Abstract
Background:
Corticotropin-releasing hormone (CRH) plays a central role in regulating the secretion of cortisol which controls a wide range of biological processes. Fetuses overexposed to cortisol have increased risks of disease in later life. DNA methylation may be the underlying association between prenatal cortisol exposure and health effects. We investigated associations between maternal CRH levels and epigenome-wide DNA methylation of cord blood in offsprings and evaluated whether these associations persisted into mid-childhood.
Methods:
We investigated mother-child pairs enrolled in the prospective Project Viva pre-birth cohort. We measured DNA methylation in 257 umbilical cord blood samples using the HumanMethylation450 Bead Chip. We tested associations of maternal CRH concentration with cord blood cells DNA methylation, adjusting the model for maternal age at enrollment, education, maternal race/ethnicity, pre-pregnancy body mass index, parity, gestational age at delivery, child sex, and cell-type composition in cord blood. We further examined the persistence of associations between maternal CRH levels and DNA methylation in children’s blood cells collected at mid-childhood (N = 239, age: 6.7–10.3 years) additionally adjusting for the children’s age at blood drawn.
Results:
Maternal CRH levels are associated with DNA methylation variability in cord blood cells at 96 individual CpG sites (False Discovery Rate < 0.05). Among the 96 CpG sites, we identified 3 CpGs located near the LEP gene. Regional analyses confirmed the association between maternal CRH and DNA methylation near LEP. Moreover, higher maternal CRH levels were associated with higher blood-cell DNA methylation of the promoter region of LEP in mid-childhood (P < 0.05, β = 0.64, SE = 0.30).
Conclusion:
In our cohort, maternal CRH was associated with DNA methylation levels in newborns at multiple loci, notably in the LEP gene promoter. The association between maternal CRH and LEP DNA methylation levels persisted into mid-childhood.
Keywords: corticotropin-releasing hormone, DNA methylation, cord blood, childhood, LEP
Background
Corticotropin-releasing hormone (CRH), a peptide that is synthesized and released from the hypothalamus, is the main central regulator in the hypothalamic-pituitary-adrenal (HPA) axis 1. During pregnancy, maternal CRH in the bloodstream is largely derived from placenta secretion and triggers secretion of maternal cortisol that activates more release of CRH from placenta 2. This feed-forward loop leads to an increase in maternal CRH 3 and a two- to four-fold higher maternal cortisol secretion in normal pregnancy 4. Placental enzymes only partially inactivate maternal cortisol, and a large amount of active cortisol are passed to the fetus 5. Fetal cortisol levels are positively correlated with maternal cortisol levels 6, and thus, maternal plasma CRH levels during pregnancy reflect the intensity of fetal cortisol exposure.
Fetal cortisol exposure leads to both beneficial and detrimental consequences: cortisol is necessary for fetal organ maturation (e.g., lung, kidney, heart) 7; however, in humans, excessively high levels of cortisol have been associated with preterm birth 6, small for gestational age 8, HPA axis dysfunction 9, and with obesity 10 and metabolic syndromes 11 later in life. Animal studies also showed adverse effects of excess cortisol that prenatal synthetic glucocorticoids treatments modify HPA axis function and increase the risk of obesity and diabetes in the offspring 12–15. These studies support the hypothesis that prenatal cortisol exposure – regulated by maternal CRH – contributes to the fetal growth and long-term development. Additionally, previous studies in our cohort have demonstrated that higher maternal CRH is associated with a higher risk of central adiposity and adiponectin levels in early childhood 16, but lower BMI in offspring 10.
DNA methylation is regarded as an important mechanism by which early life exposures may lead to long-term programming, sometimes leading to structural and functional changes that can persist later in life and have long-term effects 17,18. Both human and animal studies found DNA methylation modifications at HPA-axis genes (e.g., NR3C1, NR3C2, CRH, POMC, HSD11B2β) and modified HPA-axis function in the offspring whose mother were treated with synthetic glucocorticoids during gestation19,20. However, no prior study investigated the associations between maternal CRH and offspring DNA methylation in humans using an epigenome-wide approach.
We hypothesized that maternal CRH is associated with DNA methylation levels in cord blood, and some of the associations would last into mid-childhood. We investigated associations between maternal CRH levels and DNA methylation in cord blood cells in >450,000 sites across the genome and evaluated whether these associations persist into mid-childhood.
Methods
Study population
We recruited pregnant women into Project Viva during their first prenatal visit (initially 2128 mother-child pairs at median 9.9 gestation weeks) at eastern Massachusetts Atrius Harvard Vanguard Medical Associates practices, between 1999 and 2002. Details of this prospective pre-birth cohort have been described previously 21. We included 257 mother-child pairs in the cord blood analysis, and we included 239 mother-child pairs in mid-childhood (range: 6.7–10.3 years) DNA methylation analysis, according to availability of maternal CHR measurement and DNA methylation data that passed our sample quality control (Figure S1). We analyzed 134 mother-child pairs who had all data available at both time-points (Figure S2). All mothers provided written informed consent. The research was approved by the Institutional Review Board at Harvard Pilgrim Health Care.
Maternal corticotropin-releasing hormone (CRH) measurement
Collection of maternal blood samples, CRH measurement and data processing in this cohort has been previously described 22. Briefly, we assayed for CRH with an RIA assay kit (Peninsula Laboratories Inc., San Carlos, CA) following the manufacturer’s instructions. To eliminate bias introduced by batch effect and gestational age at CRH test, we conducted a two-stage correction for the original CRH measurements. In the first stage, we adjusted CRH levels for batch effects (adjusted CRH = e(ln(original CRH) + 0.48)). In the second stage, we additionally adjusted CRH levels for gestational age at maternal blood drawn 10 (Figure S3). Henceforward, maternal CRH refers to the fully two-stage adjusted maternal CRH.
DNA methylation assessment
Collection, processing, and storage of cord blood samples and children’s peripheral blood in mid-childhood has been previously described 23. Trained laboratory staff extracted DNA from blood samples using commercially available PureGene Kits (Fisher, catalog #: A407–4, A416–4; Qiagen, catalog #: 158908, 158912, 158924) and conducted sodium bisulfite conversion on DNA using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). We measured DNA methylation across the genome using the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) following standard manufacturer’s protocols. To reduce the influence of batch effect and to ensure the balance of sex, samples were randomly allocated to assay chips, and chips were randomly assigned to plates.
Quality control
The DNA methylation data processing is as previously described 23. We excluded 65 actual SNPs, 11648 probes on sex chromosomes, 2994 non-CpG probes, 8715 probes with poor detection (P > 0.05) for more than 1% samples, 27951 cross-reactive probes 24 and 99224 SNP-containing probes (Figure S4). We included a total of 334980 out of 485577 CpGs in the analysis. For each CpG site, methylation level is represented by the average β-value = M/ (M+ U+ ε), where M and U represent the average fluorescence intensity corresponding to the methylated and unmethylated target CpG while ε usually equals 100. The average β-value, ranging between 0 and 1, is interpreted as the proportion CpG methylated at each site. β-scale DNA methylation levels were logit transformed to M-values (M = log2(β) – log2(1 – β)) for a distribution that was more appropriate for the analysis of DNA methylation. The batch effect was not associated with the first 30 principal components of DNA methylation variance in cord blood (Figure S5) and children’s blood (plot not shown) after we performed the batch effect adjustment using ComBat approach, which indicated adequate removal of batch effects. We used ComBat to adjust for batch effect (details are described in the Supplementary materials).
