Abstract
During differentiation thousands of genes are repositioned toward or away from the nuclear envelope. These movements correlate with changes in transcription and replication timing. Here, using synthetic transcription factors (TALE-TFs) we show that transcriptional activation of endogenous genes by a viral trans-activator is sufficient to induce gene repositioning toward the nuclear interior. However, gene relocation is also induced by recruitment of an acidic peptide that decondenses chromatin without affecting transcription, indicating that nuclear reorganization is driven by chromatin remodeling rather than transcription. We identify an epigenetic memory of gene activation, which maintains central nuclear positioning through mitosis even after the TALE-TF is lost. Finally, we demonstrate that transcription activation, but not chromatin decondensation, is sufficient to change replication timing.
Radial nuclear organization of the genome is conserved in eukaryotes (1) with accumulation of heterochromatin, gene-poor and late-replicating chromatin domains near the nuclear envelope (2). Lamin associated domains (LADs) are gene-poor, show low levels of transcription and are depleted for active histone marks (3). Artificial tethering to the nuclear envelope has demonstrated that a peripheral nuclear environment is sufficient to induce transcriptional down-regulation of both reporter and some endogenous genes in somatic cells (4, 5) and during differentiation many genes change their association with components of the nuclear lamina - often associated with altered gene expression (6). However, these correlations do not determine whether re-localization relative to the nuclear periphery is a cause or a consequence of gene regulation during differentiation.
Two thirds of the genes that lose lamin B1 association during differentiation of ESCs to neural precursor cells (NPCs) are transcriptionally up-regulated. The others are more likely to be strongly activated later in differentiation (6). Ptn, Sox6 and Nrp1 are three genes that are up-regulated during ESC differentiation (7); they exhibit some of the most significant losses of LaminB1-Dam association during ESC to NPC differentiation (6) and, concomitantly, Ptn loses its peripheral nuclear position (8). In fact Ptn and Nrp1 expression begins to increase as ESCs differentiate into epiblast stem cells (EpiSCs) (Fig. 1B). Although there is no data on LADs in EpiSCs, correlated to these expression changes, fluorescence in situ hybridization (FISH) shows that Ptn, Sox6 and Nrp1 loci relocate away from the nuclear periphery and toward more central nuclear positions during the differentiation of ESCs to EpiSCs, or to NPCs (Fig. S1, Fig. 2B, p < 0.01).
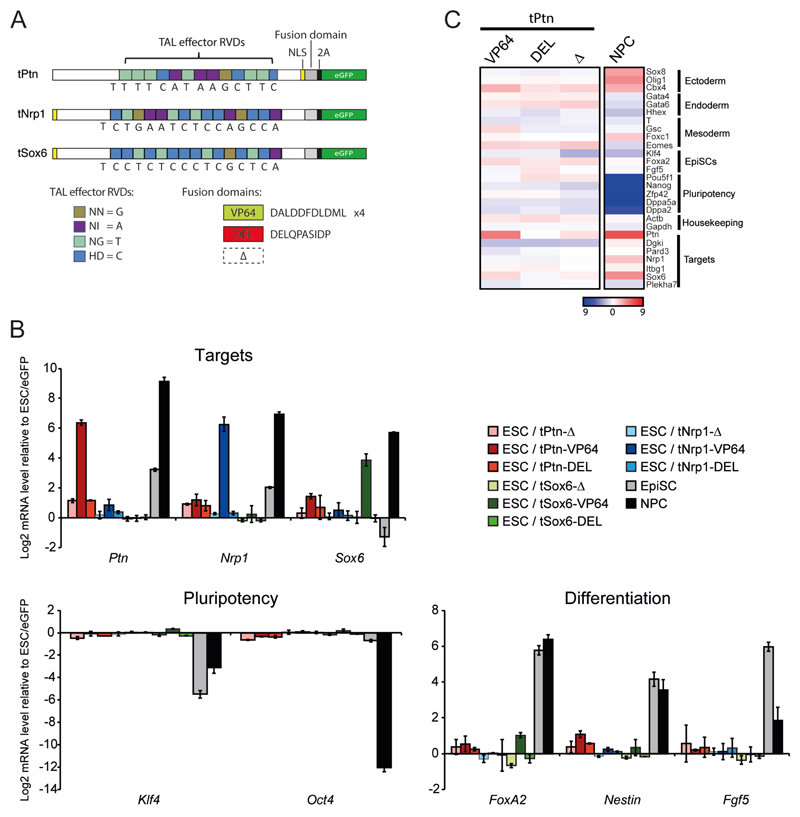
Figure. 1. TALE-TF induces transcriptional activation in mouse ESC.
