Abstract
Tetraspanins are a family of proteins possessing four transmembrane domains that help in lateral organization of plasma membrane proteins. These proteins interact with each other as well as other receptors and signaling proteins which result in functional complexes called “tetraspanin microdomains”. Tetraspanins, including CD82, play an essential role in the pathogenesis of fungal infections. Dectin-1, a receptor for the fungal cell wall carbohydrate β−1,3-glucan, is vital to host-defense against fungal infections. In the present study, we show a novel association between tetraspanin CD82 and Dectin-1 on the plasma membrane of Candida albicans-containing phagosomes independent of phagocytic ability. Deletion of CD82 in mice resulted in diminished fungicidal activity, increased C. albicans viability within macrophages, and decreased cytokine production (TNF-α, IL-1β) at both mRNA and protein level in macrophages. Additionally, CD82 organized Dectin-1 clustering in the phagocytic cup. Deletion of CD82 modulates Dectin-1 signaling, resulting in a reduction of Src and Syk phosphorylation and reactive oxygen species production. CD82 knockout mice were more susceptible to C. albicans as compared to wild-type mice. Furthermore, patient C. albicans-induced cytokine production was influenced by two human CD82 single-nucleotide polymorphisms (SNPs), while an additional CD82 SNP increased the risk for candidemia independent of cytokine production. Together, these data demonstrate that CD82 organizes the proper assembly of Dectin-1 signaling machinery in response to C. albicans.
Introduction
Invasive fungal infections are a major cause of morbidity and mortality worldwide, particularly among patients with immunodeficiency or alterations in host barrier defenses (1–3). The most prevalent fungal pathogen, Candida albicans, causes a wide-spectrum of clinical disease ranging from mucocutaneous infections of the gastrointestinal tract and vagina to life-threatening disseminated infections (4, 5). C. albicans is the fourth leading cause of nosocomial acquired blood stream infections and even with the appropriate antifungal therapy, mortality rates remain at 30–50% (6, 7). This unacceptably high mortality rate highlights the limitations of our current antifungal armamentarium and underscores the need to understand better immune responses to C. albicans to develop new therapeutic approaches.
Tetraspanins, a conserved family of integral membrane proteins, play a significant role in the pathogenesis of infections (8). Tetraspanins are expressed on most cell types and regulate a diverse range of cellular functions, including cell morphology, invasion, motility, fusion, and signaling. Tetraspanins contain four transmembrane domains, a short intracellular amino and carboxy terminus, and two extracellular loops of unequal sizes (9, 10). Furthermore, they possess multiple conserved cysteine residues, a conserved CCG motif in the larger extracellular loop, and conserved palmitoylation sites (11, 12). Tetraspanins are located on endosomal membranes and interact with various proteins to establish functional multimeric complexes known as tetraspanin-enriched microdomains (TEMs) (13). The formation of TEMs within the cell membrane and intracellular vesicles facilitates the formation of functional protein complexes enhancing signaling and other cellular functions. Defining the unique function of individual tetraspanins remains challenging due to a high degree of functional redundancy, lack of intrinsic tetraspanin catalytic domains, and insufficient tools (e.g. antibodies).
The tetraspanin CD82, also known as Kai1, is involved in regulating cancer progression and immunity (14, 15). Down-regulation of CD82 is associated with poor prognosis and tumor metastasis in multiple solid organ malignancies including breast, colon, lung, ovarian, and pancreatic cancer (14, 16–20). In addition, CD82 regulates several processes including cell adhesion, motility, and aggregation. At least 20 different tetraspanins, including CD82, are expressed on the surface of immune cells. Several of these tetraspanins (CD9, CD37, CD63, and CD81) interact with pattern-recognition receptors (PRRs) such as C-type lectin receptors, TLRs, scavenger receptors, and Fc receptors (8, 9). In dendritic cells, cell surface expression of CD82 is upregulated upon activation and associates with class II major histocompatibility complex (21), as well as other components of the antigen-processing and presentation complex (22, 23). Interestingly, both macrophages and dendritic cells specifically recruit CD82 to phagosomes upon phagocytosis of various fungi such as Aspergillus fumigatus, Cryptococcus neoformans, and C. albicans (24). However, the mechanism of CD82 in the innate immune response to fungal infections is poorly understood.
Dectin-1, a C-type lectin receptor, is expressed on the surface of innate immune cells, including macrophages and dendritic cells (25–27). Furthermore, Dectin-1 recognizes β−1,3-glucan, a fungal carbohydrate and component of the cell wall (28). Dectin-1 is critical for host-defense against C. albicans, as demonstrated by increased susceptibility to Candida infections in both mice and humans lacking functional Dectin-1 (29, 30). Activation of Dectin-1 triggers phagocytosis, cytokine secretion, and reactive oxygen species (ROS) production that are all critical for anti-Candida defense (31, 32). Although Dectin-1 can bind both soluble and particulate β−1,3-glucan, due to its ability to promote receptor clustering into “phagocytic synapses”, only the particulate form of β−1,3-glucan can trigger Dectin-1-mediated signaling (33). Clustering of Dectin-1 receptors leads to activation of Src kinase, phosphorylation of the hemi-ITAM within the cytosolic tail of Dectin-1, and subsequent recruitment and activation of spleen tyrosine kinase (Syk) (34, 35). Additionally, Dectin-1 is known to interact with the tetraspanins CD37 and CD63 (36–38). Deficiency of CD37 leads to decreased Dectin-1 cell surface localization and significantly enhances Dectin-1 mediated IL-6 production (37).
Since CD82 is recruited to fungal phagosomes, we sought to define the role of CD82 in antifungal immune responses. We demonstrate that CD82 is required for generation of a pro-inflammatory cytokine response to C. albicans in macrophages. CD82-knockout (CD82−/−) macrophages produce less TNFα and IL-1β in response to C. albicans and are impaired in their ability to control C. albicans growth. Proteomic analysis of phagosomes containing chemically defined fungal-like particles (FLPs) (purified fungal cell wall carbohydrate antigens covalently attached to polystyrene beads) revealed that CD82 associates with β−1,3-glucan and mannan containing phagosomes, suggesting interaction with Dectin-1. Association of CD82 and Dectin-1 on the fungal phagosome was confirmed using fluorescent microscopy, co-immunoprecipitation, and proximity ligation assay (PLA). Using β−1,3-glucan FLPs that specifically stimulate Dectin-1, we demonstrate that absence of CD82 impairs Dectin-1 mediated Syk and Src activation, as well as ROS production. We further demonstrate by stochastic optical reconstruction microscopy (STORM) super-resolution microscopy that CD82 regulates clustering of Dectin-1 at the phagocytic synapse and absence of CD82 impairs Dectin-1 clustering. Additionally, mice lacking CD82 have increased susceptibility to C. albicans, further highlighting the importance of CD82. Lastly, human whole blood or peripheral blood mononuclear cells (PBMCs) samples showed two single-nucleotide polymorphisms (SNPs) in CD82 which affected cytokine production and one additional CD82 SNP that increased the risk for candidemia without affecting cytokine production. Taken together our results demonstrate a mechanistic role for CD82 in antifungal immune responses in macrophages by organizing the membrane to promote clustering of Dectin-1 into signaling domains.
Materials and Methods
Animals
Mice were maintained in specific-pathogen-free facilities at Massachusetts General Hospital (MGH; Boston, MA). Macrophages were isolated from approximately equal numbers both male and female mice between 6–8 weeks of age. Primary macrophages were isolated from wildtype (WT) C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME), CD82−/− mice and Dectin-1−/− mice. CD82−/− mice were a gift from Cindy Miranti, PhD (University of Arizona Health Sciences, Tucson, AZ) and the Dectin-1−/− mice were gifted by Gordon Brown, PhD (University of Aberdeen, UK). All knock-out mice were on a C57BL/6 background. All animal studies were conducted under protocols approved by the IACUC (Institutional Animal Care and Use Committee) Subcommittee on Research Animal Care at MGH.
Cell Lines, Viral Transduction, and Cell Culture
The following cell lines were used: immortalized bone-marrow-derived macrophages (BMDMs) from C57BL/6 (gifted by Douglas Golenbock, MD, University of Massachusetts Medical School, Worcester, MA), CD82−/− and Dectin-1−/− mice (gift from Stuart Levitz, MD, University of Massachusetts Medical School, Worcester, MA). Immortalized macrophages were generated using a J2 recombinant retrovirus with v-myc and v-raf(mil) oncogenes (39).
Cells were cultured in RPMI-GlutaMax (Life Technologies, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT), 1% penicillin/streptomycin, 1% HEPES buffer, and 50 μM of β-mercaptoethanol (RPMI complete media). Puromycin was added to a final concentration of 5 μg/mL for selection of transduced cell lines. BMDM cell culture was adapted with the following modifications: cells were grown in media containing 20 ng/mL of M-CSF (Peprotech, Rocky Hill, NJ) for 7 days in tissue culture treated plate with 10 cm diameter. C. albicans (strain SC5314; ATCC, Manassas, VA) was grown in yeast extract peptone dextrose media (Sigma-Aldrich, St. Louis, MO) overnight in an orbital shaker at 30°C. The next morning yeast was washed twice in sterile PBS and heat-killed at 95°C for 30 min.
