Abstract
Human pathogens belonging to the Nakaseomyces clade include Candida glabrata sensu stricto, Candida nivariensis and Candida bracarensis. Their highly similar phenotypic characteristics often lead to misidentification by conventional laboratory methods. Therefore, limited information on the true epidemiology of the Candida glabrata species complex is available. Due to life-threatening infections caused by these species, it is crucial to supplement this knowledge. The aim of the study was to estimate the prevalence of C. bracarensis and C. nivariensis in a culture collection of C. glabrata complex isolates. The study covered 353 isolates identified by biochemical methods as C. glabrata, collected from paediatric and adult patients hospitalised at four medical centres in Southern Poland. The multiplex PCR was used to identify the strains. Further species confirmation was performed via sequencing and matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) analysis. One isolate was recognised as C. bracarensis (0.28%). To our knowledge, it is the first isolate in Poland. C. glabrata sensu stricto species has been confirmed for all the remaining isolates. No C. nivariensis was found. Our study has shown that the prevalence of C. nivariensis and C. bracarensis strains is infrequent. However, it should be emphasised that the incidence of these strains may differ locally and depend on environmental factors and the population.
Introduction
Candida glabrata species complex, including Candida glabrata sensu stricto, Candida nivariensis and Candida bracarensis, are the only human pathogens belonging to the Nakaseomyces clade (Gabaldón et al. 2013). The latter two yeast species have been described for the first time about 10 years ago, but their clinical relevance is still to be determined (Alcoba-Flórez et al. 2005; Correia et al. 2006). Several reports from different parts of the world indicate their association with bloodstream infections, invasive and oral candidiasis, urinary tract infections and vulvovaginitis (Angoulvant et al. 2016). A decreased susceptibility to some antifungal agents, observed among the isolates, is particularly important (Fujita et al. 2007; Borman et al. 2008). Although their pathogenicity has been well documented, the true epidemiology of infections caused by species belonging to C. glabrata complex is not yet fully understood. Due to little evidence for phenotypic differences between C. nivariensis, C. bracarensis and C. glabrata, these species cannot be distinguished by conventional methods (Criseo et al. 2015). It is one of the reasons why in the last years a number of molecular methods have been developed to improve identification of these microorganisms. While DNA sequencing of the fungal rDNA ITS region is considered as the reference method for confirmation of species identification, PCR-based assays, proposed by some authors, have proved to be highly efficient in the identification of C. nivariensis and C. bracarensis (Romeo et al. 2009; Enache-Angoulvant et al. 2011; Taverna et al. 2013). The advantages of these methods are their simplicity, time and cost-effectiveness, as well as accuracy, making them suitable tools for investigating Candida epidemiology at a local level.
The aim of the study was to estimate the prevalence of C. bracarensis and C. nivariensis in a culture collection of C. glabrata complex isolates, obtained from ambulatory and hospitalised patients at four different medical centres in Southern Poland, using the multiplex polymerase chain reaction (PCR) method.
Material and methods
Strains collection and phenotypic identification
The study covered 353 yeast strains collected from paediatric and adult patients (56 isolates; 15.9% and 297 isolates; 84.1%, respectively) at four medical centres in Kraków, Poland, between 2009 and 2016.
Among the adult patients, most yeast strains were isolated from the lower respiratory tract samples (endotracheal aspirate, sputum, bronchoalveolar lavage fluid) (81; 27.3%), upper respiratory tract samples (49; 16.5%), stool specimens (47; 15.8%), vulvovaginal swabs (45; 15.2%), urine (35; 11.8%) and blood (17; 5.7%). Isolates from children were obtained mainly from stool samples (36; 64.3%), vulvovaginal swabs (8; 14.3%) and urine (5; 8.9%). Antifungal susceptibility patterns of the tested isolates did not differ from epidemiological data reported in the literature, in which most of the isolates were intermediately susceptible to fluconazole.
All isolates were identified as C. glabrata by routine mycological culture on Sabouraud dextrose agar supplemented with chloramphenicol and biochemical testing, e.g. using VITEK 2 Compact automated system (bioMérieux, Marcy l’Etoile, France).
