Abstract
Receptor Interacting Serine/Threonine Kinase 1 (RIPK1) is a master regulator of signaling pathways leading to inflammation and cell death and is of medical interest as a drug target. Here, we report four patients from three unrelated families with complete RIPK1 deficiency caused by rare homozygous mutations. The patients suffered from recurrent infections, early-onset inflammatory bowel disease and progressive polyarthritis. They had immunodeficiency with lymphopenia and altered production of various cytokines revealed by whole-blood assays. In vitro, RIPK1-deficient cells showed impaired MAPK activation and cytokine secretion and were prone to necroptosis. Hematopoietic stem cell transplantation reversed cytokine production defects and resolved clinical symptoms in one patient. Thus, RIPK1 plays a critical role in the human immune system.
Primary immunodeficiencies (PIDs) are a heterogeneous group of disorders characterized by increased susceptibility to infections. In many cases PIDs are monogenic disorders that follow Mendelian inheritance and mutations in more than 300 genes have been shown to cause PIDs (1). However, in many PID patients causative mutations remain unknown. Identification of such mutations not only facilitates diagnosis of PIDs, but also can provide the fundamental knowledge about the roles of the affected proteins in the human immune system.
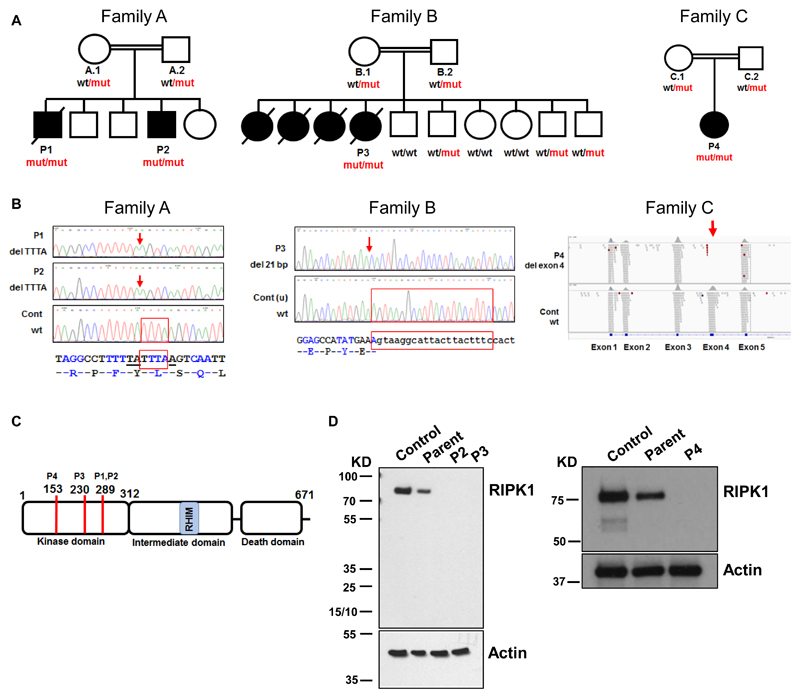
Here, we used exome sequencing to identify causative mutations in a heterogeneous cohort of PID patients with unknown genetic etiology (2). We excluded known polymorphisms (2) and studied rare variants. We noticed that four patients (P1 – P4) from three unrelated consanguineous families (Fig. 1A) had homozygous loss-of-function mutations in the same gene, RIPK1 (ENST00000380409). In patients P1 and P2 from family A, the mutation was a 4-nucleotide frameshift deletion in RIPK1 exon 6 that led to a premature stop codon (Fig. 1B). In patient P3 from family B, we found a 21-nucleotide deletion that removed one nucleotide in RIPK1 exon 4 and 20 nucleotides in the following intron (Fig. 1B). This deletion activated an alternative splice site in intron 4, so that the RIPK1 transcript lacked the last nucleotide of exon 4 and had an insertion of 48 nucleotides from the following intron (fig. S1). Patient P4 from family C had a homozygous 2,064-nucleotide deletion that completely removed RIPK1 exon 4 (Fig. 1B and fig. S2). Parents of all patients were heterozygous carriers of these RIPK1 mutations (Fig. 1A and fig. S3). Given that homozygous loss-of-function mutations in the RIPK1 gene had never been reported in humans (e.g., absent from more than 120,000 subjects in the gnomAD database (3)), such mutations in our patients are likely to be pathogenic. The RIPK1 gene encodes a 671 amino-acid serine/threonine kinase (Fig. 1C). All three mutations mapped to the N-terminal kinase domain and produced premature stop codons. The RIPK1 protein was absent in cells of patients P2, P3 and P4 (Fig. 1D). Therefore, all three homozygous mutations led to complete RIPK1 deficiency. The four patients had lymphopenia, suffered from recurrent viral, bacterial and fungal infections, early-onset inflammatory bowel disease (IBD) involving upper and lower gastrointestinal tract and developed arthritis (Tables S1 and S2, fig. S4 and Supplementary Text (2)). Therefore, these clinical features characterize RIPK1 deficiency in humans.