Covariates
Using self-administered questionnaires and medical record data, we collected information of maternal age at enrollment, education level, race/ethnicity, smoking status, parity, pre-pregnancy body mass index (BMI), last menstrual period; and children’s sex and birth weight. We estimated gestational age at delivery by subtracting the LMP date from the delivery date or by the ultrasound if the ultrasound estimation were available and differed from the LMP estimation by more than ten days. Based on U.S. national reference data 25, we calculated sex-specific birth weight-for-gestational age z-score using data on birth weight, gestational age, and child sex. We estimated the nucleated cell-type compositions (CD8 T-cells, CD4 T-cells, granulocytes, natural killer cells, B-cells, monocytes, and nucleated red blood cells) in the cord blood using a validated reference panel 26. We estimated the cell-type compositions (CD8 T-cells, CD4 T-cells, granulocytes, natural killer cells, B-cells and monocytes) in the children’s peripheral blood in mid-childhood using adult leukocyte reference panel for blood samples as implemented in minfi 27.
Leptin and body composition measurements
We measured leptin concentration in cord blood and plasma of children’s blood in mid-childhood using a radioimmunoassay (Linco Research Inc, St Charles, MO), as described previously 28. We used log-transformed leptin concentration and removed outliers. We included 235 cord blood samples and 225 children’s blood samples in our analyses. At mid-childhood, we measured children’s height (cm), weight (kg), waist and hip circumferences (cm), subscapular (SS) and triceps (TR) skinfold thicknesses (mm), as described previously 29. We calculated age-sex-adjusted BMI z-score based on the U.S. national data 30, the sum of skinfold thicknesses (SS + TR) and waist-to-hip circumference ratios.
Statistical Analysis
We used mean [standard deviations (SD)], median (IQR) or percentages of covariates to describe characteristics of participants at birth and in mid-childhood. To test for individually differentially methylated CpG sites in relation to maternal CRH levels, we fit robust linear regression models for each CpG site on the M-value scale, adjusting for potential confounders including maternal age at enrollment, education, maternal race/ethnicity, maternal smoking status (never/former/current), pre-pregnancy BMI, parity, gestational age at delivery, child sex and cord blood cell-type estimation (CD8 T-cells, CD4 T-cells, natural killer cells, B-cells, monocytes, and nucleated red blood cells), using the MASS package in R. After inclusion of cell-type estimation and other potential confounders, the genomic inflation factor (λ) for the Epigenome-Wide Analysis was 1.03, indicating that results were unlikely driven by population stratification or cryptic relatedness (Figure S6). The statistical significance of the CpG-by-CpG analysis was evaluated using an FDR adjusted level (q < 0.05). To ease biological interpretation, we presented the adjusted regression coefficient of maternal CRH on DNA methylation on the β-value scale. We also conducted a pathway analysis on the CpG sites passing the FDR using the ‘gometh’ function in ‘missMethyl’ package in R.
Subsequently, we performed a differentially methylated regional DNA methylation analysis using the DMRcate 31 package of R to examine the association between maternal CRH and differentially methylated regions (DMRs) in cord blood. We used a Gaussian kernel smoothing function to test statistics grouping significant CpG sites with bandwidth λ = 300 base pairs and scaling factor C = 2. Significance testing among DMRs was adjusted for multiple comparisons using a Stouffer adjusted P < 0.05. We visualized the DMRs and their neighborhood regions using the coMET 32 in R.
Furthermore, we explored whether the associations of maternal CRH with differentially methylated CpG sites and regions in cord blood were persistent into mid-childhood using robust linear regression among our top candidate CpGs identified in our cord blood analyses. We regarded the mean methylation across all CpGs in the DMR as the regional methylation level, since the DNA methylation of CpGs in the DMR found in the present study were significantly correlated with each other. We adjusted robust linear regression models with maternal age at enrollment, education, maternal race/ethnicity, pre-pregnancy BMI, parity, child sex, cell-type estimations and the child’s age at blood collection for DNA methylation test. We considered associations statistically significant for persistence in childhood when the P < 0.05 for the one region we had found in cord blood DMR analyses, and we used FDR to correct for number of CpG sites found in the cord blood CpG-by-CpG analyses.
Additionally, we performed two sensitivity analyses to test for bias caused by prenatal steroid treatments and preterm birth. All analyses were carried out using the R software, version 3.4.1.
Code availability
Code is available upon request.
Results
Characteristics of participants
Table 1 displays characteristics of mother-child pairs. In the participants included in the cord blood methylation analyses, the log2-transformed fully adjusted CRH levels ranged between 4.3 – 9.7. Gestational age at blood draw for the CRH test ranged between 25 – 34 gestational weeks. In the primary analysis of mother-child pairs with cord blood DNA methylation, a majority of mothers were white (73%), multiparous (55%), never smoked (68%), and college graduate (67%). Characteristics of participants included in analyses of DNA methylation measured in mid-childhood were similar to those of participants included in the cord blood analyses.
Table 1.
Characteristics of mothers and children included in analyses at birth and in mid-childhood.
| Characteristics | Cord blood | Middle childhood |
|---|---|---|
| N = 257 | N = 239 | |
| Mothers | ||
| Maternal age at enrollment, years: Mean (SD) | 32.1 (5.3) | 32.2 (5.3) |
| Pre-pregnancy BMI, kg/m2: Mean (SD) | 24.4 (4.9) | 24.7 (5.3) |
| Mothers’ race/ethnicity: N (%) | ||
| . White | 189 (73) | 162 (68) |
| . Black | 29 (11) | 41 (17) |
| . Hispanic | 21 (8) | 15 (6) |
| . Other | 18 (7) | 21 (9) |
| Nulliparous: N (%) | 115 (45) | 106 (44) |
| >= College graduate: N (%) | 173 (67) | 166 (70) |
| Smoking status: N (%) | ||
| . Never | 176 (68) | 169 (71) |
| . During pregnancy | 26 (10) | 24 (10) |
| . Former | 55 (21) | 46 (19) |
| Log2-transformed CRH corrected for technician and gestational age at CRH test, Mean (SD) | 7.1 (0.8) | 7.1 (0.9) |
| Gestational age at CRH test, weeks: Mean (SD) | 27.9 (1.3) | 27.9 (1.7) |
| Children | ||
| Gestational age at delivery, weeks: Mean (SD) | 39.7 (1.3) | 39.6 (1.4) |
| Birth weight, g: Mean (SD) | 3559.1 (514.4) | 3554.3 (528.4) |
| Birth weight (z-score)a: Mean (SD) | 0.3 (1.0) | 0.3 (1.0) |
| Age at blood samples collection | ||
| . At birth (gestational age in weeks) | 39.7 (1.3) | - |
| . In middle childhood (6.7 to 10.3 years) | - | 7.9 (0.8) |
| Female: N (%) | 126 (49) | 115 (48) |
| Children’s race/ethnicity: N (%) | ||
| . White | 178 (69) | 153 (64) |
| . Black | 34 (13) | 45 (19) |
| . Hispanic | 14 (5) | 11 (5) |
| . Other | 31 (12) | 30 (13) |
| BMI z-score: Mean (SD), n = 236 | - | 0.40 (1.3) |
| Skinfold thickness (mm): Median (IQR), n = 236 | - | 16.4 (8.7) |
| Waist-hip ratio: Median (IQR), n = 237 | - | 0.9 (0.1) |
BMI, body mass index. CRH, corticotropin-releasing hormone.