GFP-Dectin-1 (gifted by David Underhill, PhD, Cedars-Sinai Medical Center, Los Angeles, CA) and HA-CD82 were sub-cloned into pHAGE II vector, which is a fourth-generation lentiviral self-inactivating non-replicative vector. Immortalized C57BL/6 macrophages were stably transduced with GFP-Dectin-1, HA-CD82, or both GFP-Dectin-1 and HA-CD82 using the pHAGE II vector. HEK293T cells were used to produce the lentivirus as described previously (40).
ELISA and Colony-Forming Units (CFU)
Primary WT and CD82−/− macrophages were plated in tissue culture-treated 24-well plates (Greiner Bio-One, Monroe, NC) at a density of 5×105/well. Macrophages were stimulated under 4 conditions: 1) media only, 2) live C. albicans (MOI=10), 3) heat-killed C. albicans (E:T 1:10), or 4) lipopolysaccharide (LPS; 200 ng/mL). TNF-α and IL-1β expression in cell supernatants were determined by the BD™ CBA Flex Set per manufacturer’s instructions (BD Biosciences, Franklin Lakes, NJ). For CFU determination, cells were stimulated for 4 hr, washed three times with PBS then lysed with 0.02% Triton-X-100. Serial dilutions from each well were made in nanopure water and plated on YPD agar plates with ampicillin (5 μg/mL). CFU was determined manually after incubation for 24 hr at 30°C. For the CFU studies, an unpaired Mann-Whitney test was used to test statistical significance using GraphPad Prism7 software (GraphPad Software, La Jolla, CA). ELISA studies used a two-way ANOVA to test for statistical significance [(WT vs CD82−/−) × (unstimulated vs C. albicans vs LPS)]. Bonferroni’s post-hoc test was used to compare means from significant ANOVAs. A p-value <0.05 was considered significant.
PrestoBlue Assay
The PrestoBlue Assay kit (Thermo Fisher, Waltham, MA) was used to measure fungal viability in WT and CD82−/− macrophages according to manufacturer’s protocol. Cells were plated in triplicate in a 96-well plate at a density of 1×105 cells/well. WT and CD82−/− BMDM were stimulated with live C. albicans (MOI=1) for 4 hr. Cells were then lysed with NP40 to liberate phagocytosed yeast cells and centrifuged at 1500 rpm for 5 min. After incubating the lysate with the PrestoBlue reagent (1:10) for 24 hr at 30°C, fluorescence was measured using a SpectraMax i3x Multi-Mode Microplate Reader System (Molecular Devices, San Jose, CA). Statistical significance was examined by an unpaired Mann-Whitney test using GraphPad Prism7 software (GraphPad Software, La Jolla, CA). A p-value <0.05 was considered significant.
Real Time and Quantitative PCR
Gene expression of cytokines (TNF-α and IL-1β) was measured by isolating RNA using the Qiagen RNeasy mini kit. RNA was converted to complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Rochester, NY). Primers were designed using Primer-BLAST Software (NCBI). PCR of the cDNA was carried out using a S1000™ Thermal Cycler (BioRad). Quantitative PCR analysis of the cDNA was performed using iTaq™ Universal SYBR Green Supermix (BioRad). A RT-negative sample was included as a control for each experiment. A melting curve analysis was done to confirm the specificity of primers used. An unpaired t-test was used to test statistical significance of mRNA levels using GraphPad Prism7 software (GraphPad Software, La Jolla, CA). A p-value <0.05 was considered significant.
Confocal Microscopy
Macrophages were plated in Nunc Lab-Tek 8-chambered coverglass (Thermo Fisher Scientific, Rochester, NY) and allowed to adhere overnight. The cells were stimulated with heat-killed C. albicans for 1 hr (E:T 1:5) and fixed with 4% PFA at room temperature. Saponin (0.2% saponin in PBS, 0.03M sucrose, 1% BSA) was used to permeabilize cells for 10 min at room temperature. Non-specific binding in cells was prevented by incubating with blocking buffer (2% goat serum, 1% BSA, 0.1% cold fish skin gelatin, 0.1% saponin, 0.05% Tween-20 in 0.01M PBS, pH 7.2) for 1 hr. Cells were incubated overnight at 4°C with specific primary antibodies (GFP 1:300, Santa Cruz Biotechnology, Dallas, TX; HA 1:200, Cell Signaling, Danvers, MA) in dilution buffer (PBS, 0.05% Tween-20, 1% BSA, 0.1% saponin). Following three washes in dilution buffer, cells were incubated in secondary antibodies (1:500) in dilution buffer for 1 hr at room temperature. The chambered coverglass was then mounted with Vectashield Antifade Mounting Medium (Vector Laboratories, Burlingame, CA). Images were captured on a Nikon Ti-Eclipse inverted microscope equipped with a CSU-X1 confocal spinning-disk head (Yokogawa, Sugarland, TX) and Coherent 4W continuous-wave laser (Coherent, Santa Clara, CA) excited the sample. A high-magnification, high-numerical aperture objective (Nikon, 100×, 1.49 numerical aperture, oil immersion) was used. Images were obtained using an EM-CCD camera (Hamamatsu, Bridgewater, NJ). Image acquisition was performed using MetaMorph software (Molecular Devices, Downingtown, PA). Raw images were then cropped using Adobe Photoshop CS5 and assembled in Adobe Illustrator, version CS4 (Adobe Systems, San Jose, CA).
Phagosome Isolation and Proteomics
Phagosome isolation was adopted from previously described protocols (41). Briefly, macrophages were cultured in 10 cm plates at a density of 1×107 and were stimulated with FLPs (E:T = 25) for 45 min, followed by the addition of hypotonic lysis buffer (2 mM MgCl2, 6 mM β-mercaptoethanol, 10 mM HEPES) with protease inhibitor cocktail (Roche, Indianapolis, IN). Next, cells were mechanically sheared by aspirating and ejecting cell suspension through a 1cc syringe fitted with a ½-inch 26G needle for 15 cycles. The addition of 62% (w/v) sucrose solution to a final of 40% was used to adjust the hypotonic buffer. A discontinuous gradient was then constructed in high-speed ultracentrifugation tubes (Beckman Polyallomer, Brea, CA) overlaying 2 mL of 62%, lysed cell suspension at 40% (sample), 30%, 25%, and 10%. Sucrose gradients were then subjected to ultracentrifugation at 80,000 × g for 1 hr at 4°C (Beckman SW-28 rotor, L8-M ultracentrifuge, Brea, CA). Phagosomes were isolated at the interface of the 25%/10% sucrose layers and washed in PBS.
For proteomics, GFP-Dectin-1 macrophages were incubated with FLPs at a ratio of 5:1 for 1 hr. Macrophages were subjected to hypotonic lysis and mechanical shearing as described above. The phagosomes were then washed in protein-free buffer, digested and peptides resolved by LTQ-Orbitrap Velos mass spectroscopy (42). High-field asymmetric waveform ion mobility spectrometry (FAIMS) and photoionization is used to additionally capture hydrophobic proteins, classically missed with traditional ionizing techniques, increasing peptide yield by 30% (43). Proteomic data was compared to the mouse genome database. Analysis of the macrophage phagosomal protein repertoire comparing β−1,3-glucan to mannan FLP was annotated using Ingenuity software (QIAGEN Bioinformatics, Redwood City, CA).
Co-Immunoprecipitation (IP) and Western Blot
Cells were lysed with lysis buffer containing 1× Brij-58 and protease inhibitor cocktail (Roche, Indianapolis, IN). Next, cell lysates were pre-cleared with bare protein G sepharose beads. Following three washes in lysis buffer, the beads were placed in SDS-containing sample buffer with a reducing agent according to NuPage/Novex protocol. Samples were boiled for 10 min at 95°C, then separated on 4–15% NuPage Bis-Tris gels (Life Technologies, Carlsbad, CA) and transferred to PVDF membranes (Perkin Elmer, Waltham, MA). Non-specific binding was blocked with 5% BSA in 0.1% PBS-Tween 20 (Sigma-Aldrich, St. Louis, MO) followed by incubation with primary antibodies bound to Dynabeads (Invitrogen, Carlsbad, CA) shaking overnight at 4°C. For co-IP data, anti-HA-HRP (Cell Signaling, Danvers, MA) and anti-GFP-HRP (Santa Cruz Biotechnology, Dallas, TX) antibodies (1:1000) were used. For western blots, membranes were probed for either phosphorylated Src or phosphorylated Syk (Cell Signaling, Danvers, MA). Membranes were then washed thoroughly with PBS-Tween (0.1%) and developed using ECL reagents (Perkin Elmer, Waltham, MA) on Kodak BioMax XAR Film (Sigma-Aldrich, St. Louis, MO). For re-probing, blots were stripped for 30 min at 37°C with Restore PLUS stripping buffer (Thermo Fisher Scientific). Actin was used as a loading control for all western blots (Sigma-Aldrich, St. Louis, MO). Films were evenly adjusted using Adobe Photoshop CS4 (Adobe Systems, San Jose, CA). ImageJ software was used to measure band intensity. Western blots were analyzed by a two-way ANOVA to test for statistical significance [(WT vs CD82−/−) × (unstimulated vs live C. albicans vs heat killed C. albicans)]. Bonferroni’s post-hoc test was used to compare means from significant ANOVAs. A p-value <0.05 was considered significant.