Fig. 1. Mutations of the RIPK1 gene cause complete RIPK1 deficiency.
(A) Three families with the RIPK1 gene mutations: wt, wild-type allele; mut, mutant allele.○ and □ - unaffected; ● and ■ - affected. (B) Patients’ mutations. Locations of deletions are shown by red arrows; deleted nucleotides are shown by red frames. Deletion of exon 4 in P4 (right panel). (C) Domains of the RIPK1 protein. RHIM - Receptor-interacting protein (RIP) Homotypic Interaction Motif. Codons affected by mutations are shown by red lines; codon numbers and corresponding patients are indicated above. (D) Western blot assays for detection of RIPK1 protein in fibroblasts (left panel) and T cell blasts (right panel).
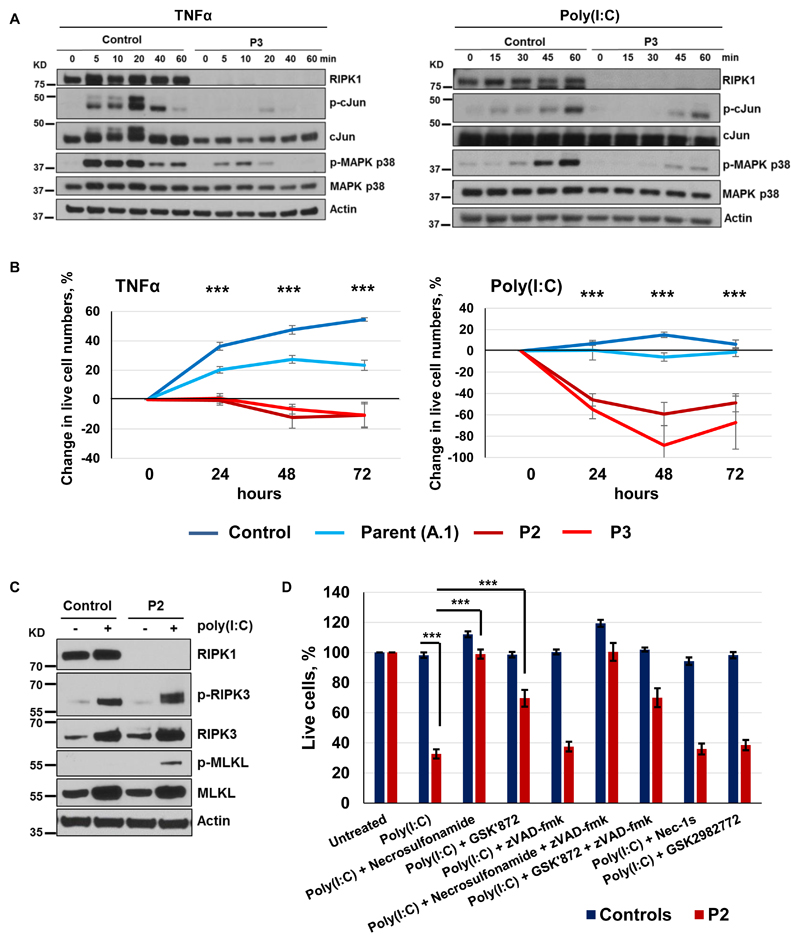
RIPK1 is a widely expressed cytosolic protein kinase controlling multiple signaling pathways leading to inflammation and apoptotic or necroptotic cell death (4, 5). RIPK1 is present in protein complexes that mediate signal transduction from cell surface receptors, including TNFR1, TLR3 and TLR4 (4, 5). Stimulation of these receptors activates the canonical NF-κB pathway and the mitogen-activated protein kinases (MAPKs). This leads to phosphorylation of NF-κB and AP-1 transcription factors, which induces expression of proinflammatory and pro-survival genes (4, 6). To assess the functioning of these signaling pathways, we stimulated patients’ skin fibroblasts with TNFα and poly(I:C) in vitro. We found that phosphorylation of MAPK p38 and the AP-1 subunit cJun was markedly reduced (Fig. 2A and fig. S5), while phosphorylation of MAPK p42/44 (ERK2/1) and NF-κB p65 was partially reduced (fig. S6). TNFα -induced secretion of cytokines IL-6 and RANTES by patients’ fibroblasts was also diminished (fig. S7). Thus, similar to studies in RIPK1-deficient mice (7–11), pro-inflammatory signaling downstream of TNFR1 and TLR3 is impaired in the patients. We next investigated whether RIPK1 deficiency affected cell viability. After stimulation with TNFα or poly(I:C), we found significantly fewer viable fibroblasts of patients than those of healthy