(A) Schematics of TAL-effector constructs specific for Ptn, Nrp1 and Sox6. DNA binding domains fused to either 4 repeats of VP16 (VP64) or DELQPASIDP peptide (13). Constructs (Δ) with the VP64 domain removed serve as a negative control. Expression of eGFP from the same construct via a self-cleaving (2A) peptide allows for isolation of cells expressing the construct by FACS. (B) Mean (+/- s.e.m.) log2 mRNA level, established by RT-qPCR, for TALE target genes (Ptn, Nrp1, Sox6), genes involved in pluripotency (Oct4, Klf4) or differentiation (FoxA2, Nestin, Fgf5) in ESCs transfected with the different TALE vectors. Expression is shown relative to eGFP transfection. Expression changes are also shown for the differentiation of ESCs to EpiSCs or NPCs. n = 3 biological replicates. (C) Heatmap showing 27 genes from the expression microarray selected to represent different differentiation states, as well as genes surrounding the regions targeted by the TAL-effectors. Log2 transformed mRNA ratios for ESCs transfected by the different tPtn constructs relative to eGFP transfection, or for cells differentiated into NPCs compared to levels in ESCs, are displayed using the color code shown.
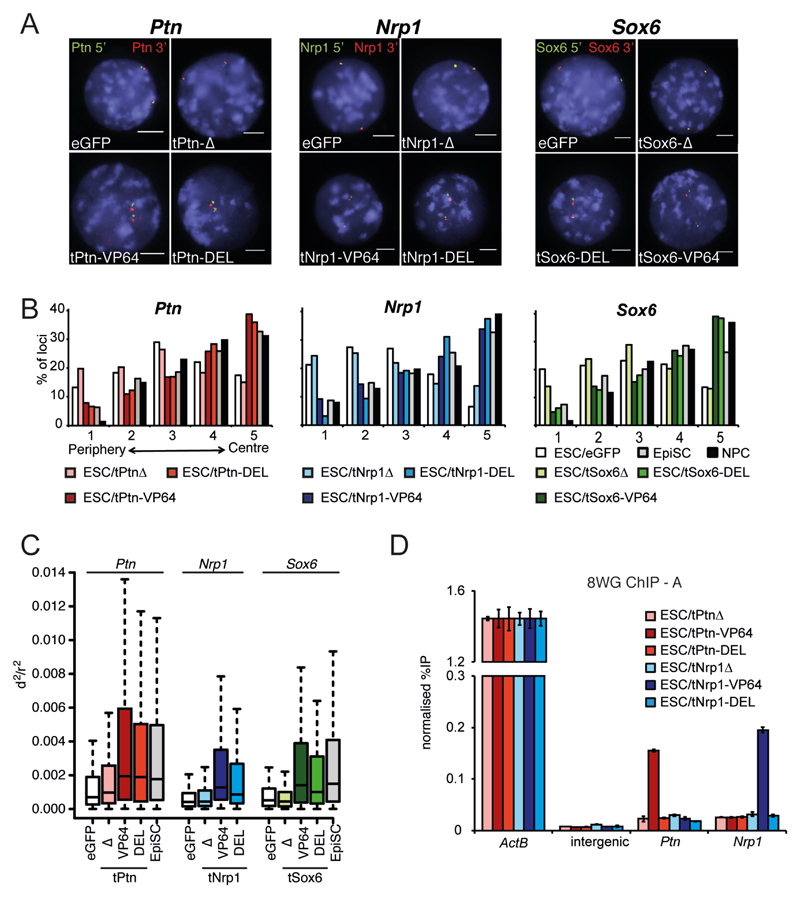
Figure. 2. Synthetic chromatin decondensation causes gene relocalization.