Lucigenin-Enhanced Chemiluminescence Assay for ROS
Macrophages from WT and CD82−/− mice were plated in a 96-well plate at a density of 5×104 cells/well. 24 hr after cells were seeded, cells were placed on ice and washed in PBS three times. Cells were incubated on ice for 10 min in lucigenin solution (0.9 mM CaCl2, 0.5 mM MgCl2, 20 mM dextrose, and 20 μM lucigenin). Live C. albicans was added and briefly centrifuged to enable cell-ligand contact. The initial reading (0 min) was taken immediately following centrifugation. The plate was read every 10 min and placed into an incubator at 37°C between each measurement. All readings were taken using the SpectraMax i3X Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA). An unpaired t-test was used to test statistical significance in GraphPad Prism7. A p-value <0.05 was considered significant.
Flow Cytometry
Flow cytometry was used to analyze the expression of Dectin-1 on the cell surface of WT and CD82−/− macrophages. Macrophages were washed in PBS containing 2% BSA and stained with anti-Dectin-1 APC conjugated antibody (1 μg/mL; R&D Systems) or an irrelevant isotype (rat-IgG2a antibody; eBioscience) for 1 hr at room temperature. Surface fluorescence was assessed on a BD FACSAria II flow cytometer and cell sorter (Bectin-Dickinson, Franklin Lakes, NJ) and analysis was performed by FlowJo software (FlowJo, LLC, Ashland, OR).
Proximity Ligation Assay
A PLA was performed using the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich, St. Louis, MO). Immortalized macrophages expressing HA-CD82 and GFP-Dectin-1 were cultured on Nunc Lab-Tek chambered coverglass (Thermo Fisher Scientific, Rochester, NY) and stained as described above. The primary antibodies used were anti-GFP and anti-HA. PLA probes anti-rabbit PLUS and anti-mouse MINUS were used with detection reagent red according to Duolink protocol. Cells were imaged using the confocal microscope as described above and ImageJ was used to quantify PLA signal. The number of positive foci were counted in 15 total cells in multiple (>5) fields of view per condition. An unpaired t-test was used to test statistical significance in GraphPad Prism7. A p-value <0.05 was considered significant.
STORM and Quantification
Immortalized WT or CD82−/− macrophages overexpressing GFP-Dectin-1 were seeded on tissue culture treated chambered coverglass (Thermo Fisher Scientific, Rochester, NY) and stimulated for 15 min with heat-killed C. albicans. The cells were then fixed with 4% PFA at room temperature and subjected to the super-resolution imaging technique Stocastic Optical Reconstruction Microscopy (STORM) using the GFP-Booster, and anti-GFP antibody conjugated to the photo-switchable dye ATTO488 (ChromoTek, Hauppauge, NY). A Nikon N-STORM microscope was used to image samples at a 60× objective using the “Perfect Focus” system. Imaging was performed in an extracellular solution containing reducing and oxygen scavengers, as specified by dSTORM protocols (44). The fluorochromes were first converted to a photo-switchable state using 488 nm at 60 mW. Once the fluorochromes were converted into a desired density of single molecules per frame, imaged continuously at 10,000–30,000 frames. Localization was acquired using an Andor 512 × 512 back-thinned EM-CCD and reconstructed with Nikon Elements Imaging Software with a 50 msec exposure time.
For cluster quantification, reconstructed STORM data was analyzed using Voronoï tessellation in SR-Tesseler software (45). Default parameters and a density factor of 1.3 were used. This method subdivides a super-resolution image into polygons based on molecular local densities. Quantification was performed on six 2D reconstructed images.
In Vivo Candidemia Study
C. albicans were grown in YPD media with 100 μg/mL ampicillin overnight at 30°C in a shaker incubator at 250 rpm. Yeast were washed and resuspended at a concentration (500,000 yeast/mL) in ice-cold PBS. Wild-type and CD82−/− mice were injected intravenously into the lateral tail vein with C. albicans (100,000 yeast). Following the C. albicans challenge, mice were monitored once daily for morbidity and mortality for up to 28 days. Mice displaying prespecified criteria for distress (inability to feed or drink, labored breathing, ruffled and/or matted fur, decreased activity, hunched posture, and shivering) were euthanized by CO2 asphyxiation. All mouse experiments were done under protocols approved by the MGH IACUC.
Identification of CD82 Human Polymorphisms
Single nucleotide polymorphisms (SNPs) within the CD82 gene and cQTLs were identified as previously described (46). Briefly, we extracted SNPs located within the CD82 gene (window size of +/−250kb around the gene) and tested for their effect on CD82 expression levels using HaploReg (47). We then extracted the association between CD82 SNPs and cytokine levels using previously published data from Human Functional Genomics Project (46). Association between CD82 SNPs and susceptibility to candidemia were extracted from previous candidemia genetic studies (48, 49).
Results
CD82 is required for control of C. albicans and cytokine secretion in macrophages
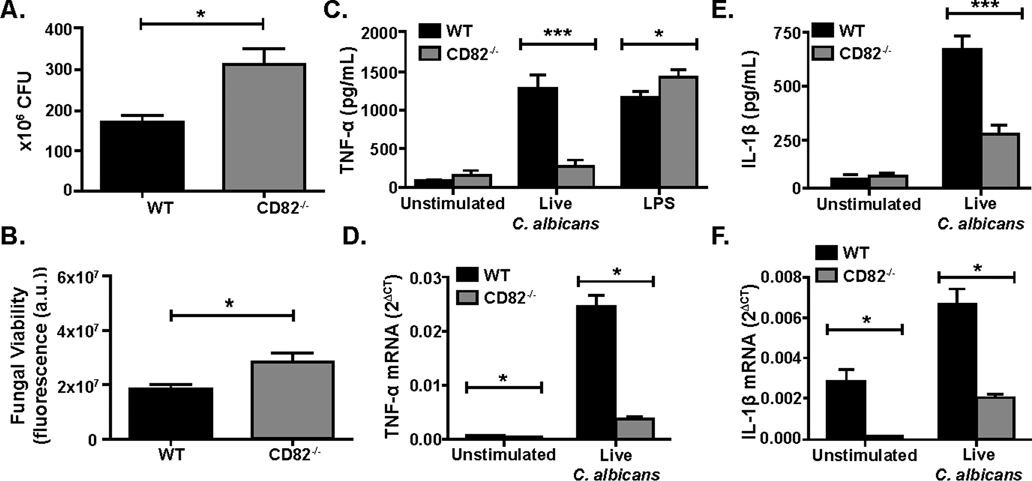
To determine the role of CD82 in the killing of C. albicans, primary BMDM from C57BL/6 WT and CD82−/− mice were stimulated with live C. albicans for 4 hr. Macrophages were then lysed to release phagocytosed yeast and either plated on agar plates for a CFU assay or added to PrestoBlue solution with YPD for a viability assay for 24 hr. There was a significant increase in fungal viability in the CD82−/− macrophages compared to WT macrophages as measured by CFU at MOI=5 after of stimulation (Fig 1A). In addition to CFU, we investigated fungal viability by PrestoBlue assay to ensure we fully accounted for the hyphal transition of C. albicans in the growth assay. Fungal viability of C. albicans in WT and CD82−/− macrophages were measured using the PrestoBlue cell viability assay after stimulation with live C. albicans for 4 hr. C. albicans from CD82−/− macrophages (MOI=5) had higher fungal viability when compared to fungi from WT macrophages (Fig 1B). These results indicate that CD82 enhances fungal killing in macrophages.
Figure 1. CD82 regulates fungal survival and cytokine production.
CD82−/− macrophages have increased fungal viability compared to WT macrophages as measured by CFU (A) and PrestoBlue (B). Decreased TNF-α (C) and IL-1β production (E) was observed in CD82−/− macrophages (gray bar) compared to WT macrophages (black bar) upon C. albicans stimulation. CD82−/− macrophages had reduced levels of TNF-α (D) and IL-1β (F) mRNA compared to WT. (*p<0.05, ***p<0.01 vs WT; n=3).
We sought to determine the role of CD82 in regulating cytokine production in response to C. albicans stimulation. Supernatants from C. albicans-stimulated primary and immortalized WT and CD82−/− macrophages (MOI=5) were analyzed for TNF-α and IL-1β cytokine production. Macrophages stimulated with live C. albicans showed reduced production of TNF-α and IL-1β in the absence of CD82 as compared to WT (Fig 1C, E). Similarly, primary CD82−/− macrophages stimulated with live C. albicans (MOI=10) showed decreased mRNA expression compared to WT macrophages (Fig. 1D, F). To determine if this change in cytokine production was specific to fungal-derived ligands, we stimulated cells with LPS and measured TNF-α production by ELISA. These data showed comparable TNF-α levels in CD82-deficient mice as compared to WT (Fig. 1C). Together, these data suggest that CD82 is required for cytokine production and gene expression in response to C. albicans.
