meaningful analysis of the risk factors for acquiring HDV. The number of individuals with HBV-HDV coinfection was too few to provide precise OR estimates, as reflected in the wide 95% CI for that group. In evaluating the test characteristics of Q-MAC for predicting western blot results in this population, we excluded 124 individuals with Q-MAC values <0.09 whose specimens did not undergo western blot testing. Based on our prior work11, 12 and results among the 121 Gambia Liver Cancer Study specimens with Q-MAC values <0.09 that we did test, it is highly likely that all of these individuals would have tested negative by anti-HDV western blot. However, given that our calculated specificity estimate was 99.4%, it is unlikely that testing those additional subjects western blot would have altered our specificity estimate to a meaningful degree.
In conclusion, HDV infection was present in this Gambian population and HBV-HDV coinfection greatly increased the risk of HCC or cirrhosis. Additional studies are needed to assess the contribution of HDV infection to HCC risk elsewhere in sub-Saharan Africa and to determine risk factors for acquiring HDV infection in this region.
Acknowledgments
Funding source: This research was supported by the Intramural Research Program of the National Institutes of Health (National Cancer Institute, Division of Cancer Epidemiology and Genetics). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations:
- aOR
adjusted odds ratio
- BMI
body mass index
- CI
confidence interval
- FBS
fetal bovine serum
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HDV
hepatitis D virus
- OR
odds ratio
- PPV
positive predictive value
- Q-MAC
quantitative microarray antibody capture
- RNA
ribonucleic acid
- RT-PCR
reverse transcriptase polymerase chain reaction
Footnotes
Conflict of interest: JSG has an equity interest in, and is a board member of Eiger BioPharmaceuticals, Inc. All other authors have no conflicts of interest to declare.
REFERENCES
- 1.Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, Artaman A, Ayele TA, Barac A, et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015;386: 1546–55. [DOI] [PubMed] [Google Scholar]
- 3.Lempp FA, Ni Y, Urban S. Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat Rev Gastroenterol Hepatol 2016. [DOI] [PubMed] [Google Scholar]
- 4.Fattovich G, Boscaro S, Noventa F, Pornaro E, Stenico D, Alberti A, Ruol A, Realdi G. Influence of hepatitis delta virus infection on progression to cirrhosis in chronic hepatitis type B. J Infect Dis 1987;155: 931–5. [DOI] [PubMed] [Google Scholar]
- 5.Wedemeyer H, Manns MP. Epidemiology, pathogenesis and management of hepatitis D: update and challenges ahead. Nat Rev Gastroenterol Hepatol 2010;7: 31–40. [DOI] [PubMed] [Google Scholar]
- 6.Ji J, Sundquist K, Sundquist J. A population-based study of hepatitis D virus as potential risk factor for hepatocellular carcinoma. J Natl Cancer Inst 2012;104: 790–2. [DOI] [PubMed] [Google Scholar]
- 7.Beguelin C, Moradpour D, Sahli R, Suter-Riniker F, Luthi A, Cavassini M, Gunthard HF, Battegay M, Bernasconi E, Schmid P, Calmy A, Braun DL, et al. Hepatitis delta-associated mortality in HIV/HBV-coinfected patients. J Hepatol 2017;66: 297–303. [DOI] [PubMed] [Google Scholar]
- 8.Abbas Z, Abbas M, Abbas S, Shazi L. Hepatitis D and hepatocellular carcinoma. World J Hepatol 2015;7: 777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaquet A, Wandeler G, Nouaman M, Ekouevi DK, Tine J, Patassi A, Coffie PA, Tanon A, Seydi M, Attia A, Dabis F. Alcohol use, viral hepatitis and liver fibrosis among HIV-positive persons in West Africa: a cross-sectional study. J Int AIDS Soc 2017;19: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Gal F, Brichler S, Sahli R, Chevret S, Gordien E. First international external quality assessment for hepatitis delta virus RNA quantification in plasma. Hepatology 2016;64: 1483–94. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Oidovsambuu O, Liu P, Grosely R, Elazar M, Winn VD, Fram B, Boa Z, Dai H, Dashtseren B, Yagaanbuyant D, Genden Z, et al. A novel quantitative microarray antibody capture assay identifies an extremely high hepatitis delta virus prevalence among hepatitis B virus-infected mongolians. Hepatology 2017;66: 1739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahale P, Aka PV, Chen XH, Liu P, Fram BJ, Wang AS, Simenel S, Tseng FC, Chen S, Edlin BR, Glenn JS, O’Brien TR. Hepatitis D Viremia Among Injection Drug Users in San Francisco. J Infect Dis 2018;217: 1902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tess BH, Levin A, Brubaker G, Shao J, Drummond JE, Alter HJ, O’Brien TR. Seroprevalence of hepatitis C virus in the general population of northwest Tanzania. Am J Trop Med Hyg 2000;62: 138–41. [DOI] [PubMed] [Google Scholar]
