Abstract
Clonal deletion of T cells specific for self-antigens in the thymus has been widely studied, primarily by approaches that focus on a single receptor (using TCR transgenes) or a single specificity (using pMHC tetramers). However, less is known about clonal deletion at the population level. Here, we report an assay that measures cleaved caspase 3 to define clonal deletion at the population level. This assay distinguishes clonal deletion from apoptotic events caused by neglect and approximates the anatomic site of deletion using CCR7. This approach showed that 78% of clonal deletion events occur in the cortex in mice. Medullary deletion events were detected at both the semi-mature and mature stages, although mature events were associated with failed Treg induction. Using this assay, we showed that bone marrow derived APC drive approximately half of deletion events at both stages. We also found that both cortical and medullary deletion rely heavily on CD28 co-stimulation. These findings demonstrate a useful strategy for studying clonal deletion within the polyclonal repertoire.
Introduction
Clonal deletion in the thymus eliminates self-reactive T cells, resulting in a T cell repertoire that is tolerant to self-peptides, but is well equipped to respond to foreign pathogens. The strength of the interaction between the T cell receptor (TCR) and self-peptide–MHC complexes is critical to this process, where strong interactions induce death by apoptosis and weak interactions promote survival and maturation. Importantly, clonal deletion occurs within discrete microenvironments within the thymus: the cortex and the medulla. The medulla is considered a specialized site for negative selection because of AIRE (autoimmune regulator)-mediated expression of tissue-specific antigens and the high concentration of dendritic cells relative to the cortex (1). However, recent studies challenged this conception, suggesting that the majority of clonal deletion occurs at the double positive (DP) stage in the thymus cortex in a process independent of the medulla (2–4). Nevertheless, the developmental stages at which thymocytes undergo clonal deletion, their corresponding anatomic locations, and the relative proportions at which they undergo clonal deletion at these developmental stages remains controversial.
The reason for this controversy lies primarily in the use of different model systems that have confounded our understanding of clonal deletion in the polyclonal repertoire. Models including the use of endogenous superantigens fail to emulate antigen-specific clonal deletion. TCR transgenics have been used extensively to study clonal deletion and more appropriately model an antigen-specific response to either exogenous peptide injection or endogenous self-peptide. However, the high frequency of a single specificity of antigen-specific T cells results in a number of nonphysiologic effects (5–7). Additionally, TCR transgenics express their TCR early during development at the double-negative (DN) stage, whereas wild-type (WT) thymocytes do not express surface TCR until the DP stage of development. This results in premature clonal deletion at anatomic locations that are not representative of the polyclonal repertoire (8–10).
More recent studies have attempted to study clonal deletion in settings more representative of the polyclonal environment. BIM deficient mice have an accumulation of self-reactive thymocytes and can therefore be utilized to enumerate T cells that were rescued from clonal deletion (3). This study demonstrated that approximately 75% of thymocytes undergo clonal deletion within the thymus cortex. However, the marked accumulation of thymocytes is not representative of the physiologic conditions in WT mice. A conditional retroviral TCR expression system (retrogenic mice), reported similar conclusions to the BIM knockout studies that approximately 85% of thymocytes undergo clonal deletion in cortex, however only 12 distinct TCRs that were negatively selected were analyzed (4). Finally, peptide–MHC class II tetramers provide a remarkable tool to study tolerance in specific clones within the polyclonal CD4+ T cell repertoire (11, 12), but because of their very small numbers, the anatomic locations at which these clones are deleted are difficult to assess and do not provide a picture at the population level.
Given this, we sought to develop a tool to evaluate clonal deletion in the polyclonal repertoire of WT mice. We designed a cleaved caspase 3-based assay and validated it by a number of different approaches. Using CCR7 to approximate anatomic location in the thymus, we delineate the fraction of thymocytes undergoing clonal deletion in the thymus cortex compared to the medulla. Surprisingly, thymocytes continue to undergo clonal deletion at the most mature stages of development in the thymus medulla. We also report a critical role for CD28-mediated co-stimulation for clonal deletion both within the thymus cortex and medulla. These findings provide a valuable strategy for studying clonal deletion under normal physiologic and pathologic conditions.
Materials and Methods
Mice
C57BL/6NCrl (B6) and B6.SJL-PtprcaPepcb/BoyCrl (B6.SJL) mice were purchased from Charles River Laboratories. C57BL/6Tg(Nr4a1-EGFP/cre)820Khog (Nur77GFP) and C57BL/6Tg(Rag2-EGFP)1Mnz (Rag2GFP) mice were described previously (13, 14). C57BL/6-Tg(Ins2-TFRC/OVA)296Wehi/WehiJ (RIP m-OVA), B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II), B6.Cg-Ptprca Pepcb Tg(TcrLCMV)1Aox/PpmJ (SMARTA), B6.129S2-H2dlAb1-Ea/J (MHC II KO), B6.129S4-Cd80tm1Shr Cd86tm2Shr/J (CD80/CD86 KO), and B6.129S2-CD28tm1Mak/J (CD28 KO) were obtained from Jackson Laboratories. C57BL/6Foxp3tm2Ayr (FOXP3GFP) were kindly provided by M. A. Farrar (University of Minnesota) and were described previously (15). CD80floxBACTg/ B6.129S4-Cd80tm1Shr Cd86tm2Shr/J, referred to as CD86 KO mice in this study, were kindly provided by R. J. Hodes (National Institutes of Health) and were described previously (16). Bone marrow chimera mice were generated by reconstituting lethally irradiated (1,000 rads) mice with 107 T cell-depleted donor bone marrow cells. Recipient mice were provided with neomycin and polymyxin B supplemented water for at least 3 weeks following irradiation and bone marrow transplantation. Chimeras were analyzed at a minimum of six weeks after reconstitution. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Minnesota. All animals were maintained under specific pathogen-free conditions at the University of Minnesota.