CD82 associates with Dectin-1 on the fungal phagosome
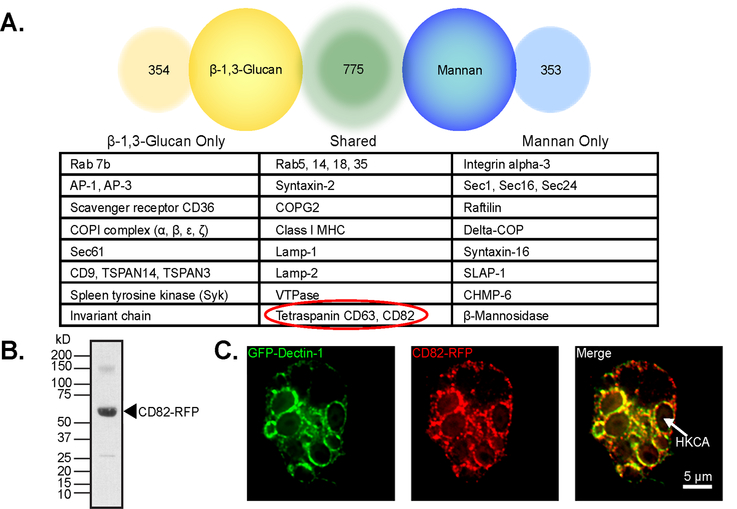
To determine whether CD82 closely associates with Dectin-1 on the fungal phagosome, we conducted a proteomic analysis of membrane proteins associated with either β−1,3-glucan or mannan FLP-containing-phagosomes. Analysis shows that of the 1482 proteins identified, 775 were shared between β−1,3-glucan beads and mannan FLPs. 354 proteins were uniquely associated with β−1,3-glucan and 353 proteins were associated with mannan FLPs. CD82 was associated with both β−1,3-glucan and mannan FLP-containing phagosomes (Fig 2A).
Figure 2. Dectin-1 and CD82 associate on the fungal phagosome.
(A) CD82 is shown in a partial list of proteomics hits from macrophages stimulated by either β−1,3-glucan or mannan FLPs. (B) Immunoblot of purified β−1,3-glucan FLP containing phagosomes, confirms CD82-mRFP1 localization to the phagosome using an α-RFP antibody. (C) Co-localization of GFP-Dectin-1 and CD82-mRFP1 was observed by confocal microscopy in immortalized macrophages. Size bar equals 5 μm. Arrow indicates a phagosome containing heat-killed C. albicans.
To confirm CD82 recruitment to β−1,3-glucan phagosomes, we stimulated macrophages overexpressing CD82-mRFP1 and GFP-Dectin-1 with β−1,3-glucan FLP and performed phagosome isolation (50, 51). Phagosomes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting for RFP. The corresponding band is to CD82-RFP (Fig 2B). To visualize the association of Dectin-1 and CD82 in the phagosome, we stimulated immortalized macrophages overexpressing CD82-mRFP1 and GFP-Dectin-1 with heat-killed C. albicans for 1 hr (E:T = 5:1). Confocal microscopy showed that Dectin-1 was recruited to the C. albicans phagosome as we have shown previously (50). Interestingly, we also see recruitment of CD82 to the fungal phagosome in addition to Dectin-1. Furthermore, we observed co-localization of Dectin-1 and CD82 on C. albicans-containing phagosomes (Fig 2C).
CD82 associates with Dectin-1
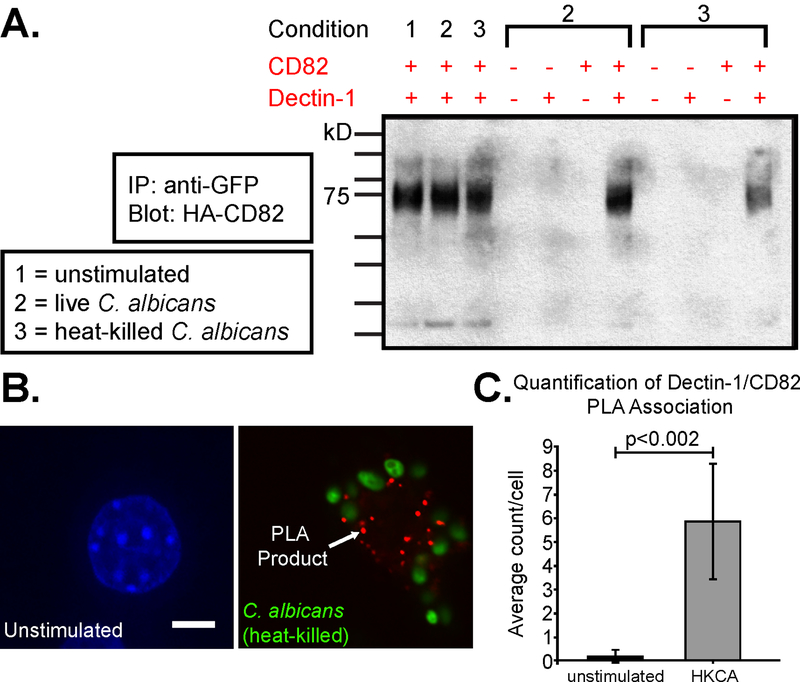
Since CD82 and Dectin-1 associate on the fungal phagosome, we next investigated whether Dectin-1 and CD82 associate on the plasma membrane at a resting state. We first performed co-IP experiments using immortalized macrophages. Since the strength and nature of the tetraspanin-protein interaction is determined by the degree of resistance to detergent conditions (52), co-IP experiments were done under mild (Brij-58) and strong (Triton X-100) lysis conditions. Interactions that are stable in 1% Triton X-100 buffers are usually considered to be strong and direct, while stability in only mild detergents indicates indirect interactions (53). Interactions between CD82 and Dectin-1 were only observed under Brij-58 lysis conditions, but not under Triton X-100 conditions, indicating an indirect interaction (data not shown). To determine specificity of co-IP lysis conditions, macrophages were modified to overexpress CD82 and/or Dectin-1 as followed: [1] WT, [2] expression of GFP-Dectin-1 only, [3] expression of HA-CD82 only, and [4] expression of both GFP-Dectin-1 and HA-CD82. Macrophages were unstimulated, stimulated by live C. albicans, or stimulated by heat-killed C. albicans. Heat-killed C. albicans was used for our molecular studies since heat-killed yeast has higher β−1,3-glucan exposure on the cell wall compared to live C. albicans (54). In macrophages lacking GFP-Dectin-1 and/or HA-CD82, no co-IP band was observed when stimulated with live or heat-killed C. albicans. Under all three stimulation conditions, macrophages expressing both Dectin-1 and CD82 showed a positive co-IP band. Importantly, GFP-Dectin-1 could be co-immunoprecipitated with HA-CD82 (data not shown), showing that the co-IP between CD82 and Dectin-1 worked in both directions (Fig 3A).
Figure 3. Dectin-1 and CD82 interact in macrophages.
(A) Dectin-1 and CD82 associate with each other as seen by co-IP of immortalized macrophages expressing HA-CD82 and GFP-Dectin-1 in resting macrophages and in cells stimulated with live and heat-killed C. albicans. (B) Representative images of the association between Dectin-1 and CD82 in stimulated macrophages expressing GFP-Dectin-1 and HA-CD82 as measured by PLA. Association was observed in macrophages stimulated by heat-killed C. albicans (HKCA). Size bar equals 5 μm. (C) Quantification of Dectin-1/CD82 PLA association confirm the number of positive co-localization per cell in unstimulated versus stimulated macrophages (p<0.002). Data are from analysis of at least of 20 cells per condition.
To confirm the association of Dectin-1 and CD82 observed by confocal microscopy and co-IP, we next used PLA, a PCR-based method which allows quantification of sites of close interaction between proteins by microscopy (55). Following 15 min of stimulation with heat-killed C. albicans, PLA showed association between CD82 and Dectin-1 (Fig 3B). However, no association was seen by PLA in a resting state, unstimulated by C. albicans, which suggests >40 nm separates CD82 and Dectin-1 in resting macrophages. The increased PLA signal in stimulated macrophages suggests that CD82 and Dectin-1 become more closely associated (<40nm) when activated by C. albicans (Fig 3C). Together, these biochemical and microscopy approaches strongly suggest an indirect association between the tetraspanin CD82 and Dectin-1 in macrophages.