Birth weight adjusted for gestational age and child sex using U.S. national reference
Genome-wide CpG-by-CpG analysis in cord blood
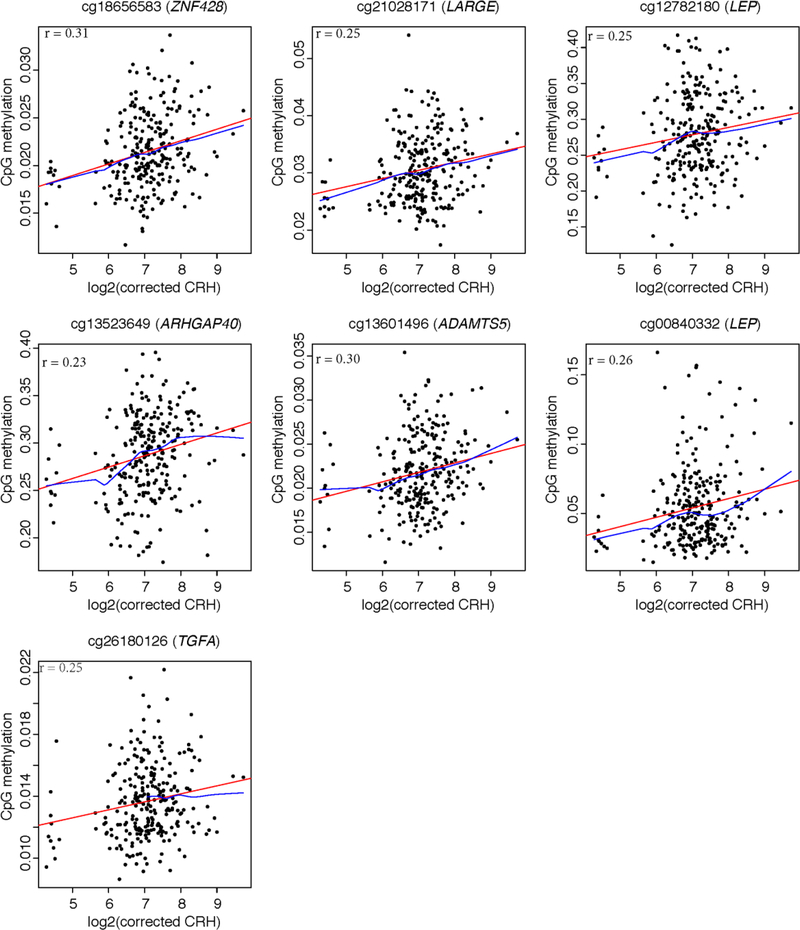
In robust linear regression models for individual CpG sites, we found 96 differentially methylated CpG sites (FDR < 0.05) in cord blood relative to maternal CRH levels (Table S1). We showed the 7 CpG sites with the strongest associations that also reached Bonferroni adjusted significant level (P < 1.49 × 10−7) in Table 2. The percent changes in cord blood DNA methylation per doubling in maternal CRH concentration (adjusted regression coefficient) ranged between 0.09% - 1.74% among the top 7 CpG sites and between 0.03% - 2.50% in absolute value among the overall 96 CpG sites. Among the 96 CpGs, cg12782180 (P = 1.32× 10−08), cg00840332 (P = 1.24 × 10−07), cg19594666 (P = 3.82 ×10−07) were annotated to leptin (LEP) gene (region within <1500 base pairs from the transcription start site (TSS)), and had 1.74%, 0.81% and 1.86% increase in the DNA methylation levels in cord blood per doubling in maternal CRH concentration.
Table 2.
CpG sites with differential DNA methylation at birth (257 cord blood samples) in relation to maternal CRH level during pregnancy (reaching P < 1.49 × 10−7).
| CpGs | Mean (SD) | β-Coefficient (SE)a % | P b | FDR b | Position c | Island c | Gene c | Region c |
|---|---|---|---|---|---|---|---|---|
| cg26180126 | 0.01 (0.00) | 0.09 (0.02) | 1.43 × 10−7 | 0.01 | chr2: 70779764 | N_Shore | TGFA | Body |
| cg12782180 | 0.28 (0.06) | 1.74 (0.31) | 1.32 × 10−8 | 1.47 × 10−3 | chr7: 127880932 | Island | LEP | TSS1500 |
| cg00840332 | 0.05 (0.03) | 0.81 (0.17) | 1.24 × 10−7 | 0.01 | chr7: 127881269 | Island | LEP | TSS200 |
| cg18656583 | 0.02 (0.00) | 0.14 (0.02) | 1.37 × 10−10 | 4.57 × 10−5 | chr19: 44124056 | Island | ZNF428 | TSS200 |
| cg13523649 | 0.29 (0.04) | 1.42 (0.26) | 8.79 × 10−8 | 0.01 | chr20: 37230741 | Island | ARHGAP40 | Body |
| cg13601496 | 0.02 (0.00) | 0.15 (0.03) | 1.20 × 10−7 | 0.01 | chr21: 28339487 | Island | ADAMTS5 | TSS200 |
| cg21028171 | 0.03 (0.01) | 0.19 (0.03) | 8.73 × 10−10 | 1.46 × 10−4 | chr22: 34316977 | Island | LARGE | TSS1500 |
Chr, chromosome; UTR, untranslated region; TSS, transcription start site. SD, standard deviation. SE, standard error of coefficient.
The coefficients are estimated according to the robust linear regression on beta value scale, adjusted for maternal age at enrollment, maternal education level, maternal race, maternal smoking status, pre-pregnant BMI, parity, gestational age at delivery, child sex, and the estimated proportions of CD8 T-cell, CD4 T-cell, natural killer cells, B-cells, monocytes, and red blood cells in cord blood. The coefficient of each CpG site represents the percent change in cord blood DNA methylation in relation to per doubling increased maternal CRH level during pregnancy.
The P values and FDRs are estimated according to the robust linear regression on M-value scale, adjusted for the same covariates with the model on beta value scale.
The annotation information are according to the R package of “IlluminaHumanMethylation450kanno.ilmn12.hg19”.
Among our top 7 differential CpG sites that reached Bonferroni adjusted significant level, we found 2 CpG sites annotated to LEP and the others were annotated to the following 5 genes: ZNF428, LARGE, ARHGAP40, ADAMTS5, TGFA. At all 7 Bonferroni significant CpG sites, greater maternal CRH was associated with higher methylation levels (Figure 1). Similarly, methylation of the majority of 96 CpG sites (FDR < 0.05) were positively correlated with maternal CRH level, such as cg07380705 (CRHBP), cg16527491 (FOXA1), cg22937891 (TRIM36), cg13641043 (PLCD3), cg01078147 (SEMA6A), cg07891473 (ADAMTS9), cg15558129 (LYPLAL1), and only eight show lower methylation levels associated with greater maternal CRH (Table S1).
Figure 1.
Scatterplots for the associations between differentially methylated CpGs (Bonferroni p < 1.49 ×10−7) and maternal corticotropin-releasing hormone level (log2-transformed CRH corrected for technician and gestational age at CRH test) during pregnancy. The red regression line was fitted with simple linear regression. The blue was non-parametric regression line with locally weighted smoothing. The Pearson correlation coefficient of each CpG site resulted from partial correlation analysis adjusted for maternal age, education, maternal race/ethnicity, gestational age, pre-pregnancy BMI, parity, child sex and cell-type estimations.
The sensitivity analyses show that our results were robust: effect sizes of associations between maternal CRH and methylation at all 96 FDR significant CpG sites were very similar in effect size and statistical significance values to our primary analyses after excluding subjects who had prenatal steroid treatment or were born at gestational age less than 37 weeks.
We conducted pathway analysis on the 96 CpGs passing FDR in the CpG-by-CpG analysis, but did not observe any GO pathways passing 5% FDR threshold. The top 10 GO terms were shown in Table S2.
Differentially Methylated Regions
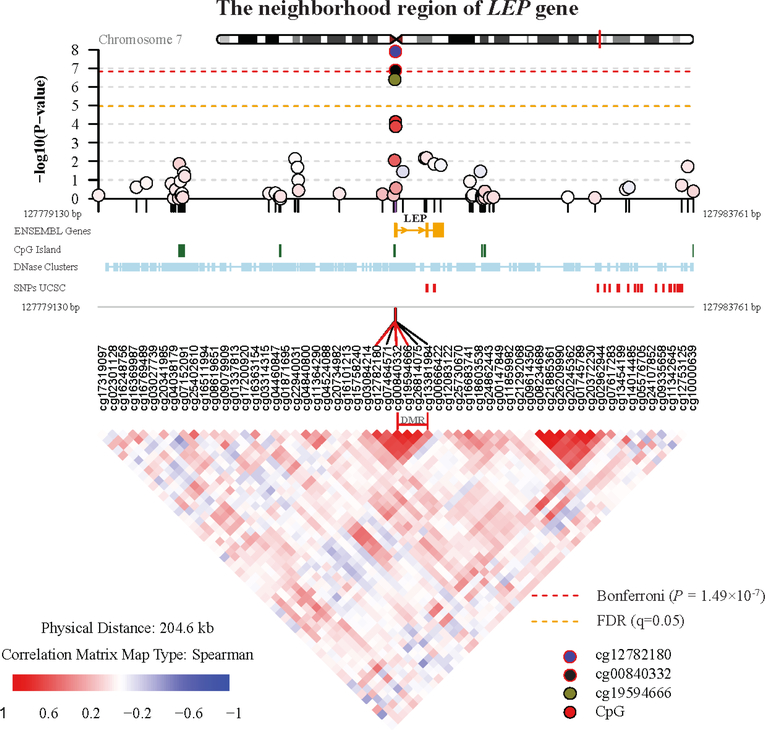
In regional DNA methylation analysis, we detected one differentially methylated region (DMR) in relation to maternal CRH exposure (Stouffer adjusted P = 0.047). This region consists of 4 consecutive CpG sites (cg00840332, cg19594666, cg26814075, cg13381984) annotated to LEP gene on chromosome 7 (genomic coordinate: chr7:127881269–127881344). This region includes two of the FDR-significant CpG sites (cg00840332, cg19594666) that we identified in CpG-by-CpG analysis. DNA methylation of each CpG site within the DMR of LEP was associated with 0.81% to 1.86% higher DNA methylation per doubling in maternal CRH concentration; the overall mean methylation of CpG sites across this region was 1.46% higher per doubling in maternal CRH level (P = 3.45 × 10−7) (Table 3). Figure 2 displays the P-value distribution of CpG sites in DMR of LEP gene and its neighborhood genomic region (upper panel), annotation of the region (middle panel) and the correlations among the CpG sites (lower panel). The methylation levels of the 4 CpG sites within the DMR of LEP were strongly and positively correlated with each other in cord blood cells (r = 0.65–0.90, P < 2.2×10−16) and mid-childhood blood cells (r = 0.66–0.91, P < 2.2×10−16); thus, we used the mean methylation of the 4 CpG sites in subsequent analyses to represent the methylation level of this region. Figure S7 displays the DNA methylation of individual subjects for the 4 CpG sites in LEP region as well as the other 6 CpG sites passing the Bonferroni significant level in CpG-by-CpG analysis. In the sex-specific analysis (Table S3), the mean levels of DNA methylation at the LEP region in cord blood were similar in males and females; moreover, the associations between maternal CRH and LEP methylation were also significant in both females and males.
Table 3.
DNA methylation of CpG sites in the differentially methylated region of LEP (chr7:127880932–127881440) at birth (cord blood) and at mid–childhood (blood) in relation to maternal CRH level during pregnancy.
| CpG | Position | Region | Island | At birth (cord blood) (N = 257) | Mid-childhood (N = 239) | ||
|---|---|---|---|---|---|---|---|
| β-Coefficient (SE)a % | P b | β-Coefficient (SE)a % | P b | ||||
| Mean DNA methylation of this region | 1.46 (0.29) | 3.45 × 10−7 | 0.64 (0.30) | 0.03 | |||
| cg00840332 | 127881269 | TSS200 | island | 0.81 (0.17) | 1.24 × 10−7 | 0.55 (0.24) | 0.02 |
| cg19594666 | 127881280 | TSS200 | island | 1.86 (0.38) | 3.82 × 10−7 | 0.70 (0.43) | 0.10 |
| cg26814075 | 127881298 | TSS200 | island | 1.51 (0.38) | 7.29 × 10−5 | 0.66 (0.35) | 0.06 |
| cg13381984 | 127881344 | 1stExon | island | 1.48 (0.38) | 1.28 × 10−4 | 0.71 (0.34) | 0.03 |
TSS, transcription start site. SD, standard deviation. SE, standard error of coefficient.
The coefficients are estimated according to the robust linear regression on beta value scale, adjusted for maternal age at enrollment, maternal education level, maternal race, maternal smoking status, pre-pregnant BMI, parity, gestational age at delivery, child sex, and the estimated proportions of CD8 T-cell, CD4 T-cell, natural killer cells, B-cells, monocytes, and red blood cells (cord blood only), age of children in years at the time of the blood draw for DNA methylation test (childhood only). The coefficient of each CpG site represents the percent change in DNA methylation in relation to per doubling increased maternal CRH level during pregnancy.