controls (P < 0.001; Fig. 2B). When we transduced patient’s fibroblasts and expressed wild-type RIPK1, this viability defect was reversed (fig. S8) and activation of the MAPK and NF-κB pathways was also rescued (fig. S9). To investigate the mechanism of cell death, we studied patient’s fibroblasts 24 hours after poly(I:C) stimulation and found increased phosphorylation of RIPK3 and Mixed Lineage Kinase Domain Like Pseudokinase (MLKL), proteins that mediate necroptosis (12, 13) (Fig. 2C). No cleavage of caspase-8 and minimal cleavage of caspase-3 were found, indicating that apoptosis was not a major death mechanism of these cells (fig. S10). Consistent with these results, MLKL inhibitor Necrosulfonamide and, to a lesser extent, RIPK3 inhibitor GSK′872 rescued patient’s cells from poly(I:C)-induced death, while pan-caspase inhibitor zVAD-fmk had no effect (Fig. 2D). As expected, RIPK1 inhibitors Nec-1s and GSK2982772 had no effect on these RIPK1-deficient cells (Fig. 2D).
Fig.2. RIPK1-deficient fibroblasts show impaired MAPK signaling and necroptosis.
(A) Primary fibroblasts were stimulated with 50 ng/mL TNFα or 100 μg/mL poly(I:C) and protein extracts were subjected to immunoblotting (N = 2). (B) Viability of primary fibroblasts after stimulation with 100 ng/mL TNFα or 20 μg/mL poly(I:C). Experiments were repeated at least three times. Differences between stimulated and unstimulated cells are shown at each time point relative to the 0 time point. P-values were calculated using two-tailed unpaired T-test, comparing combined data from healthy subjects (parent and control) vs patients (P2 and P3); graphs show mean values ± SEM. (C) Fibroblasts were stimulated with 20 μg/mL poly(I:C) for 24 hours and protein extracts were subjected to immunoblotting (N = 1). (D) Fibroblasts of P2 and controls (unrelated and parent A.1) were stimulated with 20 μg/mL poly(I:C) for 24 hours in the presence of indicated compounds and cell viability was measured (N ≥ 6); graphs show mean values ± SEM. P-values were calculated using two-tailed unpaired T-tests. *** P < 0.001.