CD82 is required for downstream Dectin-1 signaling
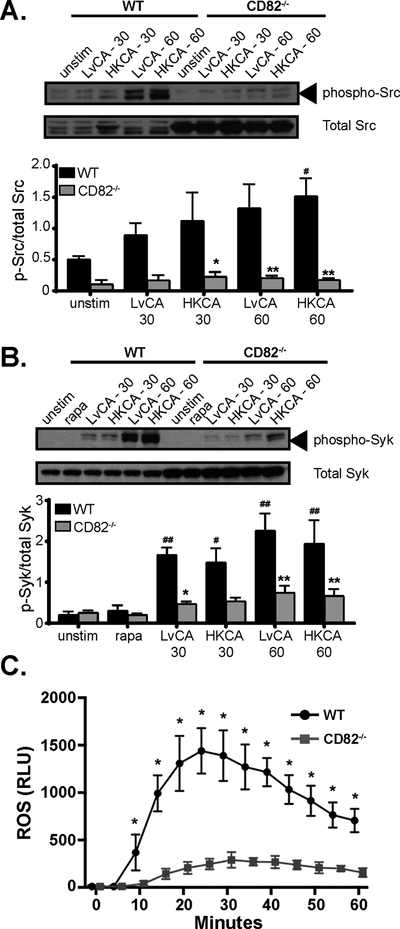
To understand the effects of CD82 association with Dectin-1, we analyzed downstream components of the Dectin-1 signaling pathway in CD82−/− macrophages. Upon stimulation of Dectin-1, Src-mediated signaling occurs in response to β−1,3-glucan binding. The ITAM motif of Dectin-1 is phosphorylated by Src kinase, resulting in the recruitment of Syk and subsequent ROS production (32, 34). Phosphorylation of Src family kinases and Syk was measured in immortalized WT and CD82−/− macrophages simulated with either live or heat-killed C. albicans. Phosphorylation of both Src family kinases and Syk started to occur 30 min after stimulation and further increased at 60 min (Fig 4A–B). In sharp contrast, CD82−/− macrophages showed little phosphorylation of Src family kinases and Syk at similar time points compared to WT macrophages. The failure of sustained Src and Syk phosphorylation in macrophages lacking CD82 corresponds with reduced ROS production (Fig 4C). Together, these observations show that CD82 is required for Dectin-1 signaling pathway and subsequent ROS production.
Figure 4. CD82 is required for downstream Dectin-1 signaling components.
Phosphorylation of Src family kinases (A) and Syk (B), which occurs downstream of Dectin-1 activation, was elevated as soon as 30 min after stimulation with live and heat-killed C. albicans (LvCA and HKCA, respectively). Immunoblotting demonstrated reduced phosphorylation of both proteins in CD82−/− macrophages when compared to total Src or Syk. Quantification was performed using ImageJ software. (C) Ameliorated ROS generation was seen in CD82−/− macrophages (curve with squares) compared to WT (curve with circles) as measured by lucigenin assay (*p<0.05, **p<0.01 vs WT; #p<0.05, ##p<0.01 vs unstimulated; Src n=3, Syk n=5).
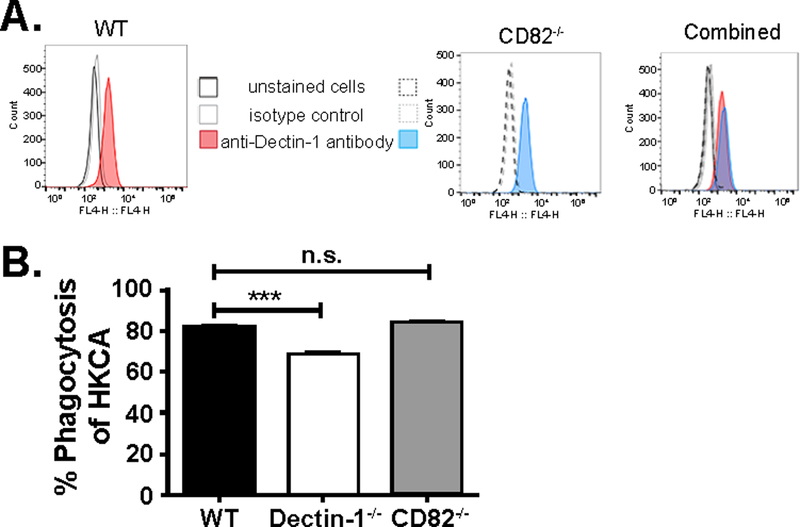
Comparable Dectin-1 expression and phagocytic uptake in wild type and CD82−/− macrophages
Since CD82 appears to play a role in the Dectin-1 signaling cascade upon stimulation by C. albicans, we next wanted to determine whether CD82−/− macrophages influenced Dectin-1 surface expression or phagocytic capacity. Analysis of Dectin-1 surface expression from primary macrophages by flow cytometry showed similar expression of Dectin-1 in CD82−/− macrophages compared to the WT control (Fig 5A). To determine whether defects in Dectin-1 signaling could be due to defective phagocytosis in CD82−/− macrophages, we compared heat-killed C. albicans labeled with Alexa-Fluor 647 phagocytosed by WT and CD82−/− macrophages. After stimulation of macrophages by heat-killed C. albicans, no difference was observed in phagocytic uptake between WT and CD82-deficient macrophages (Fig 5B). As a control, we compared phagocytosis in macrophages lacking Dectin-1 compared to WT cells. As expected, Dectin-1−/− macrophages did show a lower phagocytic uptake of heat-killed C. albicans compared to the WT cells (Fig 5B, white bar). These results show that the loss of Src and Syk phosphorylation as well as ROS production is not due to any differences in Dectin-1 expression or phagocytic uptake of C. albicans.
Figure 5. Macrophages lacking CD82 have no effect of Dectin-1 expression or phagocytosis.
(A) Similar amounts of Dectin-1 expression was seen in WT (red) and CD82−/− (blue) macrophages as measured by flow cytometry using an anti-Dectin-1 antibody. Together, these peaks overlap in the third panel (purple). (B) No significant difference was observed in phagocytosis of heat-killed C. albicans (HKCA) between WT and CD82−/− macrophages. As a control, Dectin-1−/− primary macrophages were compared to WT. Dectin-1−/− macrophages had reduced phagocytosis of HKCA. (***p<0.001 vs WT; n=3)
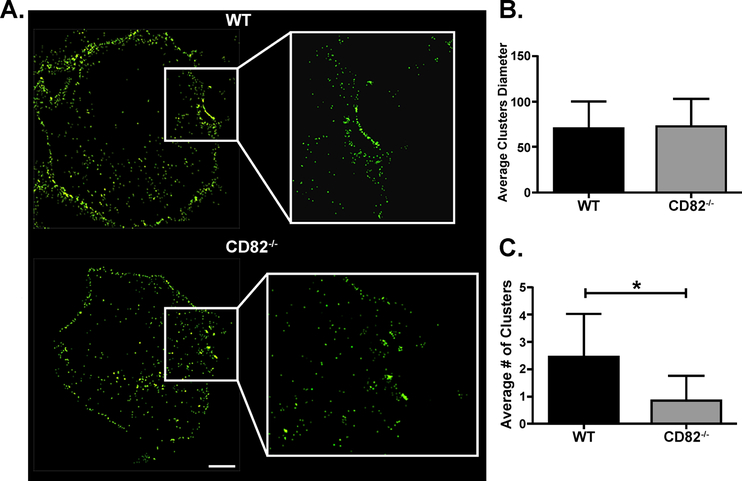
CD82 licenses Dectin-1 clustering in the phagocytic synapse
Since we observed an association between CD82 and Dectin-1, we next investigated how CD82 organizes Dectin-1 in TEMs on the plasma membrane. While we can visualize CD82 and Dectin-1 on the macrophage plasma membrane using spinning disk confocal microscopy, we were unable to analyze Dectin-1 clustering due to limitations of conventional light microscopy. Individual Dectin-1 clusters cannot be discerned in the phagocytic cup using standard epifluorescence imaging (Fig S1A). However, when the same cells are imaged at a greater resolution by STORM, individual Dectin-1 clusters can be distinguished (Fig S1B). Using STORM, we observed larger amounts of Dectin-1 clusters in the phagocytic cup in WT macrophages compared to CD82−/− macrophages (Fig 6A). Quantitative analysis performed on these Dectin-1 clusters showed that although cluster size was similar in both the WT and CD82−/− macrophages (Fig 6B), fewer Dectin-1 clusters were seen in CD82−/− macrophages compared to WT macrophages (Fig 6C). Based on these data, we conclude that CD82 organized Dectin-1 clustering in the phagocytic cup for downstream Dectin-1 signaling events.
Figure 6. CD82 regulates Dectin-1 clustering in the phagocytic cup.