The P values are estimated according to the robust linear regression on M-value scale, adjusted for the same covariates with the model on beta value scale.
Figure 2.
Regional plot of the LEP gene DMR and its neighborhood area. The upper panel shows the P values of CpGs methylation in the epigenome-wide association with maternal CRH. The middle panel provides the annotation tracks, including ENSEMBL Genes, CpG islands tracked from UCSC, DNase cluster tracked from UCSC, SNPs tracked from UCSC. The lower panel shows the correlation of methylation among the CpGs in this selected region. The CpG sites in the differentially methylated region (DMR) of LEP gene was marked.
Persistence of associations in mid-childhood blood cell DNA methylation
In the analysis on persistence, we first focus our attention on the LEP methylation because we believe the association between maternal CRH and LEP methylation is the most robust among all CpGs. Multiple significant CpGs were detected in the LEP promoter region; moreover, the CRH-associated LEP methylation was verified by regional analysis. We evaluated the persistence of the association between maternal CRH and the DMR of LEP gene in the children’s blood samples in mid-childhood. The results were summarized in Table 3. The association between maternal CRH and DNA methylation at this DMR was attenuated at mid-childhood (mean methylation beta = 0.64% per doubling of maternal CRH; P=0.03), compared to cord blood associations (mean methylation beta= 1.46% per doubling of maternal CRH). In the sex-specific analysis (Table S3), mean levels of DNA methylation at the LEP region in mid-childhood were similar, and the associations between maternal CRH and LEP methylation were significant in females but not in males. We additionally explored the persistence of the association in mid-childhood for the 96 significant CpGs identified in cord blood. We found nominal associations between maternal CRH and mid-childhood blood methylation at 12 CpG sites (unadjusted P < 0.05), yet only cg25936177 (ACAN) remained significant after accounting for multiple testing (Table S4).
Associations among maternal CRH, LEP methylation, leptin levels, birth weight and body composition in mid-childhood.
Higher maternal CRH was associated with lower leptin concentration in cord blood (Table S5), but we did not observe an association between LEP methylation and leptin concentration, both measured in cord blood. Also, lower leptin level in cord blood was correlated with lower birth weight (Table S5) as expected, but we did not found that maternal CRH or LEP methylation were associated with birth weight (Table S5). In mid-childhood, higher LEP methylation level in blood was associated with lower BMI z-score (β [se] = −3.11 [1.32], P = 0.02), log2-transformed skinfold thickness (β [se] = −1.92 [0.72], P = 0.01) and log2-transformed waist-hip ratio (β [se] = −0.18 [0.10], P = 0.07) (Table S6). The results indicated that one percent increase in LEP methylation was associated with 3.11 decrease in BMI z-score, 3.84 times smaller in skinfold thickness and 0.36 times smaller in waist-hip ratio. We did not detect a significant association between LEP methylation and leptin concentration in blood at mid-childhood (β [se] = −2.72 [11.02], P = 0.80).
Discussion
In this pre-birth prospective cohort study, we found that higher maternal CRH concentration in mid-late pregnancy was associated with higher DNA methylation of CpG sites of the LEP gene by both individual CpG-by-CpG and regional analyses, and that this association persisted into mid-childhood. We also observed that maternal CRH was associated with DNA methylation levels at many other individual CpG sites for a total of 96 CpG sites, yet few of these seemed to persist later in childhood.
To our knowledge, this is the first study reporting associations between maternal CRH and LEP DNA methylation both at birth and persisting in mid-childhood in humans. Although no prior study has described this association, one study demonstrated that maternal dexamethasone (a type of synthetic glucocorticoids) treatment induced the programming of liver steatosis in offspring rats through increased LEP DNA methylation and the subsequent decreased leptin levels 33, which is in line with our finding. Additional findings from the same team demonstrated that rats prenatally overexposed to glucocorticoids had lower leptin levels in fetal plasmas and placenta 34,35. In a human study, mothers treated with betamethasone (a type of synthetic glucocorticoids) had offspring with lower leptin levels at birth 36. These studies support the association between high glucocorticoids exposure in pregnancy and lower circulating fetal leptin levels, as was the case of our observed positive correlation between maternal CRH and fetal LEP methylation and concordant with the known role of methylation down-regulating gene expression. The differentially methylated region of LEP found in our study was located in a CpG island, a CpG-rich region, within the proximal promoter region of LEP 37 containing many regulation elements of gene expression. Although the present study could not directly address the function of methylation in this region on gene expression due to lack of RNA data, prior studies have documented the negative regulatory effect of DNA methylation level at this region on the LEP expression in human preadipocytes 37, subcutaneous adipose tissue 38, and placenta tissues 39. However, it was worth noting that we measured the DNA methylation of the white blood cells (WBCs) and use it as a proxy for other tissues that secrete leptin. The DNA methylation status of WBCs cannot fully represent the LEP DNA methylation in other tissues, such as adipose tissue. Circulating leptin in blood is mainly released from fat tissues and to a lesser extend from other tissues 40. Therefore, the blood LEP methylation may not be the most relevant tissue predicting leptin levels in the circulation. It could be one of the reasons why the association between LEP methylation and circulating leptin were not significant at both two-time points in the present study. Another reason is the complex regulation of gene transcription and protein translation. Due to lack of RNA data, we do not know either the association between LEP methylation and LEP expression or the association between LEP expression and leptin translation. Apart from DNA methylation, many other processes are involved in the regulation of gene transcription (e.g., non-coding RNA and histone modifications 41) as well as protein translation (e.g., global and gene-specific regulations 42). Such factors may influence associations between LEP methylation and leptin levels that were not captured in our study.
Leptin regulates feeding and energy balance by affecting hypothalamic function to balance food intake and body fat mass 43. Additionally, leptin serves broader roles during pregnancy. Studies found that lower leptin levels in cord blood were significantly correlated with lower birth weight-for-gestational age z-score44,45, indicating the role of leptin in fetal growth. Moreover, the prior study of Project Viva found that lower cord blood leptin levels were associated with higher BMI at early childhood (3 years old) 46. The key roles of leptin in fetal growth and long-term development were also observed in LEP DNA methylation level. One study has found that the higher DNA methylation of LEP promoter in cord blood was associated with lower birth weight 47. However, we did not find significant association between LEP methylation and birth weight-for-gestational age z-score in the present study. This inconsistency may be attributed to our relatively small sample size or differences in the two populations including potential influence of genetic variation in LEP 47.