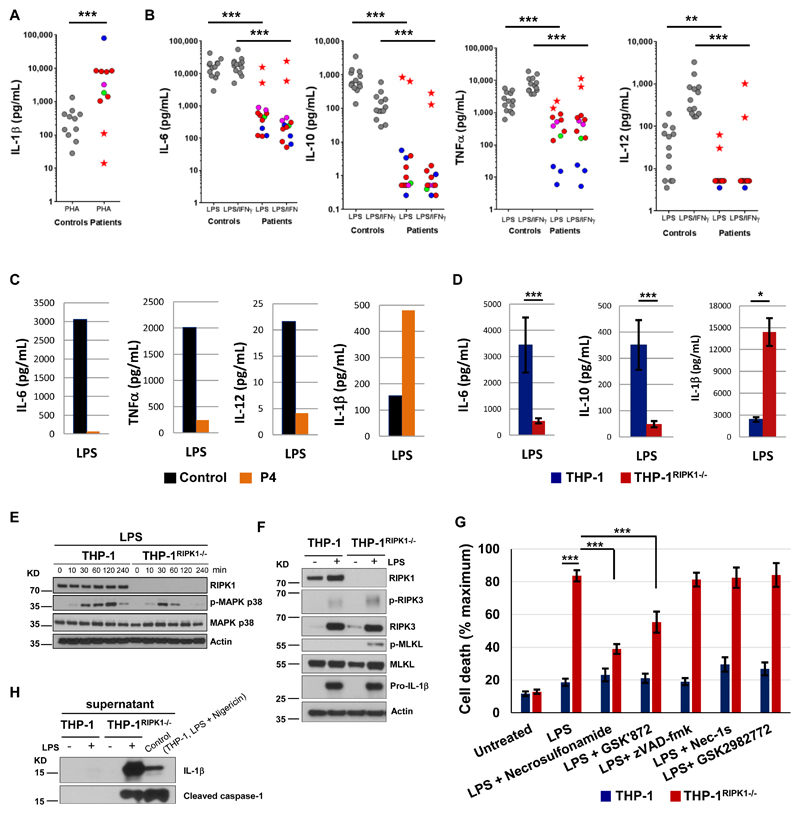
To investigate the molecular basis of the immune dysfunction responsible for the disease, we studied cytokine release in whole blood assays. After stimulation with phytohemagglutinin (PHA), patients’ blood cells produced markedly increased amounts of IL-1β (Fig. 3A). This enhanced IL-1β response to PHA normalized in patient P2 after hematopoietic stem cell transplantation (HSCT) (Fig. 3A). Also, after PHA stimulation patients’ blood produced reduced amounts of IL-17 and IFN-γ, while the production of TNFα IL-6 and IL-10 was similar to controls (fig. S11), which shows dysregulated but not generally suppressed responses of stimulated T cells. After stimulation with lipopolysaccharide (LPS), whole blood of healthy controls produced large amounts of IL-6 and IL-10 and showed strongly up-regulated production of TNFα and IL-12 upon co-stimulation with LPS and IFN-γ (Fig. 3B). These responses were dramatically reduced in the RIPK1-deficient patients, but normalized in patient P2 after HSCT (Fig. 3B). At the same time, we found normal production of IL-10, IL-6 and TNFα in patients’ whole blood assays after stimulation with TLR1/2 and TLR2/6 ligands Pam3CSK4 and Pam2CSK4 (fig. S12), consistent with the RIPK1-independent signaling downstream of these receptors (4). The production of IL-1β in whole blood of the patients was within normal range after LPS stimulation and was slightly reduced after LPS and IFN-γco-stimulation (fig. S13). Similar to whole blood assays, primary monocytes isolated from blood of patient P4 and stimulated with LPS showed reduced production of IL-6, TNFα and IL-12 (Fig. 3C). However, the production of IL-1β was increased (Fig. 3C).
Fig. 3. RIPK1-deficient immune cells show dysregulated cytokine production, impaired MAPK signaling and necroptosis.