- 14.Mullis CE, Laeyendecker O, Reynolds SJ, Ocama P, Quinn J, Boaz I, Gray RH, Kirk GD, Thomas DL, Quinn TC, Stabinski L. High frequency of false-positive hepatitis C virus enzyme-linked immunosorbent assay in Rakai, Uganda. Clin Infect Dis 2013;57: 1747–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seremba E, Ocama P, Opio CK, Kagimu M, Thomas DL, Yuan HJ, Attar N, Lee WM. Poor performance of hepatitis C antibody tests in hospital patients in Uganda. J Med Virol 2010;82: 1371–8. [DOI] [PubMed] [Google Scholar]
- 16.Mbopi-Keou FX, Ndjoyi-Mbiguino A, Talla F, Pere H, Kebe K, Matta M, Sosso MA, Belec L. Association of inconclusive sera for human immunodeficiency virus infection with malaria and Epstein-Barr virus infection in Central Africa. J Clin Microbiol 2014;52: 660–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasasira AF, Dorsey G, Kamya MR, Havlir D, Kiggundu M, Rosenthal PJ, Charlebois ED. False-positive results of enzyme immunoassays for human immunodeficiency virus in patients with uncomplicated malaria. J Clin Microbiol 2006;44: 3021–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wandeler G, Coffie PA, Kuniholm MH, Ocama P, Egger M. Issues with measuring hepatitis prevalence in resource-limited settings. Lancet 2018;391: 835–6. [DOI] [PubMed] [Google Scholar]
- 19.Stockdale AJ, Chaponda M, Beloukas A, Phillips RO, Matthews PC, Papadimitropoulos A, King S, Bonnett L, Geretti AM. Prevalence of hepatitis D virus infection in sub-Saharan Africa: a systematic review and meta-analysis. Lancet Glob Health 2017;5: e992–e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 01/03/2018 [Google Scholar]
- 21.Kirk GD, Lesi OA, Mendy M, Akano AO, Sam O, Goedert JJ, Hainaut P, Hall AJ, Whittle H, Montesano R. The Gambia Liver Cancer Study: Infection with hepatitis B and C and the risk of hepatocellular carcinoma in West Africa. Hepatology 2004;39: 211–9. [DOI] [PubMed] [Google Scholar]
- 22.Szymanska K, Lesi OA, Kirk GD, Sam O, Taniere P, Scoazec JY, Mendy M, Friesen MD, Whittle H, Montesano R, Hainaut P. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer 2004;110: 374–9. [DOI] [PubMed] [Google Scholar]
- 23.Lin HH, Lee SS, Yu ML, Chang TT, Su CW, Hu BS, Chen YS, Huang CK, Lai CH, Lin JN, Wu JC. Changing hepatitis D virus epidemiology in a hepatitis B virus endemic area with a national vaccination program. Hepatology 2015;61: 1870–9. [DOI] [PubMed] [Google Scholar]
- 24.Miettinen O Estimability and estimation in case-referent studies. Am J Epidemiol 1976;103: 226–35. [DOI] [PubMed] [Google Scholar]
- 25.Lemoine M, Shimakawa Y, Njie R, Taal M, Ndow G, Chemin I, Ghosh S, Njai HF, Jeng A, Sow A, Toure-Kane C, Mboup S, et al. Acceptability and feasibility of a screen-and-treat programme for hepatitis B virus infection in The Gambia: the Prevention of Liver Fibrosis and Cancer in Africa (PROLIFICA) study. Lancet Glob Health 2016;4: e559–67. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization, International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans, 1994. [Google Scholar]
- 27.Rockhill B, Newman B, Weinberg C. Use and misuse of population attributable fractions. Am J Public Health 1998;88: 15–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamili S, Drobeniuc J, Mixson-Hayden T, Kodani M. Delta hepatitis: Toward improved diagnostics. Hepatology 2017;66: 1716–8. [DOI] [PubMed] [Google Scholar]
- 29.Rizzetto M Targeting Hepatitis D. Semin Liver Dis 2018;38: 66–72. [DOI] [PubMed] [Google Scholar]
- 30.Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M, Schwab M, Haefeli WE, et al. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: First results of a phase Ib/IIa study. J Hepatol 2016;65: 490–8. [DOI] [PubMed] [Google Scholar]
- 31.Yurdaydin C, Keskin O, Kalkan C, Karakaya F, Caliskan A, Karatayli E, Karatayli S, Bozdayi AM, Koh C, Heller T, Idilman R, Glenn JS. Optimizing lonafarnib treatment for the management of chronic delta hepatitis: The LOWR HDV-1 study. Hepatology 2018;67: 1224–36. [DOI] [PubMed] [Google Scholar]
- 32.Viviani S, Carrieri P, Bah E, Hall AJ, Kirk GD, Mendy M, Montesano R, Plymoth A, Sam O, Van der Sande M, Whittle H, Hainaut P. 20 years into the Gambia Hepatitis Intervention Study: assessment of initial hypotheses and prospects for evaluation of protective effectiveness against liver cancer. Cancer Epidemiol Biomarkers Prev 2008;17: 3216–23. [DOI] [PubMed] [Google Scholar]