(A) FISH with probes flanking Ptn (left), Nrp1 (center) and Sox6 (right), in nuclei of ESCs transfected by eGFP or the -Δ, -VP64 and -DEL TALEs targeting Ptn (left); Nrp1 (center); and Sox6 (right). Nuclei were counter- stained with DAPI (blue). Scale bar = 5 μm. (B) Distribution of Ptn (left), Nrp1 (center) and Sox6 (right) hybridization signals across 5 concentric shells eroded from the periphery (shell 1) through to the center (shell 5) of the nucleus (Fig. S1A) in ESCs transfected by eGFP or the different TALEs constructs, or for ESCs differentiated to EpiSCs and NPCs. n= 100-150 nuclei for at least 2 biological replicates. (C) Box plots showing the distribution of normalized squared interprobe distances (d2/r2) at Ptn (left), Nrp1 (center) and Sox6 (right) in ESCs transfected by eGFP (open bars) or the corresponding TALEs. Un-transfected EpiSCs differentiated from ESCs are shown for comparison (dark gray). Shaded boxes show the median and interquartile range of the data. n = 100-150 nuclei. Statistical analysis is in Table S1. (D) ChIP for RNAPII (8WG antibody) at Actb, Ptn, and Nrp1 promoters and at an intergenic negative contro, in ESCs transfected by tPtn-Δ (light red), tPtn-VP64 (dark red), tPtn-DEL (red), tNrp1-Δ (light blue), tNrp1-VP64 (dark blue), tNrp1-DEL (blue). Enrichment is shown as mean % input bound ± SD over three technical replicates of biological replicate A (replicate B is shown in Fig. S5).
To directly address the role of transcription in nuclear re-organization we ectopically activated Ptn, Sox6 or Nrp1 in ESCs using synthetic transcription factors composed of TALE DNA binding domains with specificity for the respective gene promoters (9), fused to VP64: a tetramer of the VP16 acidic transcriptional activator (10, 11) (Fig. 1A). When transfected into ESCs, tPtn-VP64 induces expression of its target >30 – 90 fold (Fig 1B-C). Other than Ptn, only two additional genes (Il33 and Nnmt) were significantly upregulated, and genes involved in ESC pluripotency or differentiation were not significantly changed. This suggests that Ptn upregulation is not just an indirect consequence of differentiation triggered by transfection or the non-specific expression of an acidic activator (Fig. 1C, Fig. S2). Specific activation of Nrp1 or Sox6 in cells transfected by tNrp1-VP64 or tSox6-VP64, respectively, also showed no expression signature of differentiation (Fig 1B). Control plasmids lacking the VP64 domain (tPtn-Δ, tSox6-Δ and tNrp1-Δ) had almost no effect (Fig. 1B-C). Furthermore, we did not detect any changes in the expression of genes neighboring those targeted by the TALEs (Fig. 1C, Fig. S3).
As well as activating Ptn, Nrp1 or Sox6, FISH showed that tPtn-VP64, tNrp1-VP64 and tSox6-VP64 caused specific relocalization of the targeted loci toward the center of ESC nuclei compared to either control eGFP transfection (tPtn-VP64: p = 4.6 x 10-9, tNrp1-VP64: p = 5.3 x 10-14, tSox6-VP64: p = 6.4 x 10-12) or to constructs lacking the activation domain (tPtn-Δ: p = 3 x 10-6, tNrp1-Δ: p = 8.1 x 10-10, tSox6-Δ: p = 8 x 10-12) (Fig. 2A-B). The extent of this relocalisation is similar to that seen upon normal differentiation of ESCs to EpiSCs or NPCs (Fig. S1 and Fig. 2B). tPtn-VP64 and tNrp1-VP64 did not significantly affect radial positioning of Sox6 (tPtn-VP64: p = 0.56; tNrp1-VP64: p = 0.06), and tSox6-VP64 did not affect Nrp1 positioning (p = 0.08) (Fig. S4A-C). This reinforces that TALE transfection does not simply induce differentiation. We conclude that forced Ptn, Nrp1 or Sox6 expression induces relocalization of the targeted loci toward the center of the nucleus independent of differentiation. This is consistent with a report showing that targeting VP16 to LaminB1-DAM associated sequences disrupts chromatin at the nuclear periphery in somatic cells, although in that case this could not be linked to specific effects on transcription (12).