Flow Cytometry and MACS Purification
Single-cell suspensions were stained for 30 minutes at 4° C with the indicated antibodies. Antibodies purchased from BioLegend: CD19 (6D5), CD25 (PC61), CD45.1 (A20), CD45.2 (104), CD64 (X54–5/7.1), CD80 (16–10A1), CD90.1 (OX-7), NK1.1 (PK136), TCRγ/δ (GL3), , TCRβ (H57–597). Antibodies purchased from BD Biosciences: CD4 (GK1.5), CD8α (53–6.7), CD69 (H1.2F3), CD86 (GL1), CD90.2 (30-H12), H-2Kb (AF6–88.5), TCRβ (H57–597). Antibodies purchased from Thermo Fischer: CD5 (53–7.3), MHC Class II– I-A/I-E (M5/114.15.2). Antibodies purchased from eBioscience: CD25 (PC61.5), FOXP3 (NRRF-30). Staining for CCR7/CD197 (4B12; eBioscience) was performed for 30 minutes at 37°C prior to additional surface stains. For cleaved caspase-3 (Asp175) (D3E9; Cell Signaling Technologies) staining: cells were processed as quickly as possible following harvest to avoid non-specific apoptosis. Following surface stain, cells were fixed with cytofix/cytoperm (BD Biosciences) for 30 minutes at 4° C. Cells were then washed with perm/wash buffer (BD Biosciences) twice. Cells were stained with anti-cleaved caspase 3 at a 1:50 dilution for 30 minutes at 20° C. To isolate CD4SP thymocytes, we depleted DP and CD8SP thymocytes via negative enrichment using biotinylated anti-CD8α (53–6.7, eBioscience), Streptavidin MicroBeads (Miltinyi Biotec), and MACS separation columns (Miltinyi Biotec) per manufacturer’s protocol. Samples were acquired with BD LSR Fortessa X-20 (BD Biosciences) and analyzed with FlowJo version X (FlowJo LLC).
Immunofluorescence
Thymi were harvested and snap frozen in Optimal Cutting Temperature compound (Sakura Finetek). 7μm sections were fixed and permeabilized in 100% acetone for 20 minutes at 4° C. Samples were then blocked with 5% bovine serum albumin (BSA) and Fc block (anti-CD16/CD32; 2.4G2, Tonbo Biosciences) for one hour at 20°C prior to staining. Antibodies were purchased from BD Biosciences: CD11c (HL3), BioLegened: F4/80 (BM8), CD86 (GL-1), Tonbo biosciences: CD80 (16–10A1), and Vector Laboratories: Fluorescein labeled Ulex Europaeus Agglutinin I (UEA I). Blocked sections were stained with desired antibodies combined into a cocktail in 0.5% BSA and 0.1% Tween-20 (Sigma Aldrich) overnight at 4°C prior to DAPI staining. Sections were mounted using Prolong anti-fade mounting medium (Life Technologies). Images were acquired using a Leica DM6000B epifluorescent microscope 16–72 hours later.
Histo-cytometry
Histo-cytometry was performed as described previously, with modifications (17). Briefly, fluorochrome intensities of each region of interest (based on DAPI staining) were quantified using ImageJ. Data were exported into. csv format and imported into FlowJo for two-dimensional plotting. Medulla gates were drawn based on UEA I fluorescence intensity.
Statistical Analysis
Statistical analyses were performed using Prism 7 (GraphPad). Data sets were assessed for normality using D’Agostino & Pearson normality test. For comparison of two data sets, unpaired Student’s t test or unpaired Mann-Whitney test were performed. For comparison of three or more data sets, ordinary one-way ANOVA with Holm-Sidak’s or Tukey’s multiple comparisons test was used. P-values less than 0.05 were considered significant. Sample size, experimental replicates, and additional details are included in the figure legends.
Results
Identifying clonal deletion in the polyclonal repertoire
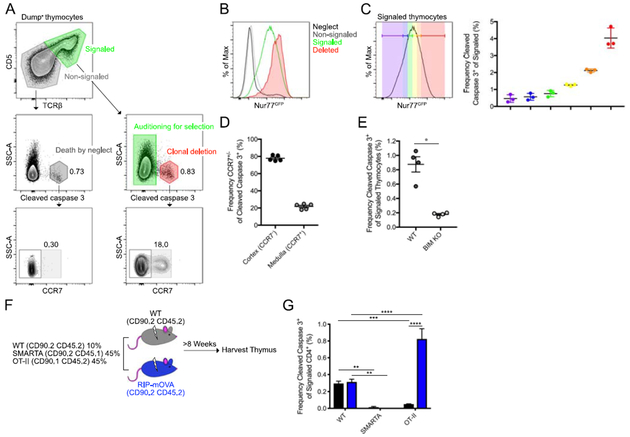
We sought to develop a flow cytometry-based assay to identify thymocytes undergoing apoptosis by clonal deletion. Cells activate a cascade of death inducers when undergoing apoptosis, including cleaved caspase 3, which can be detected by intracellular staining. However, in the thymus, caspase 3 is cleaved in cells undergoing apoptosis due either to death by neglect or clonal deletion. To differentiate between these two fates, we used CD5 and T cell receptor (TCR) β, which are upregulated upon T cell receptor signaling (18). Cleaved caspase 3+ CD5+ TCRβ+ cells (red gate, Figure 1A) potentially represent cells undergoing clonal deletion. To verify this possibility, we utilized Nur77GFP reporter mice, in which GFP expression approximates TCR signal strength (13, 19). GFP expression was high in cells presumed to be undergoing clonal deletion (CD5+TCRβ+cleaved caspase 3+), whereas cells undergoing death by neglect (CD5−TCRβ−cleaved caspase 3+) did not express GFP (Figure 1B). Additionally, GFP expression was higher in cells undergoing clonal deletion compared to the total population of signaled cells “auditioning for selection,” consistent with the affinity model for selection that proposes that strong TCR–self-peptide–MHC interactions are required to induce clonal deletion (1). In line with this, the frequency of cells undergoing clonal deletion was highest in signaled thymocytes that expressed the most GFP and decreased as GFP expression decreased (Figure 1C). We also evaluated mice lacking Bim (BIM KO), which is a proapoptotic molecule required for clonal deletion (3). The frequency of CD5+TCRβ+cleaved caspase 3+ cells was profoundly decreased in Bim KO mice (Figure 1E).
Figure 1. Cleaved caspase 3 distinguishes thymocytes undergoing clonal deletion.