(A-B) Cytokines were measured in whole blood after 24 hours stimulation using 10 μg/mL PHA (A) or 1 μg/mL LPS or 1 μg/mL LPS plus 20,000 IU/mL IFN-γ (B). To account for lymphopenia data were corrected for lymphocyte counts. Controls are shown as grey circles, patients as colored circles (P1 – magenta, P3 – blue, P4 – green, P2 before HSCT – red); P2 after HSCT – red star. P-values were calculated using two-tailed Mann-Whitney test, excluding the data of P2 after HSCT. (C) CD14+ monocytes were purified from PBMC of patient P4 (age 3 y) and a healthy adult (travel control), stimulated overnight with 5 μg/mL LPS and then cytokines were measured in supernatants. N = 1, two technical replicates, graphs show mean values. (D) THP-1 cells (2 wild-type and 7 THP-1RIPK1-/- clones) were treated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA) for 3 days, rested for 1 day and stimulated overnight with 5 μg/mL LPS. Cytokines were measured in supernatants. The data were corrected for the estimated number of live cells and show means combining data for different clones ± SEM. P-values were calculated using two-tailed unpaired T-test. (E) THP-1 cells were treated with PMA as in (D), stimulated with 1 μg/mL LPS and the extracted proteins were analyzed by immunoblotting (N = 2). (F) THP-1 cells were stimulated with 5 μg/mL LPS for 48 hours and the extracted proteins were analyzed by immunoblotting (N = 2). (G) THP-1 cells were stimulated with 5 μg/mL LPS for 48 hours in the presence of indicated compounds and cell death was measured using lactate dehydrogenase (LDH) release assay (N ≥ 4). The data show means ± SEM. P-values were calculated using two-tailed unpaired T-test. (H) Supernatants of THP-1 cells studied in (F) were analyzed by immunoblotting (N = 1). * P < 0.05, ** P < 0.01, *** P < 0.001.
We next analyzed in more detail the response of RIPK1-deficient human immune cells to LPS stimulation. We were unable to conduct further studies of primary blood cells from the patients because P1 and P3 had died, P2 had undergone HSCT and P4 was not available. We therefore used CRISPR-Cas9 technology to knock out RIPK1 in the human monocyte-like THP-1 cell line (2). After LPS stimulation, THP-1RIPK1-/- cells secreted reduced amounts of IL-6 and IL-10 and released increased amounts of IL-1β (Fig. 3D), resembling the cytokine response of patients’ cells (Fig. 3C). The impaired cytokine production was preceded by the reduced phosphorylation of MAPK p38 (Fig. 3E), mirroring the defective MAPK p38 phosphorylation in the patients’ primary fibroblasts (Fig. 2A). Activation of NF-κB p65 and other branches of the MAPK pathway was not affected in the THP-1RIPK1-/-cells (fig. S14). Differential activation of the MAPK and NF-κB pathways after LPS stimulation was reported previously (14), and our results indicate that RIPK1 deficiency in THP-1 cells preferentially alters LPS-induced MAPK p38 activation.
After LPS stimulation, THP-1RIPK1-/- cells showed increased phosphorylation of RIPK3 and MLKL and enhanced cell death (Figs. 3F and 3G). No cleavage of caspase-8 or caspase-3 was found (fig. S15). Necrosulfonamide and GSK'872, but not zVAD-fmk, reduced death of THP-1RIPK1-/- cells (Fig. 3G), again pointing at necroptosis as the main death mechanism of these cells. Necroptosis of THP-1RIPK1-/- cells was accompanied by the release of cleaved caspase-1 that we found in the supernatant together with IL-1β(Fig. 3H), suggesting concurrent activation of inflammasome. Next, we compared mechanisms that mediate IL-1β release from LPS-stimulated THP-1RIPK1-/- cells with those that mediate IL-1β release during pyroptosis of wild-type THP-1 cells stimulated by LPS and Nigericin. We found that different mechanisms are at play. After treatment with Necrosulfonamide, IL-1β release during pyroptosis was only slightly reduced, while it was completely prevented in THP-1RIPK1-/- cells, suggesting that in these cells IL-1βrelease is secondary to necroptosis (fig. S16). Of note, treatment of THP-1RIPK1-/- cells with Necrosulfonamide did not restore reduced IL-6 secretion (fig. S17), indicating that it is reduced not because of enhanced necroptosis, but is an earlier phenomenon. Taken together, these data show that human RIPK1-deficient cells have impaired pro-inflammatory signaling leading to dysregulated cytokine secretion and are prone to necroptosis, which, in myeloid cells, is accompanied by IL-1β release.