To dissect which step of gene activation is required for gene repositioning we replaced the VP64 domain of the TALE-TFs with an acidic peptide (DELQPASIDP - tPtn-DEL, tNrp1-DEL and tSox6-DEL) that is known to decondense chromatin when targeted to an artificial transgene array in somatic cells, but without activating transcription (Fig. 1B-C) (13). We confirmed the absence of significant gene expression changes in cells transfected with t-DEL constructs (Fig. 1B-C, Fig. S2). Analysis of normalized mean-squared interprobe distances (d2/r2) (14) between FISH probes flanking Ptn, Nrp1 and Sox6, revealed that DEL peptide recruitment caused significant chromatin decondensation across the targeted loci compared to eGFP control (tPtn-DEL: p = 2.4 x 10-6; tNrp1-DEL: p = 6.1 x 10-10; tSox6-DEL: p = 3.6 x 10-6; Fig. 2C and Table S1). In contrast, constructs lacking a trans-activation domain had no effect. The effect of the DEL peptide on chromatin decondensation is similar to that observed with VP64 and to that seen at Ptn and Sox6 upon the differentiation of ESCs to EpiSCs (Fig. 2C, Table S1). Chromatin decondensation at Ptn, Nrp1 or Sox6, induced by their respective t-DELs, was accompanied by relocation of the respective loci toward the nuclear center (Fig. 2B, tPtn-DEL: p = 6.22 x 10-6; tNrp1-DEL: p = 3.4 x 10-23; tSox6-DEL: p = 1.8 x 10-8). The nuclear repositioning induced by DEL peptide is similar to that induced by VP64 (tPtn-VP64: p = 0.93; tNrp1-VP64: p = 0.03; tSox6-VP64: p = 0.9). Neither tPtn nor tNrp1 affect Sox6 condensation and Nrp1 condensation is not affected by TALEs targeting Sox6 (Fig. S4D and Table S1). We also did not observe any off-target effects for TALE-DEL proteins on radial positioning (Fig. S4A-C). We confirmed that the effect of the DEL peptide was independent of the transcription machinery by chromatin immunoprecipitation (ChIP) for RNA-polymerase II (9). Whilst we observed RNAPII recruitment at Ptn and Nrp1 promoters in cells transfected by tPtn-VP64 and tNrp1-VP64 respectively, no significant enrichment was observed with tPtn-DEL or tNrp1-DEL (Fig. 2D, Fig. S5).
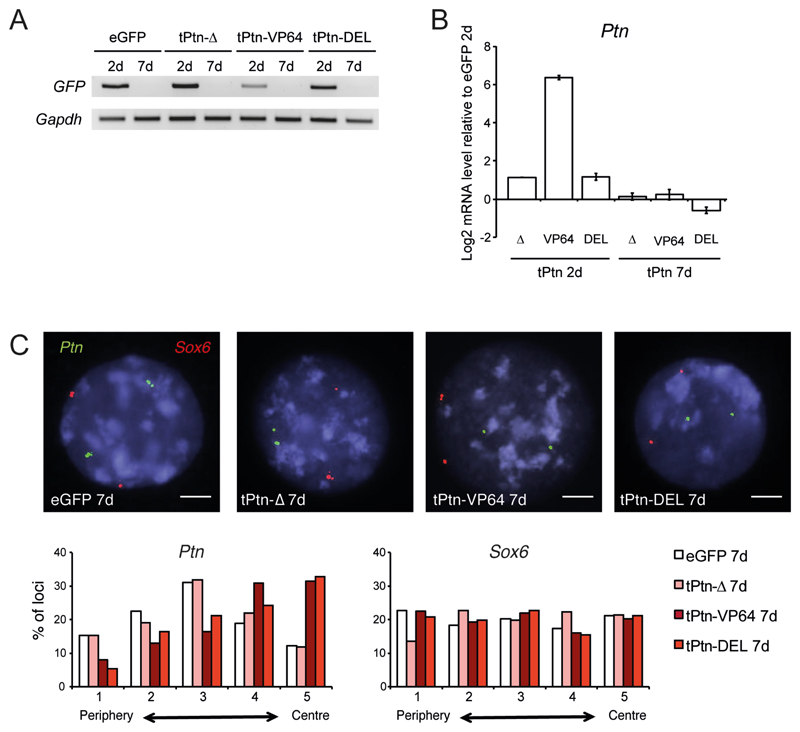
After mitosis there is stochastic re-positioning of LADs and late replicating domains to the nuclear periphery (12, 15, 16). However, it is not clear to what extent this re-establishment of nuclear organization depends on the persistence of transcriptional states or chromatin structure. We therefore examined nuclear repositioning of Ptn after loss of the original stimulus. TALE plasmid transfections of ESCs were transient; hence GFP expression from the constructs was no longer detected after 7 days in culture (Fig. 3A). Concomitant with overall loss of the tPtn-VP64 plasmid, 7 days after transfection Ptn expression had returned close to its level in ESCs, although we cannot exclude some cell to cell heterogeneity (Fig. 3B). However, Ptn remained in a more central nuclear position at this time point after both tPtn-VP64 and tPtn-DEL transfections (p = 6.5 x 10-9 and 4.73 x 10-8 relative to eGFP; Fig. 3C). This suggests that there might be epigenetic inheritance of an altered radial nuclear position – at least for the tested Ptn locus.