(A) Flow cytometry gating strategy for the identification of cells undergoing clonal deletion or death by neglect in the thymus. Signaled and non-signaled thymocytes identified by the expression of CD5 and TCRβ, excluding CD25+, NK1.1+, CD19+, and TCRγδ+ (Dump) cells (top). Clonal deletion and death by neglect identified by the expression of cleaved caspase 3 (middle) and approximate anatomic location identified by CCR7 (bottom). Numbers adjacent to outlined areas indicate percent cells in each. (B) Flow cytometry from Nur77GFP mice showing GFP expression by CD5−TCRβ−cleaved caspase 3+ (neglect) CD5−TCRβ− (non-signaled), CD5+TCRβ+ (signaled), and CD5+TCRβ+cleaved caspase 3+ (deletion) (as demonstrated in A). (C) Flow cytometry from Nur77GFP mice showing gating based on GFP expression (approximately 15% of total CD5+TCRβ+ cells per gate) (left) and frequency of cleaved caspase 3 expression among CD5+TCRβ+ (signaled) thymocytes in each gate (right). (D) Frequency of CCR7− or CCR7+ among clonally deleted cells (as shown in A). (E) Frequency of cleaved caspase 3+ cells among CD5+TCRβ+ thymocytes (as gated in A) from BIM WT or BIM KO littermates. (F) Experimental design used to generate bone marrow chimeric mice. (G) Frequency of cleaved caspase 3+ cells among CD5+TCRβ+ (signaled) thymocytes (as gated in A) from bone marrow chimeric mice (as in F). Each symbol (C, D, E) represents an individual mouse. Six- to twelve-week-old male and female mice were used. Small horizontal lines indicate the mean and error bars represent SEM. *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. Statistical significance was determined by Mann-Whitney test (E) or 2way Anova with Dunnett’s and Sidak’s multiple comparisons tests (G). Data are representative of more than ten experiments (A, B, C, D) or are pooled from two (E, G) independent experiments with at least three mice per group.
To further verify this assay, we examined TCR transgenic thymocytes undergoing either positive or negative selection. We reasoned that T cells with TCRs that do not have an epitope expressed in the thymus would not undergo clonal deletion, whereas TCRs that have an epitope that is over-expressed in the thymus would have an increased proportion of thymocytes that undergo clonal deletion. To do this, we generated 3-way mixed bone marrow chimeras that were 45% SMARTA (CD4+ TCR specific for LCMV GP61–80), 45% OT-II (CD4+ TCR specific for OVA323–339), and 10% WT bone marrow (Figure 1F and G). Congenic recipient mice were either WT or RIP-mOVA, which ectopically express OVA peptide under the control of the rat insulin promoter in the thymus. OT-II thymocytes had a high frequency of CD5+TCRβ+cleaved caspase 3+ cells in RIP-mOVA recipients, but not in WT recipients (Figure 1G). Likewise, the frequency of such cells amongst SMARTA thymocytes was quite low in either recipient (Figure 1G). Altogether, these data support the validity of this approach for identifying thymocytes undergoing clonal deletion within the polyclonal repertoire.
To determine the localization of thymocytes undergoing clonal deletion, we stained for the C-C chemokine receptor CCR7, which is required for migration to the thymus medulla (20–25) (Figure 1A). Approximately 22% of the thymocytes undergoing clonal deletion were CCR7+ (Figure 1D), which is consistent with previous reports that the majority of clonal deletion occurs in the thymus cortex (3, 4).
Mature thymocytes undergo clonal deletion
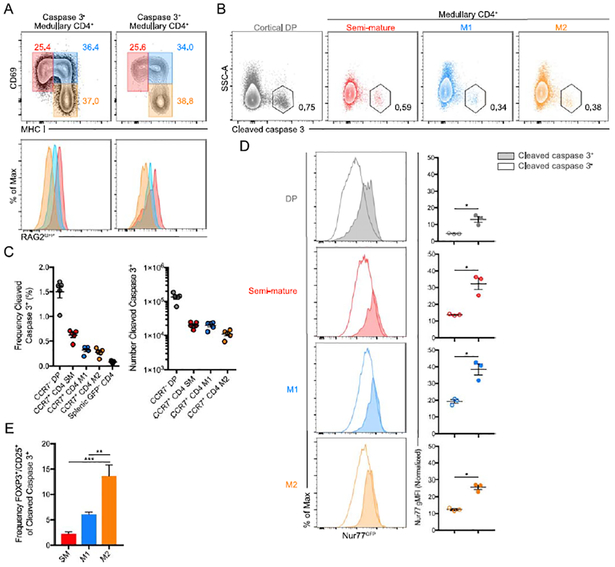
Thymocytes in the medulla undergo distinct stages of maturation, during which they become competent to produce cytokines, proliferate, and emigrate (26). A seminal study by Kishimoto and Sprent (27), showed that semi-mature SP thymocytes undergo apoptosis when stimulated with anti-CD3, whereas the most mature SP thymocytes tend to proliferate (27, 28). Thus, we expected most CCR7+CD5+TCRβ+cleaved caspase 3+ cells would be of the “semi-mature” phenotype. CD69 and MHCI staining was used to define three distinct stages of SP maturation: CD69+MHCI− semi-mature (SM), CD69+MHCI+ mature 1 (M1) and CD69−MHCI+ mature 2 (M2) (28). To our surprise, all 3 stages were present amongst CCR7+ CD5+TCRβ+cleaved caspase 3+ cells—those undergoing deletion in the medulla (Figure 2A, top), which suggests that even mature thymocytes can undergo deletion. When examining the frequency of clonal deletion events at each stage, we observed only a modest reduction from the SM to M1 to M2 stages (Figure 2B and C). To confirm that the mature phenotype deleted cells truly were later in differentiation, we performed this assay using RAG2GFP mice (where GFP decay acts as a ‘molecular timer’ from positive selection) (14). GFP decay was equivalent in clonally deleted and non-deleted thymocytes at more mature stages (Figure 2A, bottom) confirming that deletion can occur far after positive selection. To verify that these late stage clonal deletion events were due to strong TCR signals (as opposed to failed positive selection), we utilized Nur77GFP reporter mice. GFP expression was higher in thymocytes undergoing clonal deletion in all three stages of maturation (Figure 2D). These data indicate that even the most mature thymocytes can undergo TCR triggered apoptosis.
Figure 2. Mature thymocytes are susceptible to clonal deletion.
(A) Flow cytometry of total or deleted CD5+TCRβ+ medullary (CCR7+) CD4+ thymocytes from Rag2GFP mice (gated as in Figure 1A). Gates of three subsets of medullary thymocytes based on expression of CD69 and MHC I (top), and GFP expression by CD69+MHC I− (SM), CD69+MHC I+ (M1), and CD69−MHC I+ (M2) cells (bottom). Numbers adjacent to outlined areas indicate percent cells in each. (B) Flow cytometry of deleted thymocytes (as gated in Figure 1A) of cortical (CCR7−) DP or medullary (CCR7+) CD4+ subsets. Numbers adjacent to outlined areas indicate percent cleaved caspase 3+ cells in each. (C) Frequency (left) and numbers (right) of cleaved caspase 3+ thymocytes among CD5+TCRβ+ thymocyte subsets or CD4+ splenocytes from Rag2GFP mice (gated as in Figure 1A). (D) Flow cytometry of deleted or non-deleted thymocytes (as gated in Figure 1A) from Nur77GFP mice (left) and geometric mean fluorescence intensity (gMFI) normalized to controls (right). (E) Frequency of CD25+ or FOXP3+ among CD5+TCRβ+CD4+cleaved caspase 3+ thymocytes negatively enriched for CD4 using MACS purification from bone marrow chimeric mice that received FOXP3GFP bone marrow. Data are representative of more than 5 independent experiments (A, B, C, D) or 2 independent experiments (E). Each symbol (C, D) represents an individual mouse. Six- to twelve-week-old male and female mice were used. Small horizontal lines indicate the mean and error bars represent SEM. *P< 0.05, **P< 0.01, ***P< 0.001. Statistical significance was determined by one-tailed Mann-Whitney test (D) or ordinary one-way ANOVA with Tukey multiple comparisons test (E).
It was previously established that thymocytes are more susceptible to Treg induction as they mature (29). Given that Treg induction can lead to apoptosis if it falters (30), we considered the possibility that many of the late stage deletion events might be due to failed Treg induction. Consistent with this idea, we observed that the percentage of FOXP3+/CD25+ cells amongst deleted thymocytes was increased with maturation (Figure 2E).
Contribution of bone marrow APC to clonal deletion in the polyclonal repertoire
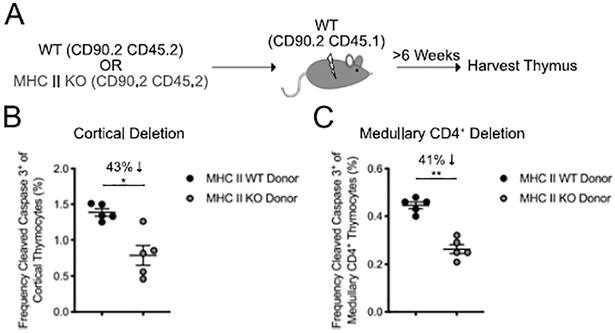
Both bone marrow-derived APCs and medullary thymic epithelial cells (mTEC) have been shown to mediate clonal deletion (31). We reasoned that this assay could be used to assess the relative contribution of bone marrow APCs to deletion in the polyclonal repertoire. For this, WT or MHC II-deficient mice (MHC II KO) were used as bone marrow donors into irradiated WT recipient mice (Figure 3A, Supplemental Figure 1). We did not assess MHC II-deficient recipients, because lack of MHC II on cortical thymic epithelial cells prevents positive selection of CD4+ T cells. Bone marrow APCs contributed to approximately 43% of clonal deletion events in the thymus cortex (Figure 3B) and 41% of clonal deletion events in the thymus medulla (Figure 3C). These data show that bone marrow APCs contribute to a substantial proportion of non-redundant clonal deletion events in the polyclonal repertoire.
Figure 3. Bone marrow derived APC contribute to approximately half of clonal deletion events.
(A) Experimental strategy for generating bone marrow chimeric mice. (B) Frequency of cleaved caspase 3+ thymocytes among CD5+TCRβ+ cortical (CCR7−) thymocytes (as gated in Figure 1A). (C) Frequency of cleaved caspase 3+ thymocytes among CD5+TCRβ+ medullary (CCR7+) CD4+ thymocytes (as gated in Figure 1A). Each symbol (B, C) represents an individual mouse. Six- to twelve-week-old female mice were used. Small horizontal lines indicate the mean and error bars represent SEM. *P< 0.05, **P< 0.01. Statistical significance was determined by Mann-Whitney test (B, C). Data are pooled from two independent experiments.
Clonal deletion is dependent on co-stimulatory molecules in both the cortex and medulla
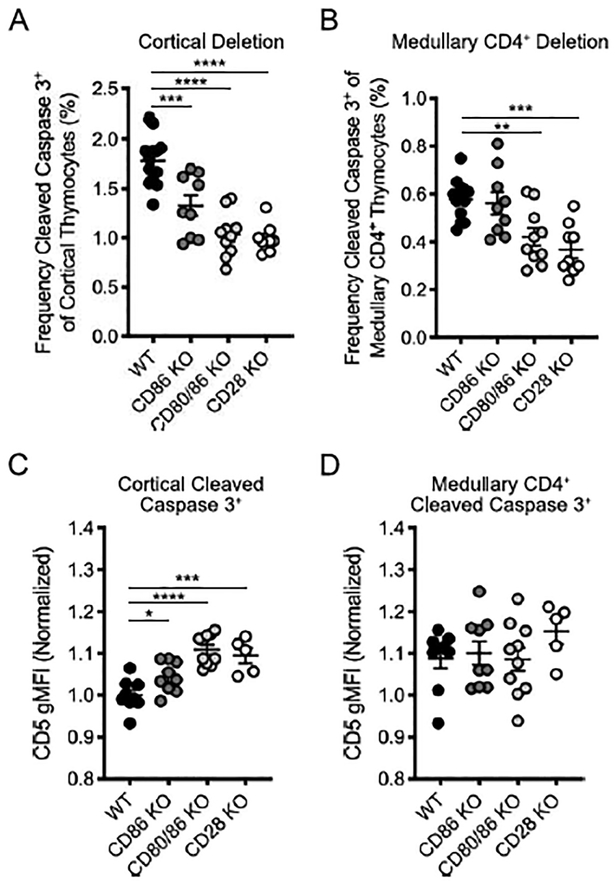
Treg and iNKT cell development and survival in the thymus require CD80 and CD86 co-stimulation via the CD28 receptor (32–36). In contrast, other agonist selected cells– CD8αα intraepithelial lymphocyte precursors (IELp)– need to avoid CD28 co-stimulation for development (37, 38). However, the role of CD28-mediated co-stimulation in clonal deletion has been controversial, primarily because TCR transgenic and superantigen systems were utilized to assess negative selection (39). To address this, we evaluated clonal deletion in CD86 KO, CD80/CD86 KO, and CD28 KO animals (Supplemental Figure 2A). Consistent with previous reports, the frequency of CD4+ and CD8+ thymocytes was increased in CD80/86 KO animals (33, 40) (Supplemental Figure 2B). CD28 co-stimulation contributed to approximately half (56%) of clonal deletion events in the cortex and a third (36%) of clonal deletion events in the medulla (Figure 4A and B), indicating that CD28-mediated co-stimulation is required for clonal deletion. Surprisingly, CD86 alone contributed to clonal deletion in the cortex, but not the medulla (Figure 4A and B), raising the possibility of a non-redundant role for CD80 and CD86 in mediating clonal deletion.
Figure 4. Co-stimulatory molecules CD80 and CD86 are required for both cortical and medullary clonal deletion.
(A) Frequency of cleaved caspase 3+ cells among CD5+TCRβ+ cortical (CCR7−) thymocytes (as gated in Figure 1A) in WT, CD86 KO, CD80/86 KO, and CD28 KO mice. (B) Frequency of cleaved caspase 3+ cells among CD5+TCRβ+ medullary (CCR7+) CD4+ thymocytes (as gated in Figure 1A). (C) CD5 gMFI of cortical deleted thymocytes (normalized to mean of WT CD5 gMFI) (from A). (D) CD5 gMFI of medullary deleted CD4+ thymocytes (normalized to mean of WT CD5 gMFI) (from B). Each symbol (A, B, C, D) represents an individual mouse. Six- to twelve-week-old male and female mice were used. Small horizontal lines indicate the mean and error bars represent SEM. *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. Statistical significance was determined by ordinary one-way ANOVA with Holm-Sidak’s multiple comparisons test (A, B, C, D). Data are pooled from at least three independent experiments (A, B, C, D).
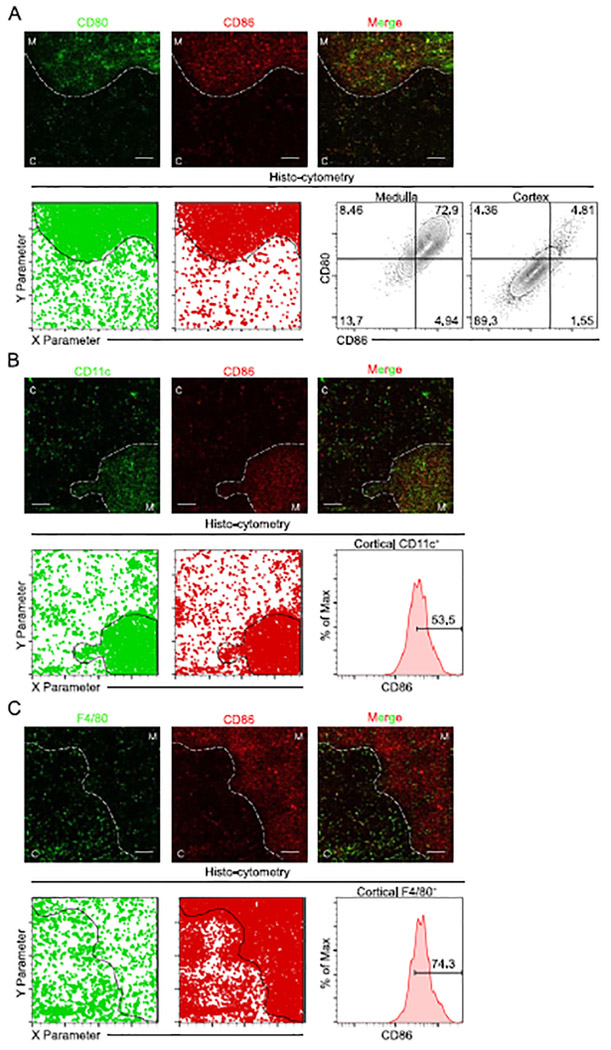
Given this, we sought to determine if CD86 was expressed in the absence of CD80 by any APC type in the thymus. Using flow cytometry of APC recovered from the thymus after collagenase digestion, we observed a substantial proportion of conventional dendritic cells (cDC) that expressed CD80 alone. However, no cDC expressed only CD86 (Supplemental Figure 2C). Furthermore, no mononuclear phagocytes (MNPs) expressed CD86 alone (Supplemental Figure 2D). To investigate the localization of CD80 and CD86, we utilized immunofluorescence staining and quantitative histo-cytometry to determine which APC subset could be mediating CD86-dependent clonal deletion events in the thymus cortex. In agreement with our findings by flow cytometry, very few cells localized within the thymus cortex or medulla expressed CD86 alone (Figure 5A, Supplemental Figure 3A). Within the cortex, CD86 expression was observed on approximately 50% of CD11c-expressing cells and on 70% of F4/80-expressing cells (Figure 5B and C, Supplemental Figure 3B and C). Therefore, CD86-mediated clonal deletion in the cortex is likely mediated by both CD11c+ and F4/80+ APC.
Figure 5. CD80 and CD86 are localized in both the thymus cortex and medulla.
(A) Immunofluorescence microscopy (top) of thymic sections from C57BL/6 mice stained for CD80 (green), and CD86 (red). White dashed lines indicate cortical-medullary border based on UEA I staining (not shown). C, cortex; M, medulla. Scale bars 100μm. Analysis of images by histo-cytometry (bottom, left). Dots represent localization of each stain as determined by histo-cytometry. Frequencies of CD80+ and CD86+ cells identified as localized in the cortex by histo-cytometry (bottom, right). Numbers indicate percent cells in each outlined area. (B) Immunofluorescence microscopy (top) of thymic sections from C57BL/6 mice stained for CD11c (green), and CD86 (red). White dashed lines indicate cortical-medullary border based on UEA I staining (not shown). C, cortex; M, medulla. Scale bars 100μm. Analysis of images by histo-cytometry (bottom, left). Dots represent localization of each stain as determined by histo-cytometry. Frequency of CD86+ cells among CD11c+ cells identified as localized in the cortex by histo-cytometry (bottom, right). Number indicates percent cells in gated area. (C) Immunofluorescence microscopy (top) of thymic sections from C57BL/6 mice stained for F4/80 (green), and CD86 (red). White dashed lines indicate cortical-medullary border based on UEA I staining (not shown). C, cortex; M, medulla. Scale bars 100μm. Analysis of images by histo-cytometry (bottom, left). Dots represent localization of each stain as determined by histo-cytometry. Frequency of CD86+ cells among F4/80+ cells identified as localized in the cortex by histo-cytometry (bottom, right). Number indicates percent cells in gated area. Six- to twelve-week-old male and female mice were used. Data are representative of at least three independent experiments.
Because we could not explain the selective requirement for CD86 in cortical deletion by identifying a cortical APC that only expressed CD86, we reasoned that cortical thymocytes may be more sensitive to and dependent on interactions with CD80/CD86+ APC for clonal deletion. Indeed, in the absence of co-stimulation, the CD5 gMFI was increased on clonally deleted thymocytes in the cortex (Figure 4C), indicating that only the most strongly signaled cells could be deleted without co-stimulation. This was not the case of medullary thymocytes (Figure 4D). Consistent with this, CD86 alone was not required for Treg development, a process that also occurs in the thymus medulla (Supplemental Figure 2E). Together, these data suggest that cortical thymocytes have a lower threshold for co-stimulation-dependent negative selection compared to medullary thymocytes.
Discussion
Defining the developmental stages and corresponding anatomic locations at which thymocytes undergo clonal deletion within the polyclonal repertoire has been challenging given current approaches. Here, we validated a cleaved caspase 3-based assay that can be used to examine clonal deletion at the population level. We made several observations regarding clonal deletion at distinct maturation stages in the thymus. Consistent with estimates that used other methods (3, 4), we observed that the majority of thymocytes underwent clonal deletion in the thymus cortex. However, thymocytes continued to undergo clonal deletion as they progressed through discrete maturation stages in the thymus medulla. Bone marrow-derived APC contributed to clonal deletion similarly in the thymus cortex and medulla. However, co-stimulatory molecule-mediated clonal deletion decreased as thymocytes emigrated from the cortex to the medulla. Thus, this approach served as a valuable tool to assess clonal deletion throughout thymocyte development.
The degree of clonal deletion that occurs in the thymus cortex (78%) supports previous studies that the majority of clonal deletion occurs within the cortex (3, 4). This likely reflects the high propensity of TCRs to interact with MHC molecules, regardless of the specific peptide (4). Signaled thymocytes that are purged of the most cross-reactive TCRs go on to upregulate CCR7 and migrate to the medulla. Upon further analysis of clonal deletion within the thymus medulla, we observed apoptotic events at each of three distinct stages of maturation (26, 28). These findings are in contrast with a previous study that suggested that only semi-mature thymocytes were susceptible to clonal deletion (41). The discrepancies in these findings may lie in the use of different model systems. Kishimoto and Sprent utilized injection of TCR cross-linking antibodies to observe thymocyte death ex vivo. However, such an approach induces extensive peripheral T cell activation, which can contribute to non-specific thymocyte death through the production of cytokines and glucocorticoid stress hormones (42, 43). To rule out non-specific deletion at these late stages of maturation, we utilized Nur77GFP reporter mice. GFP expression in deleted thymocytes was higher than non-deleted thymocytes at all three stages of maturation. Additionally, we showed that Treg and Treg precursors were enriched in apoptotic thymocytes at the most mature stages of development, supporting the notion that FOXP3 can act as a proapoptotic protein (30). These data therefore suggest that mature thymocytes indeed undergo TCR-induced apoptosis, which can involve FOXP3 induction as thymocytes reach their more mature developmental stage.
We estimate that bone marrow APC contribute to approximately 40–45% of deletion events in both the thymus cortex and medulla. Previous studies estimated that 50–70% of CD4SP thymocytes were deleted by bone marrow APC, based on CD4SP frequencies in MHC II-deficient bone marrow chimeras (44, 45). A more recent study, which based their estimates on high throughput analysis of the TCR repertoire in MHC II-deficient bone marrow chimeras, had a more conservative estimate of 30% of deletion events mediated by bone marrow APC (46). However, this study utilized a fixed TCRβ, which may not fully reflect the WT TCR repertoire. Nonetheless, all of these data support the conception that bone marrow APC play a non-redundant role in mediating clonal deletion.
We also showed a critical role for CD28-mediated co-stimulation in both the thymus cortex and medulla. The extent to which co-stimulation contributed to clonal deletion decreased as thymocytes emigrated from the cortex to the medulla. These findings are in contrast with multiple studies involving superantigen or TCR-transgenic systems that did not find a requirement for CD28 in mediating clonal deletion (39, 47–50). However, more recent studies did support a requirement for CD28 in superantigen-mediated clonal deletion (38, 51). In one of these studies, Pobezinsky et al. showed that autoreactive DN thymocytes are diverted to an alternate CD8αα IELp in the absence of CD28-mediated co-stimulation (37, 38). Conversely, this diversion into agonist subsets was not observed at other stages of thymocyte development. Other agonist-selected cells – iNKT (DP stage) and Treg (CD4SP stage) – require CD28-mediated co-stimulation for their survival and differentiation (32–36). It is unclear what signaling context distinguishes the various roles for CD28 in different thymocyte populations (39, 52).