On average, the coefficients of prenatal CRH concentration on LEP DNA methylation is 1.46% in cord blood and 0.64% in children’s blood, which are generally small effect sizes. Breton et al. have pointed out that small-magnitude effect sizes in DNA methylation are common, yet important in children’s environmental health studies 48. Although no prior study documented the effect size of prenatal CRH on LEP methylation, several studies commonly found small but significant changes in LEP methylation (generally on the scale of 0.7%−2.3%) in relation to various exposures (e.g., prenatal PM2.5 exposure 49, folic acid supplement 50, duration of breastfeeding 51). To support the functional importance of relatively small differences in the DNA methylation of LEP, we explored the association between LEP methylation and the anthropometric measurements in mid-childhood and found every 1% increase in methylation at LEP, there was a 3.11 decrease in BMI z-score, 3.84 times smaller in skinfold thickness and 0.36 times smaller in waist-hip ratio, which indicates that the small changes in methylation of LEP in relation to CRH can have strong effects on biological activity. Consistent with our findings, other studies also identified the inverse relationship between methylation at LEP promoter and children obesity 52 or indicator of obesity 53. An animal study also suggested that diet-induced obese rats had lower LEP DNA methylation in blood 54.
Among other CpG sites that reached Bonferroni significant level, we found cg13523649 (mean (SD) methylation level of 0.29 (0.04)) annotated to ARHGAP40 gene that encodes a Rho GTPase activating protein 40 involved in GTPase activator activity. Studies on ARHGAP40 gene are sparse, but the Rho GTPase activating protein family was found to be important in neuronal development 55. Interestingly, corticotropin-releasing factors modulate the brain neuronal morphology through Rho GTPases regulators in the process of stress response 56, which suggested a close relationship between Rho GTPase activating protein and corticotropin-releasing factors. These studies supported the association between maternal CRH and ARHGAP40 methylation present in our study.
Among other FDR-significant CpG sites where CRH was associated with cord blood DNA methylation, cg07380705 is annotated to Corticotropin-Releasing Hormone Binding Protein (CRHBP), which encodes a protein with important modulatory roles relative to CRH activity through binding with CRH and limiting the bioavailability of CRH 57. CRHBP is widely detected in human tissues including the placenta. Circulating plasma CRHBP in humans prevents inappropriate pituitary-adrenal stimulation caused by elevated plasma CRH during pregnancy 57. CRHBP is positively regulated by CRH concentration 58 as well as stress factors that are known to affect HPA axis activity 59. On epigenetics level, one study observed that maternal war trauma-related stress was associated with increased DNA methylation of one CRHBP CpG site (cg17448335) in placenta 60. This CpG site locates only 121bp upstream of cg07380705 that was found associated with maternal CRH levels in our present study. Moreover, the two CpG sites are all located within the same CpG island. Interestingly, CRHBP is not only involved in regulation of HPA axis activity through binding with CRH but also participates in mediating energy balance and metabolism through counteracting the thermogenic and anorectic actions of CRH 57. Our finding of CRHBP methylation changes supported the effect of maternal CRH on the energy metabolism of offspring. We also detected three CpG sites (cg16527491 FOXA1 61, cg22937891 TRIM36 62, cg13641043 PLCD363) annotated to the genes that are associated with blood pressure, and three CpG sites (cg01078147 SEMA6A 64, cg07891473 ADAMTS965, cg15558129 LYPLAL165) annotated to the genes that are linked to body mass index and waist-hip ratio. PLCD3 (Phospholipase C Delta 3) encodes a member of the phospholipase C family, which are important in vascular smooth muscle signaling. One genome-wide association study (GWAS) has demonstrated that the common variants of PLCD3 were associated with systolic blood pressure 63. LYPLAL1 (Lysophospholipase Like 1) encodes a protein involved in the regulation of hydrolase activity and lysophospholipase activity. Multiple GWAS observed the common variants of LYPLAL1 associated with neonatal anthropometric measures 66, adult waist-hip ratio 65 and adiponectin levels 67. However, among the 96 CpGs, only one (cg25936177, ACAN) has persistent association with maternal CRH in mid-childhood. The lack of significant associations in mid-childhood could be explained by a few factors: first, some CpGs detected in cord blood with low variability may be false positive findings, according to Logue et al.’s study 68; second, the effect of maternal CRH on fetal DNA methylation could be modified by many known and unknown factors between birth and mid-childhood. The effect on early life methylation may lead to changes at molecular, cellular, or tissue levels that persist through life, while the actual associations between prenatal exposure and methylation levels are not detectable because of additional modulations of methylation.
Among our strengths, this study prospectively evaluated the persistence of DNA methylation variabilities in childhood. In addition, this study is based on a well-characterized prospective cohort study, which enabled us to evaluate a large set of potential confounders. Furthermore, the regional analysis confirmed our CpG-by-CpG analyses. Lastly, we had the opportunity to analyze the potential influence of LEP DNA methylation on circulating leptin levels and anthropometric characteristics in the same dataset.
However, our study also had some important limitations. First, we only measured CRH level at one point in pregnancy, which may not comprehensively reflect maternal CRH level over the whole pregnancy and neglects the diurnal rhythm of CRH. In addition, we do not have data on gene expression measurements to assess the functionality of DNA methylation at the LEP locus; moreover, we do not have data on DNA methylation of tissues that predominantly secrete leptin, mainly adipose tissue. Many of our significant findings were at CpG sites with low (< 5%) or high (>95%) mean DNA methylation levels across all samples, which often indicated low variabilities with an increased risk of false positive 68. Moreover, we only pursued analyses of associations between maternal CRH and DNA methylation in mid-childhood blood for the CpG sites we had identified at birth; we acknowledge that other signals may emerge later in life but are not detectable in cord blood.
Conclusion
This study adds to the growing body of evidence showing that glucocorticoid exposure in pregnancy is associated with children’s health programming. Variations in DNA methylation levels of LEP promoter observed at birth might provide a potential explanation for the short- and long-term health effects of prenatal glucocorticoid exposure in offspring.
Supplementary Material
Acknowledgments
We are indebted to the Project Viva participants and staffs.
Funding
This work was supported by grants from the National Institutes of Health (R01 NR013945, R01 ES021357, R37 HD034568, K24 HD069408, R01 ES016314 and R01 HL111108).
Footnotes
Competing interests
The authors declare that they have no competing interests.
