A grand challenge in microbiology is to understand how the dormant majority lives. In natural environments, most microorganisms are not growing and instead exist in a spectrum of dormant states.
KEYWORDS: hydrogen, metabolism, mycobacteria, persistence, soil microbiology, trace gases
ABSTRACT
A grand challenge in microbiology is to understand how the dormant majority lives. In natural environments, most microorganisms are not growing and instead exist in a spectrum of dormant states. Despite this, most research on microbial metabolism continues to be growth-centric, and many overlook the fact that dormant cells require energy for maintenance. In this perspective, we discuss our research program to uncover the metabolic strategies that support microbial survival. We present two major principles underlying these studies. The first is the recent realization that microbial survival depends on previously unrecognized metabolic flexibility. The second is that new discoveries in this area depend on more sophisticated integration of approaches at the molecular, cellular, and ecosystem levels. These principles are illustrated with examples from the literature, including our own work demonstrating that bacteria can live on air, and areas for future methodological and theoretical development are highlighted.
PERSPECTIVE
It is well recognized that most microorganisms in natural environments exist in dormant rather than growing states (1). Yet, microbiology continues to be an overwhelmingly growth-centric field. This partly reflects methodological limitations: culture-dependent approaches are skewed toward relatively fast growers and culture-independent approaches often focus on the abundant few. These issues have been compounded by misconceptions about the nature and role of dormant microorganisms in different ecosystems. Many incorrectly synonymize dormancy with inactivity. In fact, dormant cells still maintain a low level of metabolic activity to maintain basal cell functions (1). Moreover, it is commonly thought that growing microorganisms are the most important players in natural ecosystems. However, many critical ecosystem services such as atmospheric gas regulation (2), antibiotic production (3), and even the maintenance of biodiversity itself (4) are mediated by dormant microorganisms.
The goal of our laboratory is to reshape understanding of how microorganisms persist in different environments. To achieve this, we combine culture-dependent and culture-independent approaches to investigate how microorganisms adapt their metabolism when limited for the electron donors and acceptors required for growth. The central theory driving this research is that microbial survival depends on previously unrecognized metabolic flexibility. To demonstrate this, we have developed an ambitious interdisciplinary research program that aims to (i) identify novel metabolic processes, (ii) resolve their molecular basis, and (iii) understand their ecological significance. In turn, we are applying these findings to address specific questions about global change, infectious disease, and biodiversity. This perspective will summarize the concepts and methodologies driving the development of this research program, while highlighting areas and opportunities for further development in this research space.
MICROORGANISMS RELY ON HIDDEN METABOLIC FLEXIBILITY TO PERSIST
There is growing evidence that the growth-centric studies have underestimated the metabolic capabilities of microorganisms. It is often thought that, during dormancy, bacteria survive primarily by downregulating the processes they use for growth. However, we have observed that aerobic bacteria often broaden their metabolic repertoire during persistence. For example, obligate heterotrophs scavenge inorganic energy sources during carbon starvation (2, 5), obligate aerobes use fermentation as a last resort during hypoxia (6), and methanotrophs will take advantage of hydrogen (H2) whenever available (7). Thus, the traditional metabolic classifications used in systematic bacteriology break down when the focus is shifted from growth to survival. Other groups have also revealed surprising metabolic flexibility of lithotrophic aerobic bacteria from a wide range of environments (8–10).
An example of the discrepancies caused by growth-centric microbiology research is the area of atmospheric H2 oxidation. It was discovered in the 1970s that soils consume large amounts of atmospheric H2 and serve as the major biogeochemical sink of this gas. However, it wasn’t until over three decades later that the bacteria and enzymes responsible were finally identified, through efforts led by Constant and Conrad (2, 11, 12). This delay reflects that previous cultivation efforts were focused on isolating chemolithoautotrophs, i.e., bacteria that grow on H2 as the sole energy source. However, the bacteria mediating atmospheric uptake in fact grow heterotrophically and switch to using H2 during dormancy. Several long-known isolates, including Mycobacterium smegmatis (2), Streptomyces avermitilis (13), and Thermomicrobium roseum (14), have since been shown to also persist on atmospheric H2. Since this breakthrough, our genetic and biochemical studies have shown this process is mediated by specialized high-affinity, oxygen-tolerant [NiFe]-hydrogenases that input electrons from atmospheric H2 oxidation into the aerobic respiratory chain (2). Deletion of these enzymes results in reduced long-term survival during starvation and hypoxia (2, 6, 13). It was ultimately the growth-centric perspective on the microbial world, rather than methodological limitations, that prevented these biogeochemically and ecologically important discoveries from being made earlier.
It is now increasingly realized that atmospheric trace gases are hidden energy sources supporting the dormant microbial majority in aerated ecosystems. Genomic surveys have shown that isolates from the nine most dominant soil phyla also encode the key enzymes for aerobic H2 oxidation (15), and three such phyla have now been experimentally shown to scavenge H2 (5, 11, 14). In parallel, metagenomic studies revealed that high-affinity hydrogenases are highly abundant in soil ecosystems (15, 16). Particularly important is the discovery that atmospheric trace gases are the main energy sources supporting communities in some of the most hostile environments of all: Antarctic dry deserts (16). Whereas it was conventionally thought that oxygenic photosynthesis drives primary production in such ecosystems, physicochemical pressures such as water limitation generally exclude phototrophs from these soils. Instead, the dominant community members of such ecosystems are actually gas-scavenging bacteria, and biogeochemical assays show they mediate rapid levels of atmospheric H2 oxidation (16). Thus, in this case, findings from pure culture about how bacteria survive carbon starvation were also extendable to the ecosystem scale.
Resource generalism provides a competitive advantage for bacteria in deprived or changeable environments. Contrary to the classical paradigms of diauxic growth, bacteria may be more competitive if they continually cometabolize a broad range of energy sources. Atmospheric trace gases may be particularly desirable: while insufficiently concentrated to support growth, they are dependable energy sources for persistence given their ubiquity and diffusibility. Metabolic flexibility may also be important for enabling organisms to maintain energy needs in highly changeable environments. For example, in a further extension of our pure culture findings (6), we have shown that facultative fermentation is critical for the stability and productivity of the predominantly aerobic communities within dynamic intertidal sand sediments (17). Going forward, we are using similar philosophies to address key questions about a range of environmentally and medically important microorganisms. For example, how does metabolic flexibility influence the microorganisms that control the production and consumption of greenhouse gases? How do bacterial pathogens remodel their metabolism to sustain energy needs in different environmental and host reservoirs?
INTEGRATIVE APPROACHES ENABLE DISCOVERY AND CHARACTERIZATION OF PERSISTENCE MECHANISMS
Despite considerable progress, much more research is needed to gain a systematic understanding of how most microorganisms persist in different environments. Through rapid developments in meta-omics technologies, researchers have developed an increasingly comprehensive knowledge of the phylogenetic diversity and functional potential of microorganisms in different biomes. However, these technologies can provide only inferences about how these organisms live, rather than the sorts of proofs derived from genetic, biochemical, or physiological approaches. Moreover, given that these technologies primarily rely on findings about genes carried by cultivated species, they have limited capacity to discover or characterize genes encoding novel function; reflecting this, over half of the genes detected in candidate phyla remain completely functionally unannotated (18). We argue that improved understanