Abstract
Even with rigorous treatments, glioblastoma multiforme (GBM) has an abysmal median survival rate, greatly due to the drug-resistant glioblastoma stem cell (GSC) population. GSCs are known to remodel their microenvironment, but the precise role of extracellular matrix components hyaluronic acid (HA) and hyaluronidases (HAases) on the GSC population is still largely unknown. Our objective was to determine how HAase can sensitize GSCs to chemotherapy drugs by disrupting the HA-CD44 signaling. GBM cell line U87-MG and patient-derived D456 cells were grown in GSC-enriching media and treated with HA or HAase. Expressions of GSC markers, HA-related genes, and drug resistance genes were measured via flow cytometry, confocal microscopy, and qRT-PCR. Proliferation after combined HAase and temozolomide (TMZ) treatment was measured via WST-8. HA supplementation promoted the expression of GSC markers and CD44 in GBM cells cultured in serum-free media. Conversely, HAase addition inhibited GSC gene expression while promoting CD44 expression. Finally, HAase sensitized GBM cells to TMZ. We propose a combined treatment of HAase and chemotherapy drugs by disrupting the stemness-promoting HA to target GSCs. This combination therapy shows promise even when temozolomide treatment alone causes resistance.
Electronic supplementary material
The online version of this article (10.1007/s12307-019-00224-2) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma (GBM), Hyaluronic acid (HA), Tumor microenvironment, Temozolomide (TMZ), Combination therapy
Introduction
Glioblastoma multiforme (GBM) is the deadliest form of brain cancer with an abysmal median progression-free survival rate of 10.6 months, despite aggressive treatments such as radiotherapy, anti-angiogenic treatments (e.g., bevacizumab), and DNA alkylating agents such as temozolomide [1]. The glioblastoma stem cell (GSC) subpopulation has been identified to have self-renewal capacity and resistance to currently available GBM treatments. For example, CD133+ GSCs were found to survive high-dose radiation therapy and accumulate in remnant tumors after resection [2]. GSCs are also thought to contribute to chemoresistance through the induction of autophagy, apoptosis, and the unfolded protein response by temozolomide (TMZ) [3]. The ineffectiveness of current GBM treatments can therefore be attributed to their failure to adequately target GSCs.
The tumor microenvironment plays a key role in controlling the fate of GBM cells. For example, GBM migration, chemoresistance, and stemness have been found to be strongly dependent on the extracellular matrix (ECM) composition [4–6]. Hyaluronan (HA) is the main component of the brain ECM, particularly in the white matter tracts that support glioma invasion, and GSCs interact with HA in their microenvironment via receptors CD44 and RHAMM [7]. HA metabolism in the ECM is controlled by HA synthases (HAS) and hyaluronaidases (HAases), and HA-CD44 signaling has been found to promote cancer cell proliferation, invasion, and chemoresistance [8, 9]. HA’s role in protecting cancer cells from chemotherapy therefore makes it an attractive molecule for targeted therapy.
Normal proliferating cells shed their HA coat via the action of HAase, and the failure to lose this coat is implicated in cancer development. Higher HA levels were correlated with worse prognosis, and cancer cells were found to induce HA production in surrounding stromal cells [10]. Cancer cells are known to actively remodel their microenvironment, manipulating the mosaic of differently sized HA molecules in the brain to suit their unique migration and invasion patterns. Since HAases break down polymeric HA, they have been found to improve physical drug delivery with HA depletion lowering tumor interstitial pressure, increasing perfusion, and reversing hypoxia [11, 12]. PH20, the human recombinant form of HAase, improved viral spreading of an oncolytic virus in GBM, and phase II and III clinical trials of PH20 plus chemotherapy in metastatic pancreatic ductal adenocarcinoma are currently underway [13, 14].
While HAases appear to successfully overcome physical drug delivery barriers in cancer, it is unknown how HAases specifically interact with GSCs. We hypothesized that HAase would sensitize GSCs to chemotherapeutics by inhibiting HA-CD44 signaling that promotes stemness. To test this hypothesis, we studied the effect of both HA and HAase on U87-MG and patient-derived GBM D456 cells enriched for GSCs. We also investigated the impact of combining HAase and TMZ treatment on proliferation.
Methods
Glioblastoma Bio Discovery Portal
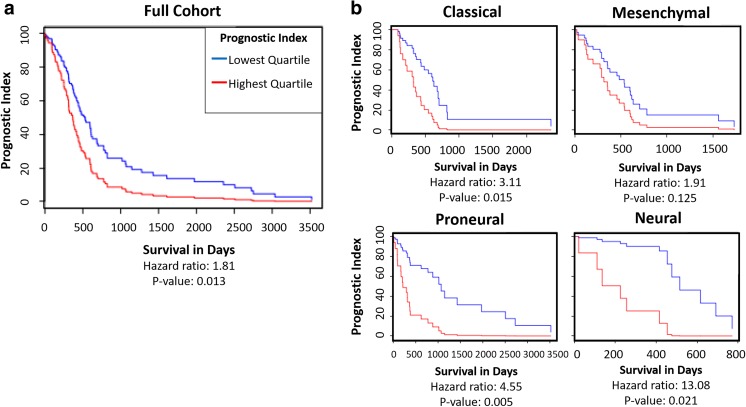
The multi-gene prognostic index for a group of HA-related genes (HAS1, HAS2, RHAMM, and CD44) was analyzed by GBM molecular subtype using the National Cancer Institute’s Glioblastoma Bio Discovery Portal software (GBM Bio-DP; https://gbm-biodp.nci.nih.gov), which accesses and visualizes data from The Cancer Genome Atlas (TCGA) [15]. 197 samples were analyzed using Cox proportional hazards model. Each sample’s prognostic index was determined by averaging individual gene expression from the Cox regression coefficient. Prognostic index was stratified into highest and lowest expression quartiles for both the entire cohort and each molecular subtype: classical, mesenchymal, proneural, and neural [16].
Cell Culture
Two glioblastoma cell types were used: U87-MG cell line (ATCC, Manassas, VA) and D456 glioblastoma cells. D456 is a proneural subtype patient-derived xenograft GBM line kindly provided by Dr. G. Yancey Gillespie (Department of Neurosurgery, University of Alabama at Birmingham) and originally established by Dr. Darrell Bigner (Duke University Medical Center). D456 is derived from a human pediatric fronto-parietal GBM directly implanted into the flank of immunocompromised mice, as previously described [17].
Both cell types were grown in either GSC-enriching sphere culture media NBE or serum containing adherent culture media MEM (for HAase-TMZ experiments) at 37 °C in a 5% CO2 environment. NBE consists of Neurobasal-A media (Gibco) supplemented with 1 mM L-glutamine (Gibco), 8 μg/mL heparin (Akron biotechnology), 0.5X B27 (Gibco), 1% Penicillin/Streptomycin (Corning), 20 ng/mL EGF (Shenandoah Biotechnology), and 20 ng/mL bFGF (Shenandoah Biotechnology). MEM is minimum essential media (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% Penicillin/Streptomycin (Corning).
HA and HAase Treatment
For HA treatment, both U87-MG and D456 cells were treated with 60 kDa sodium hyaluronate (Lifecore Biomedical) at various concentrations (i.e., 0, 20, 100, or 200 μg/mL). Cells were grown in 6-well plates with 3 mL NBE media per well and treated for a full passage length: 6 days for U87-MG and 5 days for D456 cells. For HAase treatment, both U87-MG and D456 cells were treated with HAase from bovine testes Type 1-S lyophilized powder, 400–1000 units/mg solid (Sigma) suspended in PBS at various concentrations (i.e., 0, 15, or 30 U/mL). Cells were grown in NBE media in 6-well plates for a full passage length.
Extreme Limiting Dilution Analysis (ELDA)
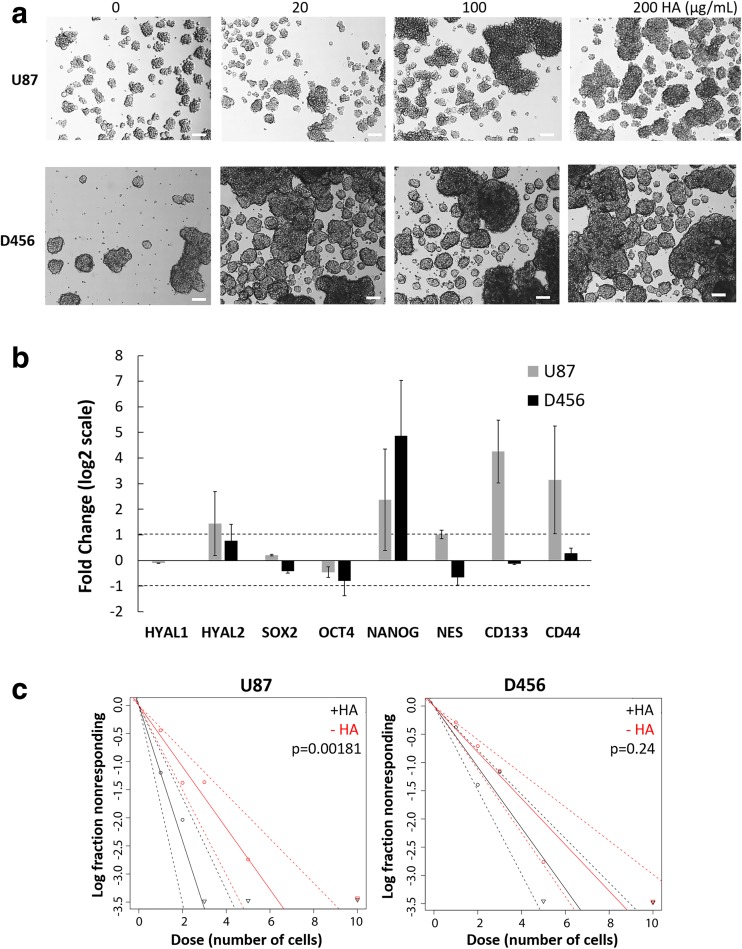
As a functional measure of stemness after HA treatment, U87-MG and D456 cells were seeded at 1, 2, 3, 5, and 10 cells/well in 96-well plates containing 100 μL of NBE media (0 μg/mL HA or 100 μg/mL HA) per well (n = 16 wells per seeding density and media condition). On day 14, the number of wells containing spheres were counted, with a sphere defined as a cell aggregate of 50 μm diameter or larger. Analysis was conducted using the ELDA software from the Walter+Eliza Hall Bioinformatics Institute of Medical Research [18].
Flow Cytometry
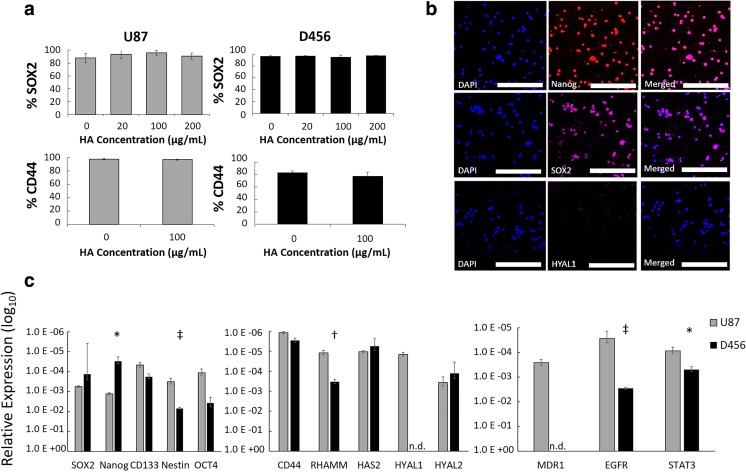
To measure SOX2- and CD44-positive populations via flow cytometry, U87-MG and D456 cells were cultured in NBE with varying concentrations of HA for a full passage. For SOX2 measurement, cell pellets were fixed and permeabilized with BD Cytofix/Cytoperm™ solution (BD) and BD Perm/Wash buffer (BD). Pellets were then stained with rabbit anti-SOX2 IgG polyclonal primary antibody (Proteintech) and goat anti-rabbit IgG Alexa Fluor 488 polyclonal secondary antibody (EMD Millipore). For CD44 measurement, cell pellets were stained with mouse anti-human IgG2b, κ CD44 APC conjugated antibody (BD Pharmingen™). Flow cytometry was performed with BD Accuri™ C6 Flow Cytometer.
Microscopy
Cell culture brightfield micrographs were obtained with VWR VistaVision microscope. Confocal microscopy was used to qualitatively examine baseline protein-level expressions of NANOG, SOX2, and HYAL1 in D456 cells cultured in NBE. Cell pellets were flash frozen in liquid nitrogen and fixed and permeabilized with BD Cytofix/Cytoperm™ solution (BD) and BD Perm/Wash buffer (BD). All cells were stained with DAPI (Invitrogen) and respective antibodies. Cells were stained with mouse anti-human IgG1,κ NANOG PE conjugated antibody (BD), mouse anti-SOX2 IgG1,κ Alexa Fluor 647 conjugated antibody (BD), or mouse anti-HYAL1 IgG1 monoclonal primary antibody (Santa Cruz Biotechnology) and rat anti-mouse IgG1 APC secondary antibody (BD). All images were obtained with Leica TCS SP2 AOBS Confocal Microscope with 4% laser power at the University of Alabama Optical Analysis Facility.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Primers were designed by retrieving nucleotide sequences from NCBI gene database for SOX2, NANOG, CD133, NES, OCT4, CD44, RHAMM, HAS2, HYAL1, HYAL2, MDR1, EGFR, and STAT3. Primer sequences are described in Supplementary Table 1. Primers were synthesized by Invitrogen.
The GeneJet RNA Purification kit (Thermo Scientific) was used for RNA isolation and RNA was quantified using Qubit 2.0 fluorometer (Life Technologies-Invitrogen, Carlsbad, CA, USA) and Qubit RNA HS Assay Kit (Invitrogen), or NanoVue Plus spectrophotometer (Biochrom, Hollison, MA). cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences) and Mastercycler Nexus Gradient Thermal Cycler (Eppendorf, Hauppauge, NY). Real-time PCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems) and the StepOnePlus Real-time PCR System (Applied Biosystems). StepOne Software (v2.3) was used for the data analysis with the ΔΔCt method.
Combined HAase-TMZ Treatment and Proliferation Assay
Combined HAase-TMZ treatment was performed in a 96-well plate. 5000 U87-MG or D456 cells were seeded per well in 100 μL NBE or MEM and cultured for 72 h prior to treatment to allow for NBE samples to form small tumorspheres and MEM samples to become confluent. At 72 h, cells were treated with varying concentrations of HAase (0 U/mL, 15 U/mL, or 100 U/mL) and temozolomide (400 μM for U87-MG and 200 μM for D456) (TMZ; Enzo ALX-420-044-M025) dissolved in DMSO (VWR International). Control wells were treated with DMSO and PBS, and media-only cell-free wells were used for OD value normalization. Cells were treated with HAase-TMZ for 48 h, at which time a WST-8 assay was performed according to manufacturer’s protocol.
Statistical Analyses
All experiments were performed with at least triplicates for each condition. Data were analyzed using a 2-tailed t-test with equal or unequal variance and ANOVA. F-test was used to determine variance prior to t-test or ANOVA. P-values less than 0.05 were considered significant.
Results
HA-Related Gene Expression in GBM is Correlated with Decreased Survival
The TCGA Bio-DP was used to compare the effects of HA-related gene expression on prognostic index of large dataset of GBM patient samples using the Cox model. The highest quartile multi-gene expression of the group of HA-related genes HAS1, HAS2, CD44, and RHAMM was found to be significantly correlated with decreased prognostic index, while the lowest quartile expression of these HA-promoting genes was significantly correlated with increased prognostic index (Fig. 1a). When the patient samples were divided into molecular subtype, classical, proneural, and neural subtypes were found to have significant hazard ratios (Fig. 1b). This analysis of existing GBM patient datasets emphasizes the importance of targeting HA-interactions in GBM in order to design more effective therapeutics.
Fig. 1.
TCGA GBM datasets show that higher expression of HA-related genes is correlated with lower survival rates. a Prognostic Index for HAS1, HAS2, RHAMM, and CD44 gene group, lowest quartile expression compared to highest quartile expression. Multi-gene prognostic indexes and hazard ratios determined with Cox model. b Prognostic Index for HAS1, HAS2, RHAMM, and CD44 gene group divided by GBM molecular subtypes
GSC Markers, HA Receptors, and Drug Resistance Genes are Highly Co-expressed in GBM
Expressions of GSC marker SOX2 and HA-receptor CD44 were measured for U87-MG and D456 cells grown in NBE using flow cytometry. Over 80% of both cell types expressed both markers with or without soluble HA addition (Fig. 2a). This established a large GSC population in both cells, as well as the potential to target HA-CD44 interactions. Confocal microscopy was also performed on dissociated D456 spheres and we found that GSC markers NANOG and SOX2 were very highly expressed, while hyaluronidase gene HYAL1 had low expression (Fig. 2b). Baseline gene expression of GSC markers, HA-related genes, and drug resistance markers was compared between the cell types via qRT-PCR (Fig. 2c). U87-MG was found to have significantly higher NES, RHAMM, and EGFR expression, while D456 had higher NANOG expression. In general, U87-MG had higher drug resistance gene expression, which suggests that it would be more resistant to chemotherapy drugs, potentially through its HA-CD44 interactions. MDR1 was not detected in D456 cells, suggesting it may show increased chemosensitivity compared to U87-MG cells.
Fig. 2.
At baseline, GSC markers, HA receptors, and drug resistance genes are highly co-expressed in GBM, with cell-line dependence. a Percent of population expressing SOX2 and CD44 within U87-MG and D456 cells treated in NBE with varying concentrations of HA for a full passage length, measured via flow cytometry (mean ± SE; n = 3). b Confocal microscopy was performed on D456 cells grown in NBE for NANOG, SOX2, and HYAL1 (scale bars = 200 μm). c qRT-PCR gene expression of (left to right) GSC markers, HA-related genes, and drug resistance genes in U87-MG and D456 cells. Reported as relative expression compared to housekeeping gene GAPDH with log10 transformation and significance noted between cell lines. HYAL1 and MDR1 were lower than detection level (n.d.) in D456 (mean ± SE; n = 3; * p < 0.05, † p < 0.01, and ‡ p < 0.001
HA Promotes GSC Sphere Formation
When GBM cells were treated with increasing concentrations of soluble HA, there was a noted increase in sphere size in both U87-MG and D456; however, spheres remained quite heterogeneous in size and shape (Fig. 3a). Since neurosphere formation is a key functional characteristic of GSCs [19], our results indicate that HA enhances the GSC microenvironment. We also quantitatively assayed how HA treatment affects gene expression of HAases, GSC markers, and the HA receptor CD44 using qRT-PCR (Fig. 3b). We found that 100 μg/mL of HA increases GSC markers, HYAL2, and CD44 in U87-MG cells. In addition, HA significantly promoted NANOG expression in both cell lines, whereas NES, CD133 (PROM1), were enhanced only in U87-MG cells. Of particular note is that U87-MG cells had relatively low baseline NANOG expression in comparison to D456, but with HA addition increased its expression 5-fold. Since CD44 was also upregulated with HA treatment, we propose that HA is maintaining the GSC microenvironment through signaling with CD44. In order to exclude that HA is impacting cell aggregation rather than sphere formation, we tested for clonogenicity using ELDA with and without 100 μg/mL HA treatment. HA either improved the sphere forming ability (i.e., clonogenicity) for U87-MG, with a significant difference in stem cell frequency between HA-treated and non-HA-treated cells (Fig. 3c). D456 similarly showed higher sphere forming ability upon treatment with HA (steeper slope in Fig. 3c), albeit not at statistically significant levels. Stronger sphere forming ability is a sign of higher self-renewal, a key property of GSCs, and non-stem GBM cells have been shown to acquire increased self-renewal capacity upon TMZ treatment measured via ELDA [20]. These data indicate that intermediate molecular weight HA in the brain microenvironment supports the GSC population in U87-MG and D456 cells.
Fig. 3.
HA promotes GSC sphere formation. a Representative micrographs of U87-MG and D456 cells treated with HA for a full passage length (scale bar = 100 μm). b qRT-PCR gene expression of U87-MG and D456 cells treated with HA for a full passage length. Represented as fold change compared to control cells. HYAL1 was not detected in D456 cells. The log2 transforms of fold changes greater than 1 or less than 1 were considered significant (mean ± SE; n = 3). c ELDA of U87-MG and D456 cells treated without (“-HA”) or with HA (100 μg/mL; “+HA”) for 14 days (dotted lines 95% CI; n = 16; p < 0.05 indicates significant change in stem cell frequency)
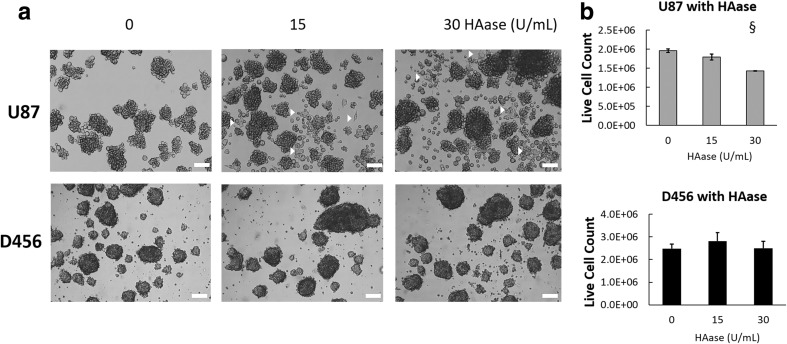
HAase Causes Differentiated Phenotype and Decreased Proliferation in U87-MG Cells
Since we found that HA is involved in preserving the GSC population, we hypothesized that HAase could disrupt the GSC microenvironment through interrupting HA-CD44 signaling. We tested this by treating U87-MG and D456 cells with varying concentrations of HAase. We found that HAase treatment resulted in an adherent morphology consistent with a differentiated phenotype in U87-MG cells, as well as notable breakdown of spheres into single cells (Fig. 4a). However, these changes were not seen in micrographs of D456 cells, presumably due to its denser and tighter spheres resisting the effects of HAase on cell-cell interactions. HAase also caused significantly decreased cell proliferation in U87-MG cells at the 30 U/mL concentration, but did not have significant growth effects on D456 cells (Fig. 4b). These data suggest that in U87-MG cells, HAase promotes an adherent, differentiated phenotype and inhibits proliferation.
Fig. 4.
HAase causes differentiated phenotype and attenuated growth in U87-MG cells, but not in D456 cells. a Representative micrographs of U87-MG and D456 cells treated with HAase for a full passage length. Arrowheads show adhered cells with spread morphology (scale bar = 100 μm). b Live cell counts of U87-MG and D456 cells treated with HAase for a full passage length. Dead population was determined with trypan blue staining prior to automatic cell counts (mean ± SE; n = 3; § p < 0.0001)
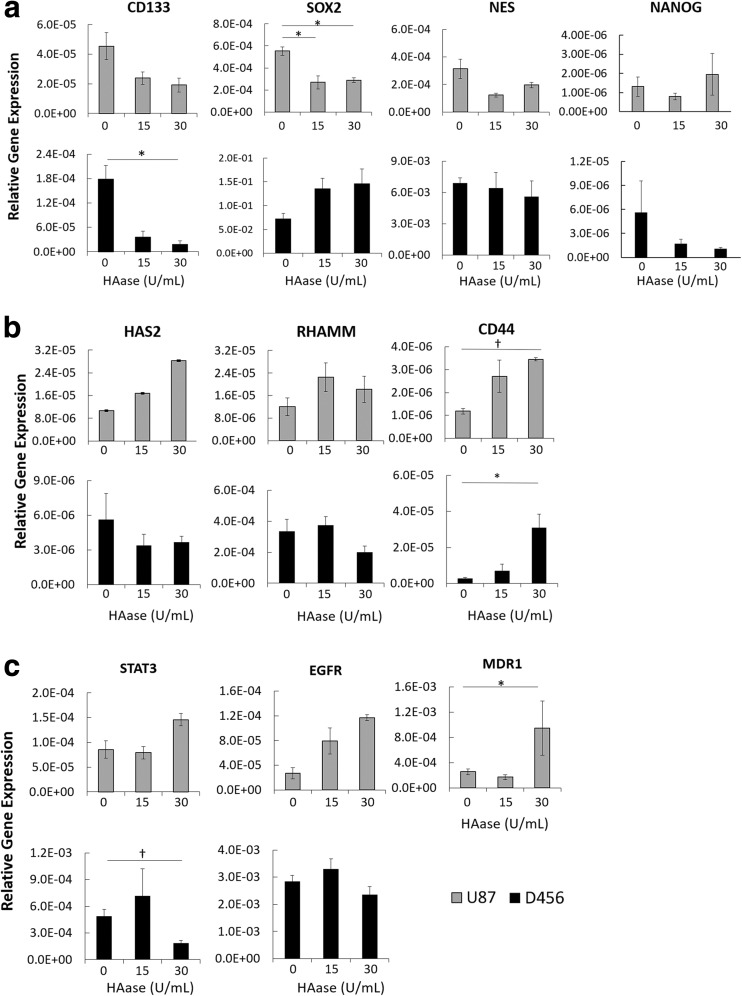
HAase Decreases Stemness Through CD44-Mediated Signaling
To assess the effects of HAase on GSCs at the genotypic level, we performed qRT-PCR on U87-MG and D456 cells treated with HAase for a full passage. We tested for stemness markers CD133, SOX2, NES, and NANOG, and found that HAase caused a significant decrease in SOX2 for U87-MG and CD133 for D456 cells (Fig. 5a). To determine the route of HAase’s impact on stemness, we measured expression of HA-related genes HAS2, RHAMM, and CD44 (Fig. 5b). In both cell lines, there was a significant increase in HA-receptor CD44 expression, suggesting that HAase has an inhibitory effect on stemness through CD44-mediated signaling. Drug resistance genes STAT3, EGFR, and MDR1 were also tested, and U87-MG showed an unexpected significant increase in MDR1 while D456 showed a very significant decrease in STAT3 (Fig. 5c). The increase in MDR1 could be explained by U87-MG’s higher drug resistance gene expression at baseline and lessened impact to HAase, compared to D456. STAT3 is regulator of gliomagenesis that is constitutively activated in GBM, and its decrease is correlated with decreased survival, invasion, angiogenesis, and immune suppression of GBM cells [21]. Overall, these data show that HAase reduces expression of GSC markers SOX2 in U87-MG cells and CD133 in D456 cells by CD44-mediated signaling, and that HAase decreases expression of the transcription factor STAT3 in D456 cells.
Fig. 5.
HAase decreases stemness through involvement of CD44 signaling. qRT-PCR gene expression of U87-MG and D456 cells treated with HAase for a full passage length. a GSC markers CD133, SOX2, NES, and NANOG. b HA-related genes HAS2, RHAMM, and CD44. c Drug resistance genes STAT3, EGFR, and MDR1. MDR1 was not detected in D456 cells. All genes reported as relative expression compared to housekeeping gene GAPDH (mean ± SE; n = 3;* p < 0.05, † p < 0.01)
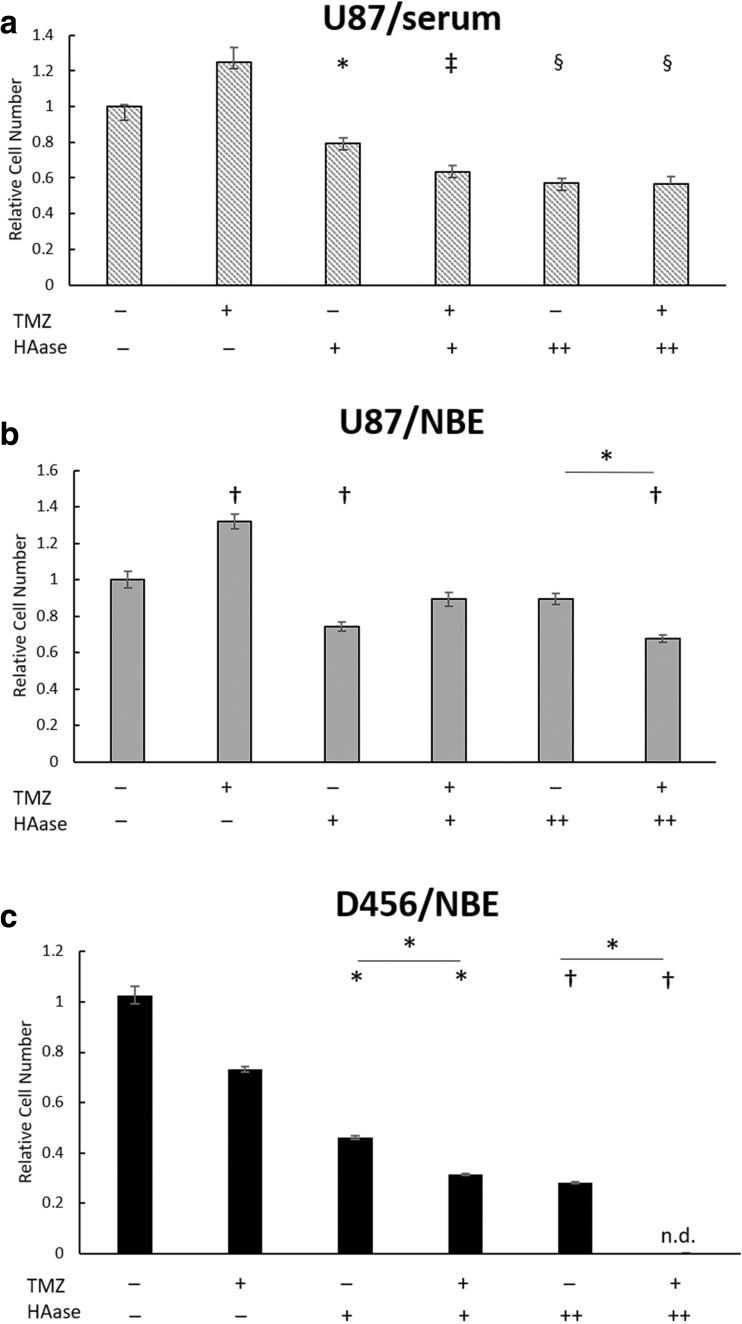
HAase is Cytotoxic to GBM Cells and Increases TMZ Sensitivity in GSC-Promoting Culture
To examine the impact of HAase combined with chemotherapy drugs, we measured proliferation of GBM cells treated with TMZ and/or HAase via WST-8. Both 200 μM and 400 μM concentrations of TMZ were tested on U87-MG cells, and due to a higher drug resistance of U87-MG to TMZ, 400 μM was chosen as the most appropriate concentration for further studies. D456 cells were sensitive to TMZ at 200 μM, so that concentration was used. We found that after 48 h of treatment, U87-MG cells grown in serum-containing media showed a cytotoxic effect of HAase alone but no significant effects when HAase was combined with TMZ (Fig. 6a). However, U87-MG cells grown in GSC-promoting NBE media showed both a cytotoxic effect of HAase alone as well as a significant combined effect with TMZ at the higher concentration 100 U/mL of HAase (Fig. 6b). Additionally, U87-MG grown in both serum and NBE showed a surprising increase in proliferation upon TMZ treatment. This may be due to TMZ causing drug resistance by converting differentiated GBM cells into highly proliferative GSCs, as has been found with TMZ primary chemotherapy in previous studies [22]. In D456 cells, there was no increased proliferative effect of TMZ alone and HAase was cytotoxic at all conditions (Fig. 6c). Additionally, a significant combined effect of HAase and TMZ at both 15 U/mL and 100 U/mL of HAase was seen for D456 cells. Overall, these data demonstrate that HAase preferentially sensitizes GSCs to TMZ.
Fig. 6.
HAase is cytotoxic to GBM cells and increases TMZ sensitivity in GSC-promoting culture. a Treatment of U87-MG cells grown in MEM media containing serum (+TMZ = 400 μM). b Treatment of U87-MG cells grown in GSC-promoting NBE media (+TMZ = 400 μM). c Treatment of D456 cells grown in GSC-promoting NBE media (+TMZ = 200 μM). Relative cell number was measured via a WST-8 assay after 48 h treatment with TMZ and/or HAase on Day 3 (+HAase = 15 U/mL; ++HAase = 100 U/mL). Both normalization and significance were determined relative to DMSO/PBS treated control wells (mean ± SE; n = 8; * p < 0.05, † p < 0.01, ‡ p < 0.001, and § p < 0.0001)
Discussion
At this time, little is known about the role of HAase in GSCs. There have been contradictory studies on the roles of HYAL1 and HYAL2 in tumor progression, but these paradoxes were attributed to dose-dependent effects [23]. Even though bovine HAase has been found in the past to improve patient survival when added to chemotherapy through degrading physical drug delivery barriers and increasing drug perfusion, much of its study was halted due to the bovine HAase causing severe allergic reactions in patients [12, 24, 25]. Specifically, in an astrocytoma study, 20% of patients had allergic reactions after being treated with bovine HAase intravenously [26]. However, a new human recombinant form of HAase (Hylenex™) has been recently created that has not caused allergic reactions and is currently being used in several clinical trials in PEGylated form [27]. This presents an opportunity to reconsider the therapeutic role of HAase in GBM, particularly on the drug-resistant GSC population.
Our data showed that HAase had a cytotoxic effect on both the established U87-MG cell line and patient-derived D456 cells grown in GSC-promoting media. When combined with TMZ, HAase had a synergistic effect only at the higher concentration of HAase (100 U/mL) for U87-MG cells, and at both low (15 U/mL) and high concentrations of HAase for D456 cells. These cell line differences can be attributed to U87-MG having significantly higher drug resistance gene expression (EGFR and MDR1) at baseline compared to D456 cells. In addition, U87-MG cells required higher doses of TMZ than D456 cells to obtain similar effects on proliferation. Therefore, D456 cells are likely more chemosensitive and responsive to the synergistic effects of HAase and TMZ than U87-MG cells.
An unexpected result was the increase in relative cell number when U87-MG was treated with TMZ alone. This could have been due to TMZ killing the non-GSC population, leaving behind only the more drug-resistant GSCs. A previous study found that TMZ failed to target the quiescent GSC-like population that sustains long-term tumor growth; the quiescent GSC-like cells are the source of highly proliferative tumor cells produced at a later time [28]. This emphasizes the importance of effective first-line chemotherapy for GBM, as improper chemotherapy regimens can increase stemness and make tumors even harder to treat. There is currently no standard of care for treatment of recurrent GBM and second-line chemotherapy has so far shown only discouraging results [29]. Adding HAase to drugs could allow GSCs to be targeted during early treatments to avoid recurrence.
As for the role of the HA receptor CD44 in HAase treatment, we found that HAase increased expression of CD44 while it decreased SOX2 for U87-MG and CD133 for D456 cells. While CD44 has previously been suggested to be a GSC marker itself, a recent study found that CD44 knockdown actually increased the stemness phenotype and increased GSC markers CD133, NES, and OCT4 [30]. This suggests that CD44 may not be an appropriate GSC marker and our observed increase in its expression did not necessarily correlate with an increase in stemness. CD44 has also been found to be essential for the catabolic function of both exogenously and endogenously expressed HYAL1 and HYAL2 [31]. In another study, HYAL2 overexpression caused CD44 to lose half of its capacity to bind exogenous HA as well as decreasing cell motility, showing that HAase disrupts formation of the HA pericellular coat through interaction with CD44 [32]. Thus, in our experiments, HAase interactions with CD44 may be forcing endogenous HA to compete for CD44 interactions, and thereby inhibiting HA’s ability to protect GSCs.
In summary, the combined HAase-TMZ treatment of GBM showed promise in both decreasing stemness and creating a synergistic therapeutic effect in vitro. Our data also warrant future study of HAase and chemotherapy combination treatment in vivo and in other cancers associated with accumulation of HA in the ECM, including the breast, lung, ovary, and gastrointestinal tract [33]. We anticipate our findings would be useful for developing better chemotherapy regimens to target the drug-resistant GSC population causing tumor recurrence.
Electronic supplementary material
(DOCX 22 kb)
Acknowledgements
The authors would like to thank Dr. G. Yancey Gillespie (University of Alabama at Birmingham) for providing the D456 patient-derived xenograft GBM line. The authors would like to further acknowledge financial support from the National Science Foundation (CBET 1604677 to Y.K. and S.R.), the University of Alabama Randall Research Scholars Program (J.S.H.), and by the Alabama Experimental Program to Stimulate Competitive Research (S.P.). S.D.G.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 2.Tamura K, Aoyagi M, Wakimoto H, Ando N, Nariai T, Yamamoto M, Ohno K. Accumulation of CD133-positive glioma cells after high-dose irradiation by Gamma Knife surgery plus external beam radiation. J Neurosurg. 2010;113:310–318. doi: 10.3171/2010.2.JNS091607. [DOI] [PubMed] [Google Scholar]
- 3.Hombach-Klonisch S, Mehrpour M, Shojaei S, Harlos C, Pitz M, Hamai A, Siemianowicz K, Likus W, Wiechec E, Toyota BD, Hoshyar R, Seyfoori A, Sepehri Z, Ande SR, Khadem F, Akbari M, Gorman AM, Samali A, Klonisch T, Ghavami S. Glioblastoma and chemoresistance to alkylating agents: involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther. 2018;184:13–41. doi: 10.1016/j.pharmthera.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Kimlin LC, Casagrande G, Virador VM. In vitro three-dimensional (3D) models in cancer research: an update. Mol Carcinog. 2013;52:167–182. doi: 10.1002/mc.21844. [DOI] [PubMed] [Google Scholar]
- 5.Herrera-Perez M, Voytik-Harbin SL, Rickus JL. Extracellular matrix properties regulate the migratory response of glioblastoma stem cells in three-dimensional culture. Tissue Eng Part A. 2015;21:2572–2582. doi: 10.1089/ten.tea.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lv D, Yu SC, Ping YF, Wu H, Zhao X, Zhang H, Cui Y, Chen B, Zhang X, Dai J, Bian XW, Yao XH. A three-dimensional collagen scaffold cell culture system for screening anti-glioma therapeutics. Oncotarget. 2016;7:56904–56914. doi: 10.18632/oncotarget.10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akiyama Y, Jung S, Salhia B, Lee S, Hubbard S, Taylor M, Mainprize T, Akaishi K, van Furth W, Rutka JT. Hyaluronate receptors mediating glioma cell migration and proliferation. J Neuro-Oncol. 2001;53:115–127. doi: 10.1023/A:1012297132047. [DOI] [PubMed] [Google Scholar]
- 8.Toole BP. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clin Cancer Res. 2009;15:7462–7468. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karbownik MS, Nowak JZ. Hyaluronan: towards novel anti-cancer therapeutics. Pharmacol Rep. 2013;65:1056–1074. doi: 10.1016/S1734-1140(13)71465-8. [DOI] [PubMed] [Google Scholar]
- 10.Stern R. Association between cancer and “acid mucopolysaccharides”: an old concept comes of age, finally. Semin Cancer Biol. 2008;18:238–243. doi: 10.1016/j.semcancer.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Shepard HM. Breaching the castle walls: Hyaluronan depletion as a therapeutic approach to cancer therapy. Front Oncol. 2015;5:192. doi: 10.3389/fonc.2015.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson CB, Shepard HM, O'Connor PM, Kadhim S, Jiang P, Osgood RJ, Bookbinder LH, Li X, Sugarman BJ, Connor RJ, Nadjsombati S, Frost GI. Enzymatic depletion of tumor hyaluronan induces antitumor responses in preclinical animal models. Mol Cancer Ther. 2010;9:3052–3064. doi: 10.1158/1535-7163.MCT-10-0470. [DOI] [PubMed] [Google Scholar]
- 13.Guedan S, Rojas JJ, Gros A, Mercade E, Cascallo M, Alemany R. Hyaluronidase expression by an oncolytic adenovirus enhances its intratumoral spread and suppresses tumor growth. Mol Ther. 2010;18:1275–1283. doi: 10.1038/mt.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong KM, Horton KJ, Coveler AL, Hingorani SR, Harris WP. Targeting the tumor stroma: the biology and clinical development of Pegylated recombinant human hyaluronidase (PEGPH20) Curr Oncol Rep. 2017;19:47. doi: 10.1007/s11912-017-0608-3. [DOI] [PubMed] [Google Scholar]
- 15.Celiku O, Johnson S, Zhao S, Camphausen K, Shankavaram U. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PLoS One. 2014;9:e101239. doi: 10.1371/journal.pone.0101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research N Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman GK, Langford CP, Coleman JM, Cassady KA, Parker JN, Markert JM, Yancey Gillespie G. Engineered herpes simplex viruses efficiently infect and kill CD133+ human glioma xenograft cells that express CD111. J Neuro-Oncol. 2009;95:199–209. doi: 10.1007/s11060-009-9926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 20.Adamski V, Hempelmann A, Flüh C, Lucius R, Synowitz M, Hattermann K, Held-Feindt J. Dormant glioblastoma cells acquire stem cell characteristics and are differentially affected by temozolomide and AT101 treatment. Oncotarget. 2017;8:108064–108078. doi: 10.18632/oncotarget.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atkinson GP, Nozell SE, Benveniste ET. NF-kappaB and STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother. 2010;10:575–586. doi: 10.1586/ern.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119–1131. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stern R, Jedrzejas MJ. Hyaluronidases: their genomics, structures, and mechanisms of action. Chem Rev. 2006;106:818–839. doi: 10.1021/cr050247k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohno N, Ohnuma T, Truog P. Effects of hyaluronidase on doxorubicin penetration into squamous carcinoma multicellular tumor spheroids and its cell lethality. J Cancer Res Clin Oncol. 1994;120:293–297. doi: 10.1007/BF01236386. [DOI] [PubMed] [Google Scholar]
- 25.Kerbel RS, St Croix B, Florenes VA, Rak J. Induction and reversal of cell adhesion-dependent multicellular drug resistance in solid breast tumors. Hum Cell. 1996;9:257–264. [PubMed] [Google Scholar]
- 26.Baumgartner G, Gomar-Höss C, Sakr L, Ulsperger E, Wogritsch C. The impact of extracellular matrix on the chemoresistance of solid tumors--experimental and clinical results of hyaluronidase as additive to cytostatic chemotherapy. Cancer Lett. 1998;131:85–99. doi: 10.1016/S0304-3835(98)00204-3. [DOI] [PubMed] [Google Scholar]
- 27.Whatcott CJ, Han H, Posner RG, Hostetter G, Von Hoff DD. Targeting the tumor microenvironment in cancer: why hyaluronidase deserves a second look. Cancer Discov. 2011;1:291–296. doi: 10.1158/2159-8290.CD-11-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallego O. Nonsurgical treatment of recurrent glioblastoma. Curr Oncol. 2015;22:e273–e281. doi: 10.3747/co.22.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HH, Liao CC, Chow NH, Huang LL, Chuang JI, Wei KC, Shin JW. Whether CD44 is an applicable marker for glioma stem cells. Am J Transl Res. 2017;9:4785–4806. [PMC free article] [PubMed] [Google Scholar]
- 31.Harada H, Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J Biol Chem. 2007;282:5597–5607. doi: 10.1074/jbc.M608358200. [DOI] [PubMed] [Google Scholar]
- 32.Duterme C, Mertens-Strijthagen J, Tammi M, Flamion B. Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J Biol Chem. 2009;284:33495–33508. doi: 10.1074/jbc.M109.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sironen RK, Tammi M, Tammi R, Auvinen PK, Anttila M, Kosma VM. Hyaluronan in human malignancies. Exp Cell Res. 2011;317:383–391. doi: 10.1016/j.yexcr.2010.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 22 kb)