Surprisingly, our results also establish a non-redundant role for CD86 in mediating clonal deletion in the cortex. We do not yet understand if this is a qualitative role for CD86 in clonal deletion (i.e. CD86 allows the deletion of certain antigen-specific populations) or a quantitative requirement for a certain total level of co-stimulatory ligand. Regarding a qualitative role, we considered the possibility that a distinct CD80−CD86+ antigen presenting cell subset was driving cortical clonal deletion. However, neither flow cytometric analysis nor immunofluorescence microscopy supported this hypothesis. Regarding a quantitative role, cortical thymocytes may require a higher total level of CD80/CD86 simulation to trigger deletion. In this case, one would predict that either CD80 or CD86 deficiency would impact cortical deletion.
In summary, we demonstrated that a cleaved caspase 3-based assay can be used to assess clonal deletion at the polyclonal level. This assay served as a valuable tool to study the location and stages of thymocyte development at which clonal deletion occurs. We found that thymocytes undergo clonal deletion even at mature stages of development in the thymus medulla and that approximately half of deletion events require co-stimulation. We expect this assay may be useful in understanding the extent to which generalized clonal deletion defects contribute to autoimmunity caused by specific genetic mutations.
Supplementary Material
Key Points.
Cleaved caspase 3 with activation analysis identifies deleted T cells in the thymus
CD28-mediated co-stimulation is critical to medullary and cortical clonal deletion
Acknowledgements
We thank J. Ding for technical assistance and R. J. Hodes for reading the manuscript and providing feedback and suggestions.
This work was supported by the National Institutes of Health Grants R37 AI39560 and P01 AI35296 (to K.A.H.), F30 AI131483, T32 AI007313, and T32 GM008244 (to E.R.B.).
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Klein L, Kyewski B, Allen PM, and Hogquist KA. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14: 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCaughtry TM, Baldwin TA, Wilken MS, and Hogquist KA. 2008. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med 205: 2575–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, and Hogquist KA. 2013. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A 110: 4679–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald BD, Bunker JJ, Erickson SA, Oh-Hora M, and Bendelac A. 2015. Crossreactive αβ T Cell Receptors Are the Predominant Targets of Thymocyte Negative Selection. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, and Lefrançois L. 2005. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6: 793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hataye J, Moon JJ, Khoruts A, Reilly C, and Jenkins MK. 2006. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312: 114–6. [DOI] [PubMed] [Google Scholar]
- 7.Bautista JL, Lio C-WJ, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, and Hsieh C-S. 2009. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat. Immunol. 10: 610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahama Y, Shores EW, and Singer A. 1992. Negative selection of precursor thymocytes before their differentiation into CD4+CD8+ cells. Science 258: 653–6. [DOI] [PubMed] [Google Scholar]
- 9.Lacorazza HD, Tucek-Szabo C, V Vasović L, Remus K, and Nikolich-Zugich J. 2001. Premature TCR alpha beta expression and signaling in early thymocytes impair thymocyte expansion and partially block their development. J. Immunol. 166: 3184–93. [DOI] [PubMed] [Google Scholar]
- 10.Erman B, Feigenbaum L, Coligan JE, and Singer A. 2002. Early TCRalpha expression generates TCRalphagamma complexes that signal the DN-to-DP transition and impair development. Nat. Immunol. 3: 564–9. [DOI] [PubMed] [Google Scholar]
- 11.Legoux FP, Lim J-B, Cauley AW, Dikiy S, Ertelt J, Mariani TJ, Sparwasser T, Way SS, and Moon JJ. 2015. CD4+ T cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity 43: 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhotra D, Linehan JL, Dileepan T, Lee YJ, Purtha WE, V Lu J, Nelson RW, Fife BT, Orr HT, and Anderson MS. 2016. Tolerance is established in polyclonal CD4+ T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 17: 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCaughtry TM, Wilken MS, and Hogquist KA. 2007. Thymic emigration revisited. J. Exp. Med. 204: 2513–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, and Rudensky AY. 2005. Regulatory T Cell Lineage Specification by the Forkhead Transcription Factor Foxp3. Immunity 22: 329–341. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M, Fujihara C, Radtke AJ, Chiang YJ, Bhatia S, Germain RN, and Hodes RJ. 2017. Co-stimulatory function in primary germinal center responses: CD40 and B7 are required on distinct antigen-presenting cells. J. Exp. Med. 214: 2795–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, and Hogquist KA. 2015. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity 43: 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutz JP, Ong CJ, Marth J, and Teh HS. 1995. Distinct differentiative stages of CD4+CD8+ thymocyte development defined by the lack of coreceptor binding in positive selection. J. Immunol. 154: 2588–99. [PubMed] [Google Scholar]
- 19.Stritesky GL, Jameson SC, and Hogquist KA. 2012. Selection of Self-Reactive T Cells in the Thymus. Annu. Rev. Immunol. 30: 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan JE, McCarthy NI, Parnell SM, White AJ, Bacon A, Serge A, Irla M, Lane PJL, Jenkinson EJ, and Jenkinson WE. 2014. Differential requirement for CCR4 and CCR7 during the development of innate and adaptive αβT cells in the adult thymus. J. Immunol. 193: 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z, Lancaster JN, Sasiponganan C, and Ehrlich LIR. 2015. CCR4 promotes medullary entry and thymocyte–dendritic cell interactions required for central tolerance. J. Exp. Med. 212: 1947–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, Seach N, Arakaki R, Hayashi Y, Kitagawa T, and Lipp M. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24: 165–177. [DOI] [PubMed] [Google Scholar]
- 23.Kwan J, and Killeen N. 2004. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol 172: 3999–4007. [DOI] [PubMed] [Google Scholar]