References:
- 1.Bonfiglio JJ, Inda C, Refojo D, Holsboer F, Arzt E, Silberstein S. The Corticotropin-Releasing Hormone Network and the Hypothalamic-Pituitary-Adrenal Axis: Molecular and Cellular Mechanisms Involved. Neuroendocrinology 2011; 94: 12–20. [DOI] [PubMed] [Google Scholar]
- 2.Sandman CA, Davis EP, Buss C, Glynn LM. Prenatal programming of human neurological function. Int J Pept 2011; 2011: 837596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emanuel RL, Robinson BG, Seely EW, Graves SW, Kohane I, Saltzman D et al. Corticotrophin releasing hormone levels in human plasma and amniotic fluid during gestation. Clin Endocrinol (Oxf) 1994; 40: 257–262. [DOI] [PubMed] [Google Scholar]
- 4.Sandman CA, Glynn L, Schetter CD, Wadhwa P, Garite T, Chicz-DeMet A et al. Elevated maternal cortisol early in pregnancy predicts third trimester levels of placental corticotropin releasing hormone (CRH): Priming the placental clock. Peptides 2006; 27: 1457–1463. [DOI] [PubMed] [Google Scholar]
- 5.Barber K, Mussin E, Taylor DK. Fetal exposure to involuntary maternal smoking and childhood respiratory disease. Ann Allergy Asthma Immunol 1996; 76: 427–30. [DOI] [PubMed] [Google Scholar]
- 6.Baibazarova E, van de Beek C, Cohen-Kettenis PT, Buitelaar J, Shelton KH, van Goozen SHM. Influence of prenatal maternal stress, maternal plasma cortisol and cortisol in the amniotic fluid on birth outcomes and child temperament at 3 months. Psychoneuroendocrinology 2013; 38: 907–915. [DOI] [PubMed] [Google Scholar]
- 7.Trainer PJ. Corticosteroids and pregnancy. Semin Reprod Med 2002; 20: 375–380. [DOI] [PubMed] [Google Scholar]
- 8.Wadhwa PD, Garite TJ, Porto M, Glynn L, Chicz-DeMet A, Dunkel-Schetter C et al. Placental corticotropin-releasing hormone (CRH), spontaneous preterm birth, and fetal growth restriction: a prospective investigation. Am J Obstet Gynecol 2004; 191: 1063–1069. [DOI] [PubMed] [Google Scholar]
- 9.Alexander N, Rosenlöcher F, Stalder T, Linke J, Distler W, Morgner J et al. Impact of antenatal synthetic glucocorticoid exposure on endocrine stress reactivity in term-born children. J Clin Endocrinol Metab 2012; 97: 3538–3544. [DOI] [PubMed] [Google Scholar]
- 10.Gillman MW, Rich-Edwards JW, Huh S, Majzoub JA, Oken E, Taveras EM et al. Maternal Corticotropin-Releasing Hormone Levels during Pregnancy and Offspring Adiposity. Obes Silver Spring Md 2006; 14: 1647–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR et al. A transgenic model of visceral obesity and the metabolic syndrome. Science 2001; 294: 2166–2170. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor DM, Blache D, Hoggard N, Brookes E, Wooding FBP, Fowden AL et al. Developmental control of plasma leptin and adipose leptin messenger ribonucleic acid in the ovine fetus during late gestation: role of glucocorticoids and thyroid hormones. Endocrinology 2007; 148: 3750–3757. [DOI] [PubMed] [Google Scholar]
- 13.McMillen IC, Adam CL, Mühlhäusler BS. Early origins of obesity: programming the appetite regulatory system. J Physiol 2005; 565: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y-C, Huang Y-H, Sheen J-M, Tain Y-L, Yu H-R, Chen C-C et al. Prenatal Dexamethasone Exposure Programs the Development of the Pancreas and the Secretion of Insulin in Rats. Pediatr Neonatol 2017; 58: 135–144. [DOI] [PubMed] [Google Scholar]
- 15.Sheen J-M, Hsieh C-S, Tain Y-L, Li S-W, Yu H-R, Chen C-C et al. Programming Effects of Prenatal Glucocorticoid Exposure with a Postnatal High-Fat Diet in Diabetes Mellitus. Int J Mol Sci 2016; 17: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasting MH, Oken E, Mantzoros CS, Rich-Edwards JW, Majzoub JA, Kleinman K et al. Maternal levels of corticotropin-releasing hormone during pregnancy in relation to adiponectin and leptin in early childhood. J Clin Endocrinol Metab 2009; 94: 1409–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boekelheide K, Blumberg B, Chapin RE, Cote I, Graziano JH, Janesick A et al. Predicting later-life outcomes of early-life exposures. Environ Health Perspect 2012; 120: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci U S A 1999; 96: 6107–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moisiadis VG, Matthews SG . Glucocorticoids and fetal programming part 2: Mechanisms. Nat Rev Endocrinol 2014; 10: 403–411. [DOI] [PubMed] [Google Scholar]
- 20.Crudo A, Petropoulos S, Moisiadis VG, Iqbal M, Kostaki A, Machnes Z et al. Prenatal synthetic glucocorticoid treatment changes DNA methylation states in male organ systems: multigenerational effects. Endocrinology 2012; 153: 3269–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D et al. Cohort profile: project viva. Int J Epidemiol 2015; 44: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich-Edwards JW, Mohllajee AP, Kleinman K, Hacker MR, Majzoub J, Wright RJ et al. Elevated Midpregnancy Corticotropin-Releasing Hormone Is Associated with Prenatal, But Not Postpartum, Maternal Depression. J Clin Endocrinol Metab 2008; 93: 1946–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardenas A, Rifas-Shiman SL, Agha G, Hivert M-F, Litonjua AA, DeMeo DL et al. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep 2017; 7. doi: 10.1038/s41598-017-00384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oken E, Kleinman KP, Rich-Edwards J, Gillman MW. A nearly continuous measure of birth weight for gestational age using a United States national reference. BMC Pediatr 2003; 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P et al. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics 2016; 11: 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D et al. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLOS ONE 2012; 7: e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavrila A, Peng C-K, Chan JL, Mietus JE, Goldberger AL, Mantzoros CS. Diurnal and ultradian dynamics of serum adiponectin in healthy men: comparison with leptin, circulating soluble leptin receptor, and cortisol patterns. J Clin Endocrinol Metab 2003; 88: 2838–2843. [DOI] [PubMed] [Google Scholar]
- 29.Boeke CE, Oken E, Kleinman KP, Rifas-Shiman SL, Taveras EM, Gillman MW. Correlations among adiposity measures in school-aged children. BMC Pediatr 2013; 13: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CDC growth charts: United States. - PubMed - NCBI. https://www.ncbi.nlm.nih.gov/pubmed/11183293 (accessed 2 Nov 2017).