Multiplex PCR-based molecular identification
A total of 353 isolates were analysed by a multiplex PCR assay described by Romeo et al. (2009) using primers targeting the ITS1 region and the 5.8S ribosomal RNA gene. Quality control of the assay was warranted by incorporating reference strains of C. glabrata (ATCC 2001), C. nivariensis (CBS 9983) and C. bracarensis (CBS 10154).
Candida bracarensis—confirmation of species identification (phenotypic and molecular analysis of the isolate)
The isolate identified as C. bracarensis by PCR analysis has been subjected to further phenotypic characterisation, including subculture on chromogenic media (CHROMagar Candida, Becton Dickinson, Franklin Lakes, NJ, USA; Candida Chromogenic LAB-AGAR ™, Biocorp, Warsaw, Poland), evaluation of fermentation and assimilation properties as well as the ability to hydrolyse urea by API Candida (bioMérieux, Marcy l’Etoile, France) and AUXAColor 2 (BioRad, Marnes-la-Coquette, France), and estimation of morphological features by culture on a nutritionally poor medium (rice extract agar) and incubation for 48 h at 27 °C.
Furthermore, MALDI-TOF MS identification using MALDI Biotyper (Bruker Daltonics, Billerica, MA, USA) was performed at the Jagiellonian Centre of Innovation (Kraków, Poland). The analysis was conducted using the IVD (in vitro diagnostic) database and scores ranging 2.00–3.00 were considered as a high-confidence identification of a microorganism to the species level.
Confirmation of species identification based on bi-directional sequencing of the PCR product and comparison of the consensus sequence with that of the C. bracarensis CBS 10154 reference strain (GenBank accession number GU1990440.1) was performed by Genomed (Warsaw, Poland). Furthermore, we analysed the sequence similarity using the BLASTn tool of the National Centre for Biotechnology Information (NCBI) (ncbi.nlm.nih.gov/BLAST/).
Susceptibility testing
In vitro susceptibility testing of the C. bracarensis isolate against amphotericin B (AMB), fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), caspofungin (CS), anidulafungin (AND) and flucytosine (5FC) was performed using strips impregnated with antifungal agent gradient, E-strips (AB BIODISK, Solna, Sweden and Liofilchem, Italy). The MIC values were determined in accordance with the manufacturer’s instructions.
Results
Multiplex PCR assay
Based on the multiplex PCR assay targeting the ITS1 region and the 5.8S ribosomal RNA gene, 352 isolates were identified as C. glabrata sensu stricto (99.7%) and one isolate (SU3498) as C. bracarensis (0.3%) (Fig. 1). No C. nivariensis were found.
Fig. 1.
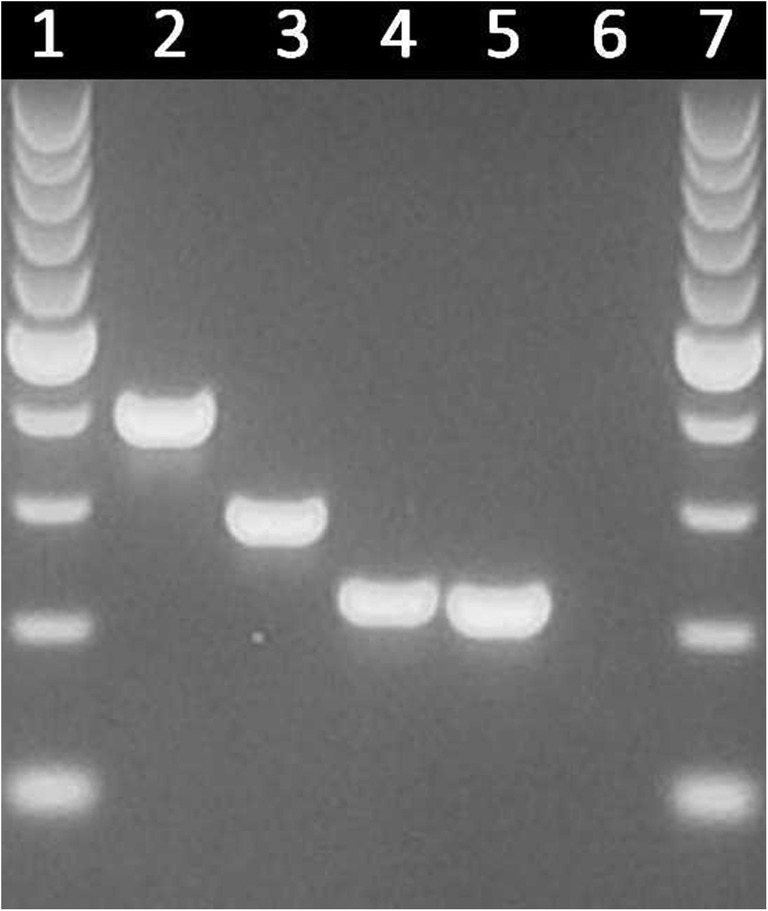
Multiplex PCR assay results of references strains and the clinical isolate identified as C. bracarensis. Lines: 1, 7—marker; 2—C. glabrata ATCC 2001; 3—C. nivariensis CBS 9983; 4—C. bracarensis CBS 10154; 5—clinical isolate SU3498; 6—negative control
Candida bracarensis—the isolate characterisation (phenotypic and molecular analysis)
The SU3498 isolate was cultured from a stool sample obtained from a 32-year-old man hospitalised at the Department of Metabolic Diseases of the University Hospital in Kraków due to peripheral neuropathy in type 1 diabetes in May 2016.
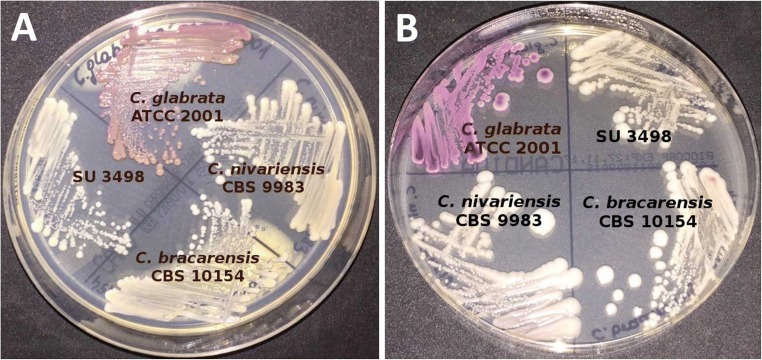
The SU3498 isolate produced small, white, shiny and smooth colonies on the Sabouraud dextrose agar. On both chromogenic media CHROMagar Candida (Becton Dickinson, Franklin Lakes, NJ, USA) and Candida Chromogenic LAB-AGAR TM (Biocorp, Warsaw, Poland), it also formed white colonies (Fig. 2). The isolate produced slow-growing colonies, better at 30 ± 2 °C than at 35 ± 2 °C, and did not form either hyphae or pseudohyphae after incubation on rice extract agar.
Fig. 2.
The appearance of C. glabrata ATCC 2001, C. nivariensis CBS 9983, C. bracarensis CBS 10154 and clinical isolate SU3498 after 48 h of incubation at 35 °C on CHROMagar Candida (Becton Dickinson, Franklin Lakes, NJ, USA) (a) and Candida Chromogenic LAB-AGAR ™ (Biocorp, Warsaw, Poland) (b)
The tested isolate fermented and assimilated glucose and trehalose but did not hydrolyse urea. We observed the inability of this isolate to assimilate maltose, sucrose, galactose, lactose, raffinose, inositol, cellobiose, adonitol, melezitose, xylose and arabinose.
Proteomic identification by the MALDI-TOF MS method has confirmed that the SU3498 isolate belongs to the species C. bracarensis with a score value of 2.1.
The consensus sequence of the PCR product (GenBank accession number MH729062) showed 98% similarity to that of C. bracarensis type strain CBS 10154 (GenBank accession number GU1990440.1). The BLAST search of all available nucleotide sequences revealed that this sequence was identical to that of the C. bracarensis strain OAHPPFR1227 (GenBank accession number JN882338.1) as well as uncultured fungus clone CMH366 (GenBank accession number KF800457.1), isolated from an environmental sample (Rittenour et al. 2014).
The SU3498 isolate has been deposited at the Polish Collection of Microorganisms under the number PCM 2995.
Susceptibility testing
Antifungal susceptibility test results were as follows: amphotericin B (MIC = 0.19 mg/L), flucytosine (MIC = 0.25 mg/L), fluconazole (MIC = 3 mg/L), voriconazole (MIC = 0.125 mg/L), itraconazole (MIC ≥32 mg/L), posaconazole (MIC ≥32 mg/L), anidulafungin (MIC = 0.004 mg/L) and caspofungin (MIC = 0.125 mg/L).
Discussion
Although more than 17 different species of Candida have been reported to be pathogenic to humans, about of 90% of invasive candidiasis are caused by five species—C. albicans, C. glabrata, C. parapsilosis, C. tropicalis and C. krusei (Pfaller and Diekema 2007). C. albicans remains the predominant species recovered from clinical specimens worldwide. However, in the last decades, non-albicans species have become increasingly prevalent. C. glabrata currently ranks second as the causative agent of candidiasis in North America and Europe. This changing epidemiology is associated with widespread use and prolonged prophylaxis of azole antifungal agents in the growing population of high-risk patients. Despite C. glabrata lacks a number of virulence factors (hyphal growth, ability to secrete proteases), it has enormous adaptability to different host niches and intrinsically low susceptibility to azoles. Therefore, infections caused by these yeasts are difficult to eradicate and associated with significant mortality (Rodrigues et al. 2014; Kołaczkowska and Kołaczkowski 2016).
C. glabrata is taxonomically classified to Nakaseomyces clade together with three environmental species: C. castellii, Nakaseomyces delphensis (syn. Kluyveromyces delphensis) and Nakaseomyces bacillisporus (syn. Kluyveromyces bacillisporus) (Kurtzman 2003). In the last decade, two new species, C. nivariensis and C. bracarensis, isolated from samples taken from patients with pathological conditions, were added to this clade. These emerging pathogens have phenotypic characteristics similar to those of C. glabrata, making them difficult to identify by routine laboratory analysis. Conventional phenotypic-based methods of identification, such as the Vitek 2 system, have proven ineffective to differentiate isolates of C. glabrata complex (Alcoba-Flórez et al. 2005; Correia et al. 2006; Hou et al. 2017), as was the case in the present study, where SU3498 isolate was “misidentified” as C. glabrata.
For the first time, C. nivariensis has been reported in a Spanish hospital in 2005. The strains were isolated from blood, bronchoalveolar lavage fluid (BALF) and urine of three patients over a 3-year study period (Alcoba-Flórez et al. 2005). Current knowledge indicates that this species occurs worldwide and the number of published cases reaches 108 in Europe (Spain, the UK, France, Poland) (Borman et al. 2008; Gorton et al. 2013; López-Soria et al. 2013; Parmeland et al. 2013; Swoboda-Kopeć et al. 2014; Aznar-Marin et al. 2016; Angoulvant et al. 2016), 31 in Asia (Japan, Indonesia, India, China, Malaysia) (Fujita et al. 2007; Wahyuningsih et al. 2008; Chowdhary et al. 2010; Sharma et al. 2013; Li et al. 2014; Tay et al. 2014; Feng et al. 2015; Hou et al. 2017), 11 in Australia (Lockhart et al. 2009; Pinto et al. 2011), four in Latin America (Argentina and Brazil) (Morales-López et al. 2016; Figueiredo-Carvalho et al. 2016) and one case in Africa (Burkina Faso) (Sanata et al. 2014).
Candida nivariensis was found to be uncommon among clinical Candida isolates. Most studies have reported only isolated clinical cases at medical centres.
Reports on large collections of yeast strains, acquired under surveillance programmes, both national and global, have shown a low prevalence of C. nivariensis of around 0.12% (Angoulvant et al. 2016; Hou et al. 2017). To date, most isolates of C. nivariensis have been reported from Europe, mainly France (68 cases), the UK (16) and Poland (13).
It is worth noting that C. nivariensis strains have not been detected among 353 C. glabrata isolates obtained from patients hospitalised at the above mentioned four medical centres in Kraków, while Swoboda-Kopeć et al. have found 13 C. nivariensis isolates among 224 (5.8%) C. glabrata complex strains collected from patients of the Warsaw Medical University Clinical Hospital (Swoboda-Kopeć et al. 2014). This is particularly significant, as these cities are only about 300 km from each other. However, Warsaw is the capital of Poland and the population in this agglomeration is about three times larger and more varied than that of Kraków. The volume of commuting in Warsaw is also higher and international trade and travel can potentially contribute to the occurrence of new pathogens (Neiderud 2015).
The number of reported C. bracarensis isolates is even lower than that of C. nivariensis. The first two strains were isolated as the causative agent of a case of vulvovaginal candidiasis (Portugal, 2006) and from a blood culture in the UK in 2003 (Correia et al. 2006). To date, a total of 10 C. bracarensis strains have been isolated in Europe (Portugal, the UK, France, Spain) (Correia et al. 2006; Cendejas-Bueno et al. 2010; Lacroix et al. 2014; Angoulvant et al. 2016), seven in North America (USA and Canada) (Bishop et al. 2008; Lockhart et al. 2009; Warren et al. 2010), two in Asia (China) and two in Latin America (Argentina) (Li et al. 2014; Hou et al. 2017; Dudiuk et al. 2017). We have recently isolated C. bracarensis from the stool of a patient with peripheral neuropathy caused by diabetes mellitus. Although C. nivariensis and C. bracarensis are reported as a part of the commensal flora of the gastrointestinal or genitourinary tract mucous membranes, the cases of fungaemia caused by these species confirm their pathogenic significance to humans. It should be noted that risk factors for invasive fungal infections, especially candidaemia, include also diabetes mellitus.
It is worth mentioning that the presence of C. nivariensis and C. bracarensis strains was confirmed also in the environment. The D1–D2 sequences of the first three Spanish C. nivariensis isolates revealed a high level of similarity (99%) with the sequence of a Candida sp. AF313362 belonging to a strain isolated by Lachance et al. (2001) from Hibiscus sp. flowers in the Northern Territories (Canada). Eleven strains of C. nivariensis have been also isolated from the phylloplane of sugarcane leaf in Thailand (Limtong and Koowadjanakul 2012; Limtong et al. 2014). The analysed sequence of C. bracarensis strain described in this study was identical to the uncultured fungus clone CMH366 (GenBank accession number KF800457.1) isolated from an environmental sample (air and dust) collected in the homes of asthmatic children, located in Kansas City (Rittenour et al. 2014). Candidiasis is most often an endogenous infection; however, exogenous infections have been also reported. Therefore, it can be assumed that C. nivariensis and C. bracarensis distributed in the environment could be a potential source of infections, especially in high-risk patients.
Varying susceptibility of C. nivariensis and C. bracarensis strains to antifungal agents is of particular concern. Several isolates have been reported either resistant or with reduced susceptibility to azoles or amphotericin B (Bishop et al. 2008; Lockhart et al. 2009). The C. bracarensis isolate described in our study had itraconazole and posaconazole MICs ≥ 32 mg/L, and fluconazole MIC = 3 mg/L. Transfer of resistance does not occur among yeasts and the acquisition of resistance can be observed mainly in restricted clinical settings, due to prolonged azole treatment. Therefore, species identification is generally sufficient to predict their drug susceptibility (Cornet et al. 2011). Empirical therapy is a common practice and the selection of antifungal agents is usually based on prediction of the most likely pathogens and their susceptibility patterns. Accurate identification of new species is becoming increasingly important for clinical management.
In the light of increasing frequency of isolation of cryptic species belonging to the Nakaseomyces clade from human samples, and decreased susceptibility to antifungals observed in some isolates, further studies on geographical distribution of C. nivariensis and C. bracarensis and clinical characterisation of infections caused by these species are required. The true incidence of C. nivariensis and C. bracarensis remains unknown. Further isolates have been reported from different parts of the world, indicating that these pathogens are widely distributed and play an important role in human infections. Published data suggest that C. nivariensis and C. bracarensis seem to be prevalent more locally. The best approach to the treatment of Candida infections, ensuring patients’ best health outcomes, is early detection and correct identification of the species in order to administer appropriate antifungal therapy.
Funding information
Scientific work financially was supported by the Jagiellonian University Medical College with founds from maintenance of the research potential of the JU MC Department of Pharmaceutical Microbiology (No. K/DSC/002884).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Contributor Information
Marianna Małek, Phone: +48 12 620 57 50, Email: marianna.tokarczyk@uj.edu.pl.
Paulina Mrowiec, Phone: +48 12 620 57 50, Email: paulina.mrowiec@uj.edu.pl.
References
- Alcoba-Flórez J, Méndez-Alvarez S, Cano J, Guarro J, Pérez-Roth E, del Pilar Arévalo M. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol. 2005;43(8):4107–4111. doi: 10.1128/JCM.43.8.4107-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angoulvant A, Guitard J, Hennequin C. Old and new pathogenic Nakaseomyces species: epidemiology, biology, identification, pathogenicity and antifungal resistance. FEMS Yeast Res. 2016;16(2):fov114. doi: 10.1093/femsyr/fov114. [DOI] [PubMed] [Google Scholar]
- Aznar-Marin P, Galan-Sanchez F, Marin-Casanova P, García-Martos P, Rodríguez-Iglesias M. Candida nivariensis as a new emergent agent of vulvovaginal candidiasis: description of cases and review of published studies. Mycopathologia. 2016;181(5–6):445–449. doi: 10.1007/s11046-015-9978-y. [DOI] [PubMed] [Google Scholar]
- Bishop JA, Chase N, Magill SS, Kurtzman CP, Fiandaca MJ, Merz WG. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J Clin Microbiol. 2008;46(2):443–446. doi: 10.1128/JCM.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J Clin Microbiol. 2008;46(3):933–938. doi: 10.1128/JCM.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendejas-Bueno E, Gomez-Lopez A, Mellado E, Rodriguez-Tudela JL, Cuenca-Estrella M. Identification of pathogenic rare yeast species in clinical samples: comparison between phenotypical and molecular methods. J Clin Microbiol. 2010;48(5):1895–1899. doi: 10.1128/JCM.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Randhawa HS, Khan ZU, Ahmad S, Juneja S, Sharma B, Roy P, Sundar G, Joseph L. First isolations in India of Candida nivariensis, a globally emerging opportunistic pathogen. Med Mycol. 2010;48(2):416–420. doi: 10.3109/13693780903114231. [DOI] [PubMed] [Google Scholar]
- Cornet M, Sendid B, Fradin C, Gaillardin C, Poulain D, Nguyen HV. Molecular identification of closely related Candida species using two ribosomal intergenic spacer fingerprinting methods. J Mol Diagn. 2011;13(1):12–22. doi: 10.1016/j.jmoldx.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A, Sampaio P, James S, Pais C. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol. 2006;56(Pt 1):313–317. doi: 10.1099/ijs.0.64076-0. [DOI] [PubMed] [Google Scholar]
- Criseo G, Scordino F, Romeo O. Current methods for identifying clinically important cryptic Candida species. J Microbiol Methods. 2015;111:50–56. doi: 10.1016/j.mimet.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Dudiuk C, Morales-López SE, Podesta V, Macedo D, Leonardelli F, Vitale RG, Tosello ME, Cabeza MS, Biasoli M, Gamarra S, Garcia-Effron G. Multiplex PCR designed to differentiate species within the Candida glabrata complex. Rev Iberoam Micol. 2017;34(1):43–45. doi: 10.1016/j.riam.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Enache-Angoulvant A, Guitard J, Grenouillet F, Martin T, Durrens P, Fairhead C, Hennequin C. Rapid discrimination between Candida glabrata, Candida nivariensis, and Candida bracarensis by use of a singleplex PCR. J Clin Microbiol. 2011;49(9):3375–3379. doi: 10.1128/JCM.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Ling B, Yang X, Liao W, Pan W, Yao Z. Molecular identification of Candida species isolated from onychomycosis in Shanghai, China. Mycopathologia. 2015;180(5–6):365–371. doi: 10.1007/s11046-015-9927-9. [DOI] [PubMed] [Google Scholar]
- Figueiredo-Carvalho MH, Ramos Lde S, Barbedo LS, Chaves AL, Muramoto IA, Santos AL, Almeida-Paes R, Zancopé-Oliveira RM. First description of Candida nivariensis in Brazil: antifungal susceptibility profile and potential virulence attributes. Mem Inst Oswaldo Cruz. 2016;111(1):51–58. doi: 10.1590/0074-02760150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Senda Y, Okusi T, Ota Y, Takada H, Yamada K, Kawano M. Catheter-related fungemia due to fluconazole-resistant Candida nivariensis. J Clin Microbiol. 2007;45(10):3459–3461. doi: 10.1128/JCM.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, Arnaise S, Boisnard S, Aguileta G, Atanasova R, Bouchier C, Couloux A, Creno S, Almeida Cruz J, Devillers H, Enache-Angoulvant A, Guitard J, Jaouen L, Ma L, Marck C, Neuvéglise C, Pelletier E, Pinard A, Poulain J, Recoquillay J, Westhof E, Wincker P, Dujon B, Hennequin C, Fairhead C. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14(14):623. doi: 10.1186/1471-2164-14-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorton RL, Jones GL, Kibbler CC, Collier S. Candida nivariensis isolated from a renal transplant patient with persistent candiduria - molecular identification using ITS PCR and MALDI-TOF. Med Mycol Case Rep. 2013;25(2):156–158. doi: 10.1016/j.mmcr.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xiao M, Chen SC, Wang H, Yu SY, Fan X, Kong F, Xu YC. Identification and antifungal susceptibility profiles of Candida nivariensis and Candida bracarensis in a multi-center Chinese collection of yeasts. Front Microbiol. 2017;19(8):5. doi: 10.3389/fmicb.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kołaczkowska A, Kołaczkowski M. Drug resistance mechanisms and their regulation in non-albicans Candida species. J Antimicrob Chemother. 2016;71(6):1438–1450. doi: 10.1093/jac/dkv445. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4(3):233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Starmer WT, Rosa CA, Bowles JM, Barker JS, Janzen DH. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001;1(1):1–8. doi: 10.1111/j.1567-1364.2001.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Lacroix C, Gicquel A, Sendid B, Meyer J, Accoceberry I, François N, Morio F, Desoubeaux G, Chandenier J, Kauffmann-Lacroix C, Hennequin C, Guitard J, Nassif X, Bougnoux ME. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for the identification of Candida species. Clin Microbiol Infect. 2014;20(2):153–158. doi: 10.1111/1469-0691.12210. [DOI] [PubMed] [Google Scholar]
- Li J, Shan Y, Fan S, Liu X. Prevalence of Candida nivariensis and Candida bracarensis in vulvovaginal candidiasis. Mycopathologia. 2014;178(3–4):279–283. doi: 10.1007/s11046-014-9800-2. [DOI] [PubMed] [Google Scholar]
- Limtong S, Koowadjanakul N. Yeasts from phylloplane and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol. 2012;28(12):3323–3335. doi: 10.1007/s11274-012-1144-9. [DOI] [PubMed] [Google Scholar]
- Limtong S, Kaewwichian R, Yongmanitchai W, Kawasaki H. Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol. 2014;30(6):1785–1796. doi: 10.1007/s11274-014-1602-7. [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Messer SA, Gherna M, Bishop JA, Merz WG, Pfaller MA, Diekema DJ. Identification of Candida nivariensis and Candida bracarensis in a large global collection of Candida glabrata isolates: comparison to the literature. J Clin Microbiol. 2009;47(4):1216–1217. doi: 10.1128/JCM.02315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Soria LM, Bereciartua E, Santamaría M, Soria LM, Hernández-Almaraz JL, Mularoni A, Nieto J, Montejo M. First case report of catheter-related fungemia by Candida nivariensis in the Iberian Peninsula. Rev Iberoam Micol. 2013;30(1):69–71. doi: 10.1016/j.riam.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Morales-López SE, Taverna CG, Bosco-Borgeat ME, Maldonado I, Vivot W, Szusz W, Garcia-Effron G, Córdoba SB. Candida glabrata species complex prevalence and antifungal susceptibility testing in a culture collection: first description of Candida nivariensis in Argentina. Mycopathologia. 2016;181(11–12):871–878. doi: 10.1007/s11046-016-0052-1. [DOI] [PubMed] [Google Scholar]