RIPK1 has been extensively studied in mouse models (7–9, 15–18) and its inhibitors are considered for the treatment of acute and chronic organ injury, including stroke, myocardial infarction and renal ischemia–reperfusion injury (19, 20), but the phenotype associated with RIPK1 deficiency in humans was unknown. While the Ripk1-knockout mice displayed systemic inflammation and cell death in multiple tissues and died during the postnatal period (7–9), the clinical presentation of our RIPK1-deficient patients was less severe. Nevertheless, they suffered from immunodeficiency, gut inflammation and progressive polyarthritis. During infection, activation of the pattern-recognition receptors TLR3 and TLR4 and stimulation of TNFR1 by secreted TNFα induces pro-inflammatory effects. Consistent with the established role of RIPK1 in the signal-transducing protein complexes assembled downstream of these receptors (4–7, 10, 11), we found impaired pro-inflammatory signaling in RIPK1-deficient cells and reduced production of multiple cytokines. These defects, as well as lymphopenia, likely explain susceptibility to infections in the patients.
Our data show that LPS stimulation of RIPK1-deficient monocytes resulted in the increased necroptosis and IL-1β release, similar to the previous observation in the RIPK1-deficient monocytes transdifferentiated from immortalized human B cells (21). Furthermore, we found high IL-1β levels after PHA stimulation of the patients’ whole blood. This may suggest that IL-1β production by T cells, which was reported recently (22, 23), is also increased in the context of RIPK1 deficiency. Alternatively, dysregulated secretion of T cell factors after PHA stimulation may have augmented the IL-1β production by patients’ monocytes in whole blood. IL-1β is a pro-inflammatory cytokine involved in the pathogenesis of arthritis and IBD (24, 25). Another cytokine dysregulated in our patients, IL-10, is essential for balancing immune response in the gut and impaired signaling in the IL-10 pathway has been associated with IBD (26, 27). Therefore, it is likely that low secretion of IL-10 and increased IL-1βproduction contributed to the pathogenesis of arthritis and IBD in our RIPK1-deficient patients. Accordingly, treatment with IL-1 inhibitors may be considered in RIPK1 deficiency, although none of our patients received such therapy. Interestingly, while patients P1, P2 and P3 developed severe IBD in the first months of life, P4 had no IBD signs up until the age of 4 years. Therefore, genetically determined RIPK1 deficiency was not the sole cause of IBD in these patients. Rather, it led to the dysregulated cytokine production, which set the immune system in a predisposition mode, while additional factors, e.g. distinct microbiomes, likely affected progression to IBD in these patients. This scenario resembles the adult-onset IBD, where immune predisposition is determined by multiple common genetic polymorphisms, while progression to clinical disease is driven by environmental factors (28).
The intestinal epithelium provides a physical barrier and participates in maintaining immune homeostasis in the gut. Mice genetically deficient in Ripk1 in the intestinal epithelial cells (IEC) developed severe lethal intestinal pathology due to FADD–caspase-8-mediated apoptosis of IEC (9, 29). Histological examination of gastrointestinal biopsies from P1 and P2 showed only occasional cells with apoptotic morphology and cells positive for cleaved caspase-3 (fig. S18). Such cells were also present in biopsies from children with idiopathic IBD and histologically normal biopsies (fig. S18). Thus, in contrast to the mouse model, no extensive IEC apoptosis was found, suggesting that it is not a characteristic feature of RIPK1 deficiency in humans. Likewise, skin disorders were not typical in our patients, indicating no special protective role of human RIPK1 in keratinocytes, in contrast to mice with epidermis-specific RIPK1 knockout that developed severe skin inflammation (9, 30). Given that HSCT in patient P2 resolved IBD and arthritis and reduced the frequency of infections (2), it is likely that dysfunction of the immune system rather than dysfunction of other cell types was critical for disease development.
In summary, our findings indicate that RIPK1 has a more narrow function in humans than in mice, with the effects of RIPK1 deficiency being largely confined to the immune system. Accordingly, HSCT performed at young age can be an effective treatment in RIPK1-deficient patients.
Supplementary Materials
Acknowledgments
We thank S. Prothero, who participated in the patients’ treatment.