(A) Macrophages were stimulated with heat-killed C. albicans for 15 min then fixed using PFA. Lower Dectin-1 clustering was observed in CD82−/− macrophages compared to WT as seen by high resolution STORM. (B) Cluster size was not different between WT and CD82−/− macrophages. (C) In a cluster analysis of 20 cells, CD82−/− macrophages had less clusters compared to WT macrophages. (*p<0.05 vs WT; n=3)
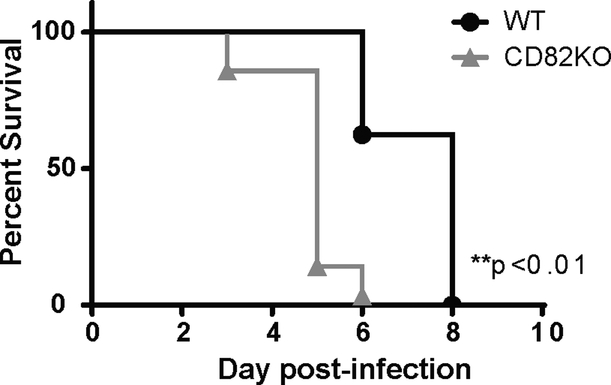
Deletion of CD82 increases susceptibility to C. albicans in mice
To confirm the relevance of in vitro findings, we next investigated susceptibility of WT and CD82−/− mice to C. albicans bloodstream infection in an in vivo model of systemic candidiasis. Mice were intravenously infected with a lethal dose (1×105 yeast) of C. albicans and survival was monitored daily for 28 days. As expected, C. albicans challenge led to 100% mortality by day 8 in WT mice. On the contrary, mice lacking CD82 had significantly reduced survival, as seen by 100% mortality by day 6 after infection (Fig 7). These data demonstrate that the absence of CD82 leads to increase susceptibility to C. albicans infection.
Figure 7. CD82 knockout mice are more susceptible to C. albicans infection.
WT and CD82−/− mice were intravenously injected with C. albicans (1×105 cfu) and survival was monitored. The data are presented as Kaplan-Meier survival curves (WT n=8; CD82−/− n=7).
CD82 polymorphisms affect Candida-induced cytokine responses and candidemia susceptibility
To test if CD82 is important for susceptibility to Candida infections in patients, we investigated CD82 polymorphisms for their association with Candida-induced cytokine production and susceptibility to candidemia. We identified two CD82 SNPs that affected IL-1β, IL-6 and IL-17 cytokine production after stimulation of human whole blood or PBMCs with C. albicans: rs12282890 and rs1429553 (Table I). Furthermore, the CD82 SNP rs7932712 was associated with an increased risk for candidemia but did not affect cytokine production. Overall, these polymorphisms indicate a key role for CD82 in human host defense and susceptibility to Candida infection.
Table I.
List of CD82 SNPs associated with cytokine changes and candidemia susceptibility
| SNPs | eQTL p-value |
Cytokine Affected | Affected Sample | cQTL p-value |
Candidemia Susceptibility p-value |
Risk Allele Frequency | Candidemia Susceptibility Odds Ratio |
|---|---|---|---|---|---|---|---|
| rs12282890 | 6.22E-06 | IL-1β | Candida blastospores, (HK), WB, 48 hr | 0.0122 | NS | n/a | n/a |
| rs12282890 | 6.22E-06 | IL-17 | C. albicans blastospores, PBMC, 7 days | 0.0471 | NS | n/a | n/a |
| rs1429553 | 1.11E-03 | IL-6 | C. albicans hyphae, PBMC, 24 hr | 0.0018 | NS | n/a | n/a |
| rs7932712 | 0.0014 | none | n/a | NS | 0.0215 | C, 0.71 | 1.49 |
List of CD82 SNPs in the 500 FG cohort for Candida-induced cytokines and candidemia susceptibility in whole blood (WB) or peripheral blood mononuclear cell (PBMC) samples. Two polymorphisms were associated with changes to IL-1β, IL-17, and IL-6 cytokines. The SNP rs7932712 was associated with an elevated susceptibility to develop candidemia. eQTL, SNPs correlated with expression level of CD82; cQTL, SNPs correlated with cytokine levels; NS, SNPs were tested but are not associated with the phenotype.
Discussion
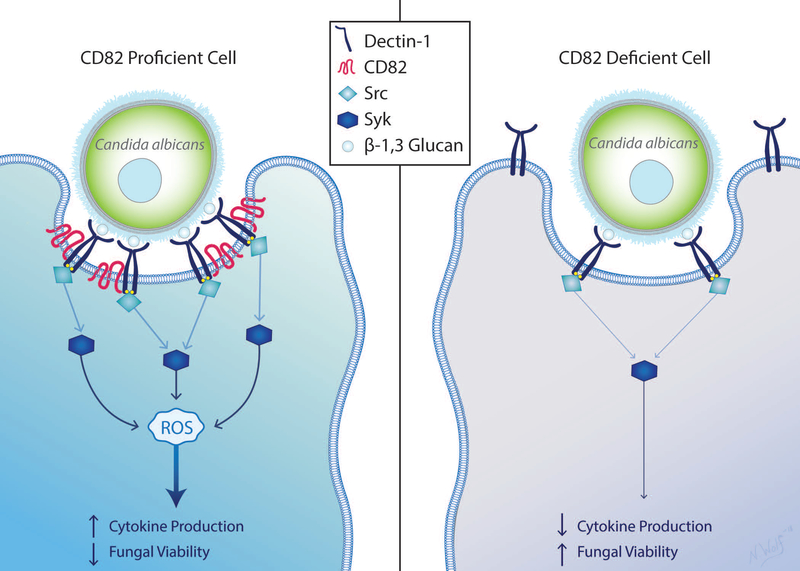
Our data show a novel mechanism for Dectin-1 activation by C. albicans mediated by the tetraspanin CD82 (Fig 8). The present study demonstrates that CD82 associates with Dectin-1 on the plasma membrane to organize Dectin-1 into TEMs in the phagocytic cup. Dectin-1 clustering via CD82 facilitates downstream Dectin-1 signaling, resulting in robust phosphorylation of Src family kinases and Syk as well as subsequent production of ROS. Additionally, deletion of CD82 in macrophages leads to reduced ROS production, cytokine production and mRNA expression of TNF-α and IL-1β. Macrophages deficient in CD82 are less efficient at fungal killing and therefore there is higher replication within these macrophages. Furthermore, absence of CD82 in mice led to greater susceptibility to C. albicans as compared to CD82-proficient mice. We propose this mechanism serves to mediate antifungal immunity in macrophages.
Figure 8. Schematic representation of CD82 and Dectin-1 interaction after C. albicans activation.
In CD82-proficient cells, Dectin-1 is activated by β−1,3 glucan on the cell wall of C. albicans. Subsequent activation of Src then Syk leads to production of ROS, elevated cytokines and reduction in fungal viability. Presence of CD82 contributes to Dectin-1 molecular organization in the membrane. Cells lacking CD82 have reduced Dectin-1 clustering in the phagocytic cup, which contributes to reduced Syk phosphorylation, ROS and cytokine production. CD82-deficiency leads to higher fungal burden.
Tetraspanins play a diverse range of functions through the interaction with membrane proteins (such as integrins, cadherins, metalloproteinases) and intracellular vesicles (10, 13, 56). Indeed, a recent study showed an essential role of CD82 in cell-cell adhesion by downregulating Snail, an E-cadherin repressor; thereby suppressing metastasis in epithelial cancer cells (56). Not only are tetraspanins implicated in cancer, but also in interactions with fungal pathogens. CD37 and CD63 have been shown to interact with fungal PRRs (36, 37), providing insight into the organization of fungal PRR and downstream signaling. Additionally, recruitment of tetraspanins CD63 and CD82 have been reported to macrophage phagosomes containing fungi, such as C. albicans, A. fumigatus, and C. neoformans (24, 40). Although previous reports showed the kinetics of tetraspanin recruitment to the phagosome prior to acidification, there has been no insight into which fungal PRRs specifically associated with tetraspanins. This observation could be due to the complex composition of carbohydrates and proteins on the surface of the fungal cell wall (34). Due to the heterogenous composition of the fungal cell wall, it has been difficult to dissect specific associations between fungal PRRs, tetraspanins and other proteins. To address this problem, we used FLPs coated with discrete, purified fungal carbohydrates (57) to perform proteomic analyses and observed tetraspanin CD82 associated with both β−1,3-glucan and mannan FLPs. These results implicate a specific association with Dectin-1 and CD82.
The association between CD82 and Dectin-1 was further confirmed by co-IP, fluorescent microscopy, phagosome isolation, and PLA. In our co-IP studies, association between CD82 and Dectin-1 is only seen using Brij-58 lysis buffer, a mild detergent, thereby classifying the nature of CD82-Dectin-1 interaction as indirect. While an association was observed in resting macrophages using co-IP, we did not observe an association between CD82 and Dectin-1 on the plasma membrane of resting cells using PLA. This difference could be due to the interaction between CD82 and Dectin-1 is greater than the theoretical maximum distance of 30–40 nm to create a signal with PLA (58). CD82 has been shown to interact with other membrane proteins, including CD63 and class II major histocompatibility complex (40). Furthermore, it has been well demonstrated that tetraspanins interact with each other as well as other transmembrane proteins (i.e. integrins, immunoglobulin, receptors for cytokines and growth factors, etc.) (9, 10, 13, 59). CD82 may act indirectly through these other protein interactions to facilitate Dectin-1 signaling at the phagocytic cup. Indeed, Dectin-1 has been shown to interact with the tetraspanins CD63 (36) and CD37 (37, 38). Deletion of CD37 resulted in reduced Dectin-1 cell surface localization (37). Further investigation into CD82-Dectin-1 interaction with other transmembrane or tetraspanin proteins is warranted. In many conditions, loss of a single tetraspanin fails to reveal a specific phenotype. However, our studies revealed a non-redundant role of CD82 in Dectin-1-mediated signaling in macrophages.