Figure. 3. Central nuclear positioning is maintained after the loss of gene induction.
(A) RT-PCR of Gfp mRNA in ESC, 2 (2d) or 7 days (7d) after transfection with tPtn-Δ, tPtn-VP64 and tPtn-DEL. Gapdh serves as a constitutively expressed control. (B) Mean (+/- s.e.m.) log2 mRNA level for Ptn in ESCs 2 (left) or 7 days (right) after transfection with tPtn-Δ, tPtn-VP64 and tPtn-DEL. Expression is shown relative to eGFP transfection (n = 3; 2 biological and 1 technical replicates). (C) FISH at Ptn (green) and Sox6 (red), in ESCs 7 days after transfection by eGFP, tPtn-Δ, tPtn-VP64 or tPtn-DEL. Scale bar = 5 μm. Histograms below quantify the nuclear distribution of Ptn (left) and Sox6 (right) signals - as in Fig. 2B - for ESCs 7 days after transfection by eGFP (open bars), tPtn-Δ (light red), tPtn-VP64 (dark red) and and tPtn-DEL (red). n = 100-150 nuclei for each condition for each of at least 2 biological replicates.
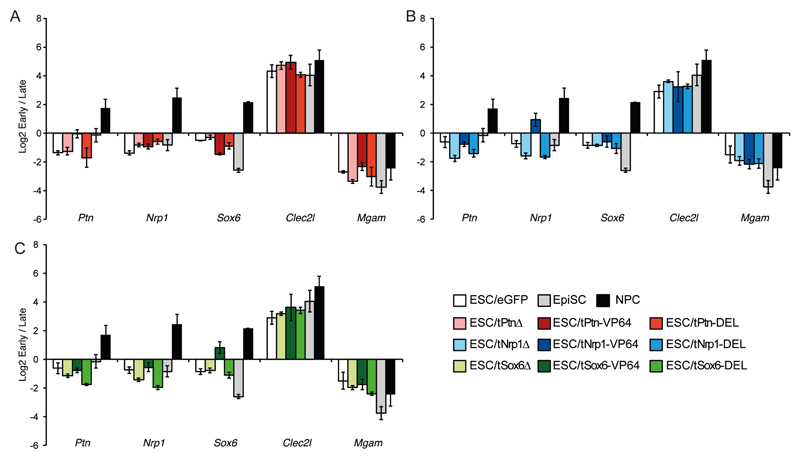
Late replicating DNA is concentrated around the nuclear periphery; transcription activation and gene relocation away from the nuclear envelope often correlate with a shift to an earlier replication time during S phase (8, 17, 18). Indeed, replication timing of Ptn, Nrp1 and Sox6 shifts from late to early S-phase during ESC to NPC differentiation (18). We confirmed the late replication timing of Ptn in undifferentiated ESCs and its progressive shift to earlier time points during differentiation (Fig. 4). Nrp1 and Sox6 remain late replicating in EpiSCs but become early replicating in NPCs. Clec2l and Mgam regions serve as early and late replicating controls respectively and their replication timing remains unchanged during ESC differentiation.