Funding: S.N. was supported by the Wellcome Trust (095198/Z/10/Z), MRC (MR/M012328/1) and the ERC Starting grant (260477). S.N., D.K., and R.D. are supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. H.W. is supported by the Deutsche Forschungsgemeinschaft (WA 1025/31-1). This work was also supported by the MRC/EPSRC Newcastle Molecular Pathology Node.
Footnotes
Author contributions: D.C.-L., D.E., C.W., O.P., M.M., M.G., A.A., E.G., D.S. and H.W. performed cell experiments and analyzed the data. V.P. analyzed exome data. J.C. performed sequencing. M.A., P.D.A., M.S.AlZ., D.K., E.AlI., B.A. and S.H. looked after the patients and collected patients’ data. C.M.B. analyzed biopsies. R.D. and L.C.-G. performed cytokine analyses. S.N. planned the experiments, analyzed the data and wrote the first draft of the manuscript.
Competing interests: Authors declare no competing interests.
Data and materials availability: Cells are available upon signing a Material Transfer Agreement. Sequence data for the RIPK1 gene region are available in Sequence Read Archive under the accession number SRP136541. Whole exome data are available upon signing a Data Transfer Agreement.
References
- 1.Bousfiha A, et al. The 2017 IUIS Phenotypic Classification for Primary Immunodeficiencies. J Clin Immunol. 2018;38:129. doi: 10.1007/s10875-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Materials and Methods are available as Supplementary Materials
- 3.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14:727. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 5.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 6.Najjar M, et al. RIPK1 and RIPK3 Kinases Promote Cell-Death-Independent Inflammation by Toll-like Receptor 4. Immunity. 2016;45:46. doi: 10.1016/j.immuni.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelliher MA, et al. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 8.Rickard JA, et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell. 2014;157:1175. doi: 10.1016/j.cell.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Dannappel M, et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513:90. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TH, et al. The death domain kinase RIP1 is essential for tumor necrosis factor alpha signaling to p38 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:8377. doi: 10.1128/MCB.23.22.8377-8385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meylan E, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 12.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 14.Gottschalk RA, et al. Distinct NF-kappaB and MAPK Activation Thresholds Uncouple Steady-State Microbe Sensing from Anti-pathogen Inflammatory Responses. Cell Syst. 2016;2:378. doi: 10.1016/j.cels.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 16.Polykratis A, et al. Cutting edge: RIPK1 Kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J Immunol. 2014;193:1539. doi: 10.4049/jimmunol.1400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser WJ, et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc Natl Acad Sci U S A. 2014;111:7753. doi: 10.1073/pnas.1401857111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roderick JE, et al. Hematopoietic RIPK1 deficiency results in bone marrow failure caused by apoptosis and RIPK3-mediated necroptosis. Proc Natl Acad Sci U S A. 2014;111:14436. doi: 10.1073/pnas.1409389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, et al. Role of necroptosis in the pathogenesis of solid organ injury. Cell Death Dis. 2015;6:e1975. doi: 10.1038/cddis.2015.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaidt MM, et al. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 2016;44:833. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Martin BN, et al. T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nat Immunol. 2016;17:583. doi: 10.1038/ni.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arbore G, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells. Science. 2016;352:aad1210. doi: 10.1126/science.aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schett G, Dayer JM, Manger B. Interleukin-1 function and role in rheumatic disease. Nat Rev Rheumatol. 2016;12:14. doi: 10.1038/nrrheum.2016.166. [DOI] [PubMed] [Google Scholar]
- 25.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 27.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi N, et al. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature. 2014;513:95. doi: 10.1038/nature13706. [DOI] [PubMed] [Google Scholar]
- 30.Lin J, et al. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540:124. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plagnol V, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng C, Baum BJ. All human EF1alpha promoters are not equal: markedly affect gene expression in constructs from different sources. Int J Med Sci. 2014;11:404. doi: 10.7150/ijms.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatorje EJ, et al. Paediatric reference values for the peripheral T cell compartment. Scand J Immunol. 2012;75:436. doi: 10.1111/j.1365-3083.2012.02671.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.