Since CD82 creates signaling domains on the plasma membrane and association between CD82 and Dectin-1 was corroborated using several techniques, we next examined the consequences of CD82 deletion on Dectin-1 and associated clustering. In order for Dectin-1 to signal properly, it must cluster in the “phagocytic synapse” though particulate binding of an organism containing β−1,3-glucan (33). Without Dectin-1 clustering, cells have decreased Syk phosphorylation and ROS generation. While we were unable to investigate individual Dectin-1 clustering using traditional epifluorescence techniques, use of the super-resolution technique STORM showed that CD82 mediates Dectin-1 clustering in the phagocytic synapse for Src and Syk phosphorylation and ROS production. Taken together, these techniques demonstrate an interaction between CD82 and Dectin-1 on the phagosome. Deletion of CD82 from macrophages reduced Dectin-1 signaling events and in turn, increased fungal burden within macrophages. Tetraspanins are implicated in a variety of membrane organization-related processes, therefore future studies should investigate changes in sensitivity to membrane stressors in CD82-deficient cells (10, 13). Although the cardiovascular system, eye, bone and metabolism are impacted with deletion of CD82, mice are viable and have no physiological abnormalities (60–63). Therefore, to demonstrate the relevance of our in vitro findings, we used an in vivo model of systemic candidiasis to show that mice lacking CD82 have enhanced susceptibility to Candida compared to wild-type mice. Furthermore, by incorporating human data, we show that human polymorphisms in CD82 result in modifications of cytokine production and increased susceptibility to candidemia. These data provide critical insight and a novel therapeutic target for the prevention and treatment of Candida infections.
In this study, we show a novel interaction between tetraspanin CD82 and Dectin-1. Here, we show the role of CD82 in the organization of Dectin-1 clustering in the phagocytic cup. Deletion of CD82 disrupts macrophage response to fungal pathogens, such as C. albicans, leading to increased fungal viability and decrease host survival. In support of this hypothesis, we see higher metabolism of C. albicans and higher colony forming units in macrophages. Interestingly, we observed multiple CD82 SNPs in whole blood or PBMC from patients associated with modulation of cytokine production or increased risk of Candida infections. From these findings, we suggest CD82 as a potential new target in modulating the host response to fungal pathogens, including C. albicans, whereby deletion of CD82 reduces the inflammatory response to the pathogen. These results add to the body of literature that illustrates the growing role of tetraspanins in the interaction between the host and fungal pathogens.
Supplementary Material
Key Points:
Dectin-1 clustering via CD82 enables robust downstream Dectin-1 signaling
Lack of CD82 reduces macrophage fungal killing and survival of Candida infection
CD82 SNPs modulate cytokine production or risk of Candida infections
Acknowledgements
We thank Dr. Gordon Brown, Dr. Douglas Golenbock and Dr. Stuart Levitz for reagents and Nicole Wolf for assistance with the artwork. Illustration (Figure 8/visual abstract) by Nicole Wolf, MS, ©2018. Printed with permission. We also thank Hazen Babcock for useful discussions in image analysis for STORM.
Grant support: This work was supported by the National Institutes of Health Grants 5RO1 AI092084 and 1R01 AI097519 (to J.M.V.), 1R01 AI132638 (to M.K.M) T32 A1007061–35 (to J.L.R.), and a KL2/Catalyst Medical Research Investigator Training award (an appointed KL2 award) from Harvard Catalyst | The Harvard Clinical Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health [NIH], award KL2 TR001100) (to J.L.R.), T32 HL116275 (to M.B.F.). M.G.N. was supported by a Spinoza grant of the Netherlands Organization for Scientific Research.
Abbreviations used in article:
- BMDM
bone marrow-derived macrophages
- CFU
colony forming units
- FLP
fungal-like particles
- HKCA
heat-killed Candida albicans
- IP
immunoprecipitation
- PLA
proximity ligation assay
- PRR
pattern-recognition receptor
- ROS
reactive oxygen species
- SNP
single-nucleotide polymorphism
- STORM
stochastic optical reconstruction microscopy
- Syk
spleen tyrosine kinase
- TEM
tetraspanin-enriched microdomains
- WT
wild-type
References
- 1.Menzin J, Meyers JL, Friedman M, Korn JR, Perfect JR, Langston AA, Danna RP, and Papadopoulos G. 2011. The economic costs to United States hospitals of invasive fungal infections in transplant patients. American journal of infection control 39: e15–20. [DOI] [PubMed] [Google Scholar]
- 2.Miceli MH, Diaz JA, and Lee SA. 2011. Emerging opportunistic yeast infections. The Lancet. Infectious diseases 11: 142–151. [DOI] [PubMed] [Google Scholar]
- 3.Lanternier F, Cypowyj S, Picard C, Bustamante J, Lortholary O, Casanova JL, and Puel A. 2013. Primary immunodeficiencies underlying fungal infections. Current opinion in pediatrics 25: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderone RA, and Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol 9: 327–335. [DOI] [PubMed] [Google Scholar]
- 5.Nobile CJ, and Johnson AD. 2015. Candida albicans Biofilms and Human Disease. Annu Rev Microbiol 69: 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, and Castanheira M. 2016. Nosocomial Candidiasis: Antifungal Stewardship and the Importance of Rapid Diagnosis. Medical mycology 54: 1–22. [DOI] [PubMed] [Google Scholar]
- 7.Pfaller MA, and Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36: 1–53. [DOI] [PubMed] [Google Scholar]
- 8.van Spriel AB, and Figdor CG. 2010. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect 12: 106–112. [DOI] [PubMed] [Google Scholar]
- 9.Charrin S, Jouannet S, Boucheix C, and Rubinstein E. 2014. Tetraspanins at a glance. Journal of cell science 127: 3641–3648. [DOI] [PubMed] [Google Scholar]
- 10.Charrin S, le Naour F, Silvie O, Milhiet P-E, Boucheix C, and Rubinstein E. 2009. Lateral organization of membrane proteins: tetraspanins spin their web. Biochemical Journal 420: 133–154. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, and Hemler ME. 2002. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell 13: 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berditchevski F, Odintsova E, Sawada S, and Gilbert E. 2002. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. The Journal of biological chemistry 277: 36991–37000. [DOI] [PubMed] [Google Scholar]
- 13.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, and Sanchez-Madrid F. 2009. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 19: 434–446. [DOI] [PubMed] [Google Scholar]
- 14.Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, and Barrett JC. 1995. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 268: 884–886. [DOI] [PubMed] [Google Scholar]
- 15.Iwata S, Kobayashi H, Miyake-Nishijima R, Sasaki T, Souta-Kuribara A, Nori M, Hosono O, Kawasaki H, Tanaka H, and Morimoto C. 2002. Distinctive signaling pathways through CD82 and beta1 integrins in human T cells. European journal of immunology 32: 1328–1337. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q, Yang Y, Wu S, Li W, Zhang N, Dong X, and Ou Y. 2015. Evaluation of the correlation of KAI1/CD82, CD44, MMP7 and beta-catenin in the prediction of prognosis and metastasis in colorectal carcinoma. Diagnostic pathology 10: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi M, Taki T, Ieki Y, Huang CL, Higashiyama M, and Miyake M. 1996. Correlation of KAI1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer research 56: 1751–1755. [PubMed] [Google Scholar]
- 18.Sho M, Adachi M, Taki T, Hashida H, Konishi T, Huang C.-l., Ikeda N, Nakajima Y, Kanehiro H, Hisanaga M, Nakano H, and Miyake M. 1998. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. International Journal of Cancer 79: 509–516. [DOI] [PubMed] [Google Scholar]
- 19.Huang CI, Kohno N, Ogawa E, Adachi M, Taki T, and Miyake M. 1998. Correlation of reduction in MRP-1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am J Pathol 153: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houle CD, Ding XY, Foley JF, Afshari CA, Barrett JC, and Davis BJ. 2002. Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol Oncol 86: 69–78. [DOI] [PubMed] [Google Scholar]
- 21.Kropshofer H, Spindeldreher S, Rohn TA, Platania N, Grygar C, Daniel N, Wolpl A, Langen H, Horejsi V, and Vogt AB. 2002. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nature immunology 3: 61–68. [DOI] [PubMed] [Google Scholar]
- 22.Hammond C, Denzin LK, Pan M, Griffith JM, Geuze HJ, and Cresswell P. 1998. The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J Immunol 161: 3282–3291. [PubMed] [Google Scholar]
- 23.Poloso NJ, Denzin LK, and Roche PA. 2006. CDw78 defines MHC class II-peptide complexes that require Ii chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J Immunol 177: 5451–5458. [DOI] [PubMed] [Google Scholar]