- 31.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015; 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin TC, Yet I, Tsai P-C, Bell JT. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics 2015; 16: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiao M-M, Huang L-T, Chen C-J, Sheen J-M, Tain Y-L, Chen C-C et al. Melatonin in the Regulation of Liver Steatosis following Prenatal Glucocorticoid Exposure. BioMed Res Int 2014; 2014. doi: 10.1155/2014/942172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugden MC, Langdown ML, Munns MJ, Holness MJ. Maternal glucocorticoid treatment modulates placental leptin and leptin receptor expression and materno-fetal leptin physiology during late pregnancy, and elicits hypertension associated with hyperleptinaemia in the early-growth-retarded adult offspring. Eur J Endocrinol 2001; 145: 529–539. [DOI] [PubMed] [Google Scholar]
- 35.Smith JT, Waddell BJ. Leptin receptor expression in the rat placenta: changes in ob-ra, ob-rb, and ob-re with gestational age and suppression by glucocorticoids. Biol Reprod 2002; 67: 1204–1210. [DOI] [PubMed] [Google Scholar]
- 36.Marinoni E, Letizia C, Ciardo F, Corona G, Moscarini M, Iorio RD. Effects of prenatal betamethasone administration on leptin and adiponectin concentrations in maternal and fetal circulation. Am J Obstet Gynecol 2008; 199: 141.e1–141.e6. [DOI] [PubMed] [Google Scholar]
- 37.Melzner I, Scott V, Dorsch K, Fischer P, Wabitsch M, Brüderlein S et al. Leptin Gene Expression in Human Preadipocytes Is Switched on by Maturation-induced Demethylation of Distinct CpGs in Its Proximal Promoter. J Biol Chem 2002; 277: 45420–45427. [DOI] [PubMed] [Google Scholar]
- 38.Houde A-A, Légaré C, Biron S, Lescelleur O, Biertho L, Marceau S et al. Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL-cholesterol levels in severely obese men and women. BMC Med Genet 2015; 16. doi: 10.1186/s12881-015-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouchard L, Thibault S, Guay S-P, Santure M, Monpetit A, St-Pierre J et al. Leptin Gene Epigenetic Adaptation to Impaired Glucose Metabolism During Pregnancy. Diabetes Care 2010; 33: 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dardeno TA, Chou SH, Moon H-S, Chamberland JP, Fiorenza CG, Mantzoros CS. Leptin in Human Physiology and Therapeutics. Front Neuroendocrinol 2010; 31: 377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity 2010; 105: 4–13. [DOI] [PubMed] [Google Scholar]
- 42.Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009; 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahima RS, Flier JS. Leptin. Annu Rev Physiol 2000; 62: 413–437. [DOI] [PubMed] [Google Scholar]
- 44.Ren R-X, Shen Y. A meta-analysis of relationship between birth weight and cord blood leptin levels in newborns. World J Pediatr WJP 2010; 6: 311–316. [DOI] [PubMed] [Google Scholar]
- 45.Kadakia R, Zheng Y, Zhang Z, Zhang W, Hou L, Josefson JL. Maternal pre-pregnancy BMI downregulates neonatal cord blood LEP methylation. Pediatr Obes 2017; 12: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantzoros CS, Rifas-Shiman SL, Williams CJ, Fargnoli JL, Kelesidis T, Gillman MW. Cord blood leptin and adiponectin as predictors of adiposity in children at 3 years of age: a prospective cohort study. Pediatrics 2009; 123: 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesseur C, Armstrong DA, Paquette AG, Koestler DC, Padbury JF, Marsit CJ. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol Cell Endocrinol 2013; 381: 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC et al. Small-Magnitude Effect Sizes in Epigenetic End Points are Important in Children’s Environmental Health Studies: The Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ Health Perspect 2017; 125: 511–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saenen ND, Vrijens K, Janssen BG, Roels HA, Neven KY, Vanden Berghe W et al. Lower Placental Leptin Promoter Methylation in Association with Fine Particulate Matter Air Pollution during Pregnancy and Placental Nitrosative Stress at Birth in the ENVIRONAGE Cohort. Environ Health Perspect 2017; 125: 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauwels S, Ghosh M, Duca RC, Bekaert B, Freson K, Huybrechts I et al. Dietary and supplemental maternal methyl-group donor intake and cord blood DNA methylation. Epigenetics 2016; 12: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obermann-Borst SA, Eilers PHC, Tobi EW, Jong FH de, Slagboom PE, Heijmans BT et al. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr Res 2013; 74: 344–349. [DOI] [PubMed] [Google Scholar]
- 52.García-Cardona MC, Huang F, García-Vivas JM, López-Camarillo C, Navarro BE del R, Olivos EN et al. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int J Obes 2014; 38: 1457–1465. [DOI] [PubMed] [Google Scholar]
- 53.Dunstan J, Bressler JP, Moran TH, Pollak JS, Hirsch AG, Bailey-Davis L et al. Associations of LEP, CRH, ICAM-1, and LINE-1 methylation, measured in saliva, with waist circumference, body mass index, and percent body fat in mid-childhood. Clin Epigenetics 2017; 9. doi: 10.1186/s13148-017-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milagro FI, Campión J, García-Díaz DF, Goyenechea E, Paternain L, Martínez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem 2009; 65: 1–9. [DOI] [PubMed] [Google Scholar]
- 55.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol 2003; 13: 13–22. [DOI] [PubMed] [Google Scholar]
- 56.Swinny JD, Valentino RJ. Corticotropin-releasing factor promotes growth of brain norepinephrine neuronal processes through Rho GTPase regulators of the actin cytoskeleton in rat. Eur J Neurosci 2006; 24: 2481–2490. [DOI] [PubMed] [Google Scholar]
- 57.Westphal N J CRH-BP: the regulation and function of a phylogenetically conserved binding protein. Front Biosci 2006; 11: 1878. [DOI] [PubMed] [Google Scholar]
- 58.McClennen SJ, Seasholtz AF. Transcriptional regulation of corticotropin-releasing hormone-binding protein gene expression in astrocyte cultures. Endocrinology 1999; 140: 4095–4103. [DOI] [PubMed] [Google Scholar]
- 59 **.Regulation of Pituitary Corticotropin-Releasing Hormone-Binding Protein Messenger Ribonucleic Acid Levels by Restraint Stress and Adrenalectomy | Endocrinology | Oxford Academic; https://academic.oup.com/endo/article/139/11/4435/2986805 This work is supported by National Institutes of Health Grant DK-42730 (to A.F.S) and by a Young Investigator Award (to A.F.S.) from the National Alliance for Research on Schizophrenia and Depression. (accessed 5 Feb 2018). [DOI] [PubMed] [Google Scholar]
- 60.Kertes DA, Kamin HS, Hughes DA, Rodney NC, Bhatt S, Mulligan CJ. Prenatal Maternal Stress Predicts Methylation of Genes Regulating the Hypothalamic-Pituitary-Adrenocortical System in Mothers and Newborns in the Democratic Republic of Congo. Child Dev 2016; 87: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salvi E, Wang Z, Rizzi F, Gong Y, McDonough CW, Padmanabhan S et al. Genome-Wide and Gene-Based Meta-Analyses Identify Novel Loci Influencing Blood Pressure Response to HydrochlorothiazideNovelty and Significance. Hypertension 2017; 69: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wain LV, Vaez A, Jansen R, Joehanes R, Van P der M, Erzurumluoglu AM et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertens Dallas Tex 1979 2017. doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 2009; 41: 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salinas YD, Wang L, DeWan AT. Multiethnic genome-wide association study identifies ethnic-specific associations with body mass index in Hispanics and African Americans. BMC Genet 2016; 17: 78–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L et al. Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet 2017; 13: e1006528–e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urbanek M, Hayes MG, Armstrong LL, Morrison J, Lowe LP, Badon SE et al. The chromosome 3q25 genomic region is associated with measures of adiposity in newborns in a multi-ethnic genome-wide association study. Hum Mol Genet 2013; 22: 3583–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dastani Z, Hivert M-F, Timpson N, Perry JRB, Yuan X, Scott RA et al. Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals. PLoS Genet 2012; 8. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Logue MW, Smith AK, Wolf EJ, Maniates H, Stone A, Schichman SA et al. The correlation of methylation levels measured using Illumina 450K and EPIC BeadChips in blood samples. Epigenomics 2017; 9: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.