Figure. 4. Synthetic activation of transcription but not nuclear re-positioning shifts replication timing.
Mean (+/- s.e.m.) log2 ratio of early: late S phase fraction for Ptn, Nrp1, Sox6, Clec2l and Mgam, in ESCs after transfection with eGFP (white), tPtn-Δ (light gray), tPtn-VP64 (dark red), tPtn-DEL (light red), tNrp1-Δ (light blue), tNrp1-VP64 (dark blue), tNrp1-DEL (blue), tSox6-Δ (light green), tSox6-VP64 (dark green), tSox6-DEL (green). For comparison the changes in replication timing during the differentiation of ESCs to EpiSCs or NPCs are included.
To dissect the relationships between transcription, chromatin decondensation, nuclear positioning and replication timing we measured the effect of the different TALE constructs on the replication timings of Ptn, Nrp1 and Sox6 in ESCs (9). TALEs lacking an activation domain (Δ) had no effect, but TALE-VP64 mediated activation is sufficient to advance the replication timing of the targeted locus (Fig. 4A-C), with no detectable off target effects on the other tested loci. In contrast the DEL peptide, which decondenses chromatin (Fig. 2C) and relocates (Fig. 2B) the targeted locus without inducing expression (Fig. 1B-C) or recruiting RNAPII (Fig. 2D), has no effect on replication timing (Fig. 4A-C). This demonstrates that neither chromatin decondensation nor nuclear relocalization are sufficient to shift replication time and suggests that the switch to early replication requires the activation of gene expression. This conclusion is reinforced by the analysis of replication timing 7 days after tPtn-VP64 transient transfection by which time Ptn activation is lost. At this time point Ptn had returned to a late replicating state (Fig. S6), even though it retained a central radial nuclear position (Fig. 3C). We therefore conclude that altered replication timing is likely to be a direct consequence of transcription.
Our data suggest that the global nuclear reorganization observed during early embryogenesis could be the consequence of chromatin regulation. This could happen concomitantly with transcription changes, as for Ptn or Nrp1 that are both repositioned (Fig. S1, Fig. 2B) and upregulated (Fig. 1B) in EpiSCs, or it could occur independently, as for Sox6, which we find decondenses and relocates in EpiSCs without any detectable transcriptional changes. A full dissection of chromatin events mediated by the DEL peptide, and those present at targeted genes after loss of the original TAL-TF would give further insight into the mechanisms involved.
Supplementary Material
One Sentence Summary.
We elucidate cause and effect relationships between transcription, chromatin decondensation, replication timing and nuclear positioning.
Acknowledgments
P.T was a recipient of a Marie Curie Intra European Fellowship. A.J.W. was a recipient of Sir Henry Wellcome and Sir Henry Dale Fellowships from the Wellcome Trust. We are grateful to Elisabeth Freyer for FACs analysis, Anestis Tsakiridis and Alessandra Lavigni for cell culture assistance and Paul Perry for help with image analysis. We thank Pradeepa Madapura-Marulasiddappa and Nick Gilbert for their insightful comments. The work was supported by the UK Medical Research Council and by an ERC advanced grant (249956). The GEO Accession number for microarray expression data is: GSE62379
References
- 1.Meister P, Taddei A. Building silent compartments at the nuclear periphery: a recurrent theme. Current opinion in genetics & development. 2013;23:96–103. doi: 10.1016/j.gde.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nature reviews. Genetics. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 3.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 4.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS genetics. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 6.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS biology. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Materials and methods are available as supplementary materials at Science Online.
- 10.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell stem cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter AE, Memedula S, Plutz MJ, Belmont AS. Common effects of acidic activators on large-scale chromatin structure and transcription. Molecular and cellular biology. 2005;25:958–968. doi: 10.1128/MCB.25.3.958-968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Molecular cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Molecular cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 16.Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Current biology : CB. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 17.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. Journal of cell science. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 18.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome research. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 20.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nature biotechnology. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 21.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & development. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Human molecular genetics. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 24.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 25.Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM. Genome-scale analysis of replication timing: from bench to bioinformatics. Nature protocols. 2011;6:870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.