- 24.Nitta T, Nitta S, Lei Y, Lipp M, and Takahama Y. 2009. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc. Natl. Acad. Sci. 106: 17129–17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno T, Saito F, Gray DHD, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, and Takahama Y. 2004. CCR7 signals are essential for cortex–medulla migration of developing thymocytes. J. Exp. Med. 200: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogquist KA, Xing Y, Hsu F-C, and Shapiro VS. 2015. T Cell Adolescence: Maturation Events Beyond Positive Selection. J. Immunol. 195: 1351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishimoto H, and Sprent J. 1997. Negative selection in the thymus includes semimature T cells. J. Exp. Med. 185: 263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing Y, Wang X, Jameson SC, and Hogquist KA. 2016. Late stages of T cell maturation in the thymus involve NF-[kappa] B and tonic type I interferon signaling. Nat. Immunol. 17: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wirnsberger G, Mair F, and Klein L. 2009. Regulatory T cell differentiation of thymocytes does not require a dedicated antigen-presenting cell but is under T cell-intrinsic developmental control. Proc. Natl. Acad. Sci. 106: 10278–10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai X, Erman B, Alag A, Mu J, Kimura M, Katz G, Guinter T, McCaughtry T, Etzensperger R, and Feigenbaum L. 2013. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity 38: 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breed ER, Lee ST, and Hogquist KA. 2018. Directing T cell fate: How thymic antigen presenting cells coordinate thymocyte selection. Semin. Cell Dev. Biol. 84: 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams JA, Zhang J, Jeon H, Nitta T, Ohigashi I, Klug D, Kruhlak MJ, Choudhury B, Sharrow SO, Granger L, Adams A, Eckhaus MA, Jenkinson SR, Richie ER, Gress RE, Takahama Y, and Hodes RJ. 2014. Thymic Medullary Epithelium and Thymocyte Self-Tolerance Require Cooperation between CD28-CD80/86 and CD40-CD40L Costimulatory Pathways. J. Immunol. 192: 630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams JA, Lumsden JM, Yu X, Feigenbaum L, Zhang J, Steinberg SM, and Hodes RJ. 2008. Regulation of thymic NKT cell development by the B7-CD28 costimulatory pathway. J. Immunol. 181: 907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lio C-WJ, Dodson LF, Deppong CM, Hsieh C-S, and Green JM. 2010. CD28 facilitates the generation of Foxp3(−) cytokine responsive regulatory T cell precursors. J. Immunol. 184: 6007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vang KB, Yang J, Pagán AJ, Li L-X, Wang J, Green JM, Beg AA, and Farrar MA. 2010. Cutting edge: CD28 and c-Rel-dependent pathways initiate regulatory T cell development. J. Immunol. 184: 4074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strom JAB, Tooley J, Ye SK, Subudhi XX, Zheng TBQ, Tang KJ, Henriksen EK, and Boden AJ. 2003. Regulatory T Cells + CD25 + Homeostasis of CD4 Cutting Edge: CD28 Controls Peripheral. J Immunol Ref. 171: 3348–3352. [DOI] [PubMed] [Google Scholar]
- 37.Ruscher R, Kummer RL, Lee YJ, Jameson SC, and Hogquist KA. 2017. CD8αα intraepithelial lymphocytes arise from two main thymic precursors. Nat. Immunol. 18: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L, Park J-H, and Singer A. 2012. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol. 13: 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein L, Robey EA, and Hsieh C-S. 2019. Central CD4+ T cell tolerance: deletion versus regulatory T cell differentiation. Nat. Rev. Immunol. 19: 7–18. [DOI] [PubMed] [Google Scholar]
- 40.Vacchio M, Williams J, and Hodes R. 2005. A novel role for CD28 in thymic selection: elimination of CD28/B7 interactions increases positive selection. Eur. J. Immunol. 35: 418–427. [DOI] [PubMed] [Google Scholar]
- 41.Kishimoto H, Cai Z, Brunmark A, Jackson MR, Peterson PA, and Sprent J. 1996. Differing roles for B7 and intercellular adhesion molecule-1 in negative selection of thymocytes. J. Exp. Med. 184: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogquist KA, Baldwin TA, and Jameson SC. 2005. Central tolerance: learning self-control in the thymus. Nat. Rev. Immunol. 5: 772–82. [DOI] [PubMed] [Google Scholar]
- 43.Brewer JA, Kanagawa O, Sleckman BP, and Muglia LJ. 2002. Thymocyte apoptosis induced by T cell activation is mediated by glucocorticoids in vivo. J. Immunol. 169: 1837–43. [DOI] [PubMed] [Google Scholar]
- 44.van Meerwijk JPM, Marguerat S, Lees RK, Germain RN, Fowlkes BJ, and MacDonald HR. 1997. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 185: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinterberger M, Aichinger M, da Costa OP, Voehringer D, Hoffmann R, and Klein L. 2010. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nat. Immunol. 11: 512–519. [DOI] [PubMed] [Google Scholar]
- 46.Perry JSA, Lio C-WJ, Kau AL, Nutsch K, Yang Z, Gordon JI, Murphy KM, and Hsieh C-S. 2014. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41: 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan R, Teh SJ, Ledbetter JA, Linsley PS, and Teh HS. 1992. B7 costimulates proliferation of CD4–8+ T lymphocytes but is not required for the deletion of immature CD4+8+ thymocytes. J. Immunol. 149: 3217–24. [PubMed] [Google Scholar]
- 48.Page DM, Kane LP, Allison JP, and Hedrick SM. 1993. Two signals are required for negative selection of CD4+CD8+ thymocytes. J. Immunol. 151: 1868–80. [PubMed] [Google Scholar]
- 49.Walunas TL, Sperling AI, Khattri R, Thompson CB, and Bluestone JA. 1996. CD28 expression is not essential for positive and negative selection of thymocytes or peripheral T cell tolerance. J. Immunol. 156: 1006–13. [PubMed] [Google Scholar]
- 50.Dautigny N, Le Campion A, and Lucas B. 1999. Timing and casting for actors of thymic negative selection. J. Immunol. 162: 1294–302. [PubMed] [Google Scholar]
- 51.Buhlmann JE, Elkin SK, and Sharpe AH. 2003. A role for the B7–1/B7–2:CD28/CTLA-4 pathway during negative selection. J. Immunol. 170: 5421–8. [DOI] [PubMed] [Google Scholar]
- 52.Collette Y, Benziane A, Razanajaona D, and Olive D. 1998. Distinct regulation of T-cell death by CD28 depending on both its aggregation and T-cell receptor triggering: a role for Fas-FasL. Blood 92: 1350–63. [PubMed] [Google Scholar]
- 53.Gross JA, Callas E, and Allison JP. 1992. Identification and distribution of the costimulatory receptor CD28 in the mouse. J. Immunol. 149: 380–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.