- Neiderud CJ. How urbanization affects the epidemiology of emerging infectious diseases. Infect Ecol Epidemiol. 2015;24(5):27060. doi: 10.3402/iee.v5.27060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeland L, Gazon M, Guerin C, Argaud L, Lehot JJ, Bastien O, Allaouchiche B, Michallet M, Picot S, Bienvenu AL, Study Group Candida albicans and non-Candida albicans fungemia in an institutional hospital during a decade. Med Mycol. 2013;51(1):33–37. doi: 10.3109/13693786.2012.686673. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One. 2011;6(10):e25712. doi: 10.1371/journal.pone.0025712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenour WR, Ciaccio CE, Barnes CS, Kashon ML, Lemons AR, Beezhold DH, Green BJ. Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ Sci Process Impacts. 2014;16(1):33–43. doi: 10.1039/C3EM00441D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CF, Silva S, Henriques M. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2014;33(5):673–688. doi: 10.1007/s10096-013-2009-3. [DOI] [PubMed] [Google Scholar]
- Romeo O, Scordino F, Pernice I, Lo Passo C, Criseo G. A multiplex PCR protocol for rapid identification of Candida glabrata and its phylogenetically related species Candida nivariensis and Candida bracarensis. J Microbiol Methods. 2009;79(1):117–120. doi: 10.1016/j.mimet.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Sanata B, Salam OA, Ibrahim S, Adama Z, Mamoudou C, Simplice KD, Jacques S, Robert GT, Christophe H. Digestive fungal flora in asymptomatic subjects in Bobo-Dioulasso, Burkina Faso. Asian Pac J Trop Biomed. 2014;4(8):659–662. doi: 10.12980/APJTB.4.201414B27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Wankhede S, Muralidhar S, Prakash A, Singh PK, Kathuria S, Kumar DA, Khan N, Randhawa HS, Meis JF, Chowdhary A. Candida nivariensis as an etiologic agent of vulvovaginal candidiasis in a tertiary care hospital of New Delhi, India. Diagn Microbiol Infect Dis. 2013;76(1):46–50. doi: 10.1016/j.diagmicrobio.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Swoboda-Kopeć E, Sikora M, Golas M, Piskorska K, Gozdowski D, Netsvyetayeva I. Candida nivariensis in comparison to different phenotypes of Candida glabrata. Mycoses. 2014;57(12):747–753. doi: 10.1111/myc.12264. [DOI] [PubMed] [Google Scholar]
- Taverna CG, Bosco-Borgeat ME, Murisengo OA, Davel G, Boité MC, Cupolillo E, Canteros CE. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem Inst Oswaldo Cruz. 2013;108(2):178–185. doi: 10.1590/0074-0276108022013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay ST, Lotfalikhani A, Sabet NS, Ponnampalavanar S, Sulaiman S, Na SL, Ng KP. Occurrence and characterization of Candida nivariensis from a culture collection of Candida glabrata clinical isolates in Malaysia. Mycopathologia. 2014;178(3–4):307–314. doi: 10.1007/s11046-014-9778-9. [DOI] [PubMed] [Google Scholar]
- Wahyuningsih R, SahBandar IN, Theelen B, Hagen F, Poot G, Meis JF, Rozalyani A, Sjam R, Widodo D, Djauzi S, Boekhout T. Candida nivariensis isolated from an Indonesian human immunodeficiency virus-infected patient suffering from oropharyngeal candidiasis. J Clin Microbiol. 2008;46(1):388–391. doi: 10.1128/JCM.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TA, McTaggart L, Richardson SE, Zhang SX. Candida bracarensis bloodstream infection in an immunocompromised patient. J Clin Microbiol. 2010;48(12):4677–4679. doi: 10.1128/JCM.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]