- 24.Artavanis-Tsakonas K, Kasperkovitz PV, Papa E, Cardenas ML, Khan NS, Van der Veen AG, Ploegh HL, and Vyas JM. 2011. The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infection and immunity 79: 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown GD, and Gordon S. 2001. Immune recognition. A new receptor for beta-glucans. Nature 413: 36–37. [DOI] [PubMed] [Google Scholar]
- 26.Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, Akira S, Adachi Y, Ohno N, Kinjo T, Nakamura K, Kawakami K, and Iwakura Y. 2007. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nature immunology 8: 39–46. [DOI] [PubMed] [Google Scholar]
- 27.Sancho D, and Reis e Sousa C. 2012. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annual review of immunology 30: 491–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Netea MG, and Marodi L. 2010. Innate immune mechanisms for recognition and uptake of Candida species. Trends in immunology 31: 346–353. [DOI] [PubMed] [Google Scholar]
- 29.Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D’Angelo C, Pierini A, Pitzurra L, Falzetti F, Carotti A, Perruccio K, Latge JP, Rodrigues F, Velardi A, Aversa F, Romani L, and Carvalho A. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 116: 5394–5402. [DOI] [PubMed] [Google Scholar]
- 30.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morre SA, Vriend G, Williams DL, Perfect JR, Joosten LA, Wijmenga C, van der Meer JW, Adema GJ, Kullberg BJ, Brown GD, and Netea MG. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. The New England journal of medicine 361: 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodridge HS, Shimada T, Wolf AJ, Hsu YM, Becker CA, Lin X, and Underhill DM. 2009. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J Immunol 182: 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, and Reis e Sousa C. 2005. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 22: 507–517. [DOI] [PubMed] [Google Scholar]
- 33.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, and Underhill DM. 2011. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 472: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill DM, Rossnagle E, Lowell CA, and Simmons RM. 2005. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood 106: 2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond RA, Saijo S, Iwakura Y, and Brown GD. 2011. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. European journal of immunology 41: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, Teillaud JL, and Mordoh J. 2004. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood 104: 1183–1190. [DOI] [PubMed] [Google Scholar]
- 37.Meyer-Wentrup F, Figdor CG, Ansems M, Brossart P, Wright MD, Adema GJ, and van Spriel AB. 2007. Dectin-1 interaction with tetraspanin CD37 inhibits IL-6 production. J Immunol 178: 154–162. [DOI] [PubMed] [Google Scholar]
- 38.van Spriel AB, Sofi M, Gartlan KH, van der Schaaf A, Verschueren I, Torensma R, Raymakers RA, Loveland BE, Netea MG, Adema GJ, Wright MD, and Figdor CG. 2009. The tetraspanin protein CD37 regulates IgA responses and anti-fungal immunity. PLoS Pathog 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, and Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature immunology 9: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artavanis-Tsakonas K, Love JC, Ploegh HL, and Vyas JM. 2006. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc Natl Acad Sci U S A 103: 15945–15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansour MK, Tam JM, Khan NS, Seward M, Davids PJ, Puranam S, Sokolovska A, Sykes DB, Dagher Z, Becker C, Tanne A, Reedy JL, Stuart LM, and Vyas JM. 2013. Dectin-1 activation controls maturation of beta-1,3-glucan-containing phagosomes. The Journal of biological chemistry 288: 16043–16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagag A, Jault JM, Sidahmed-Adrar N, Refregiers M, Giuliani A, and Le Naour F. 2013. Characterization of hydrophobic peptides in the presence of detergent by photoionization mass spectrometry. PLoS One 8: e79033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagag A, Giuliani A, Canon F, Refregiers M, and Le Naour F. 2011. Separation of peptides from detergents using ion mobility spectrometry. Rapid communications in mass spectrometry : RCM 25: 3436–3440. [DOI] [PubMed] [Google Scholar]
- 44.Bates M, Jones SA, and Zhuang X. 2013. Stochastic optical reconstruction microscopy (STORM): a method for superresolution fluorescence imaging. Cold Spring Harbor protocols 2013: 498–520. [DOI] [PubMed] [Google Scholar]
- 45.Levet F, Hosy E, Kechkar A, Butler C, Beghin A, Choquet D, and Sibarita JB. 2015. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat Methods 12: 1065–1071. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, Deelen P, Ricano-Ponce I, Schoffelen T, Jansen AFM, Swertz MA, Withoff S, van de Vosse E, van Deuren M, van de Veerdonk F, Zhernakova A, van der Meer JWM, Xavier RJ, Franke L, Joosten LAB, Wijmenga C, Kumar V, and Netea MG. 2016. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell 167: 1099–1110.e1014. [DOI] [PubMed] [Google Scholar]
- 47.Ward LD, and Kellis M. 2012. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research 40: D930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar V, Cheng SC, Johnson MD, Smeekens SP, Wojtowicz A, Giamarellos-Bourboulis E, Karjalainen J, Franke L, Withoff S, Plantinga TS, van de Veerdonk FL, van der Meer JWM, Joosten LAB, Bochud PY, Marchetti O, Perfect JR, Xavier R, Kullberg BJ, Wijmenga C, and Netea MG. 2014. Immunochip SNP array identifies novel genetic variants conferring susceptibility to candidaemia. Nature communications 5: 4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matzaraki V, Gresnigt MS, Jaeger M, Ricano-Ponce I, Johnson MD, Oosting M, Franke L, Withoff S, Perfect JR, Joosten LAB, Kullberg BJ, van de Veerdonk FL, Jonkers I, Li Y, Wijmenga C, Netea MG, and Kumar V. 2017. An integrative genomics approach identifies novel pathways that influence candidaemia susceptibility. PLoS One 12: e0180824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tam JM, Mansour MK, Khan NS, Seward M, Puranam S, Tanne A, Sokolovska A, Becker CE, Acharya M, Baird MA, Choi AM, Davidson MW, Segal BH, Lacy-Hulbert A, Stuart LM, Xavier RJ, and Vyas JM. 2014. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis 210: 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, Ye P, Morales F, Kocks C, Bader JS, Desjardins M, and Ezekowitz RA. 2007. A systems biology analysis of the Drosophila phagosome. Nature 445: 95–101. [DOI] [PubMed] [Google Scholar]
- 52.Claas C, Stipp CS, and Hemler ME. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. The Journal of biological chemistry 276: 7974–7984. [DOI] [PubMed] [Google Scholar]
- 53.Schuck S, Honsho M, Ekroos K, Shevchenko A, and Simons K. 2003. Resistance of cell membranes to different detergents. Proceedings of the National Academy of Sciences 100: 5795–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler RT, and Fink GR. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagchi S, Fredriksson R, and Wallen-Mackenzie A. 2015. In Situ Proximity Ligation Assay (PLA). Methods in molecular biology (Clifton, N.J.) 1318: 149–159. [DOI] [PubMed] [Google Scholar]
- 56.Lee MS, Byun HJ, Lee J, Jeoung DI, Kim YM, and Lee H. 2018. Tetraspanin CD82 represses Sp1-mediated Snail expression and the resultant E-cadherin expression interrupts nuclear signaling of beta-catenin by increasing its membrane localization. Cell Signal 52: 83–94. [DOI] [PubMed] [Google Scholar]
- 57.Tam JM, Mansour MK, Khan NS, Yoder NC, and Vyas JM. 2012. Use of fungal derived polysaccharide-conjugated particles to probe Dectin-1 responses in innate immunity. Integrative biology : quantitative biosciences from nano to macro 4: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, and Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. [DOI] [PubMed] [Google Scholar]
- 59.Boucheix C, and Rubinstein E. 2001. Tetraspanins. Cellular and molecular life sciences : CMLS 58: 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Risinger JI, Custer M, Feigenbaum L, Simpson RM, Hoover SB, Webster JD, Chandramouli GV, Tessarollo L, and Barrett JC. 2014. Normal viability of Kai1/Cd82 deficient mice. Molecular carcinogenesis 53: 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Q, Zhang F, Richardson MM, Roy NH, Rodgers W, Liu Y, Zhao W, Fu C, Ding Y, Huang C, Chen Y, Sun Y, Ding L, Hu Y, Ma JX, Boulton ME, Pasula S, Wren JD, Tanaka S, Huang X, Thali M, Hammerling GJ, and Zhang XA. 2014. CD82 restrains pathological angiogenesis by altering lipid raft clustering and CD44 trafficking in endothelial cells. Circulation 130: 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uchtmann K, Park ER, Bergsma A, Segula J, Edick MJ, and Miranti CK. 2015. Homozygous loss of mouse tetraspanin CD82 enhances integrin alphaIIbbeta3 expression and clot retraction in platelets. Exp Cell Res 339: 261–269. [DOI] [PubMed] [Google Scholar]
- 63.Bergsma A, Ganguly SS, Dick D, Williams BO, and Miranti CK. 2018. Global deletion of tetraspanin CD82 attenuates bone growth and enhances bone marrow adipogenesis. Bone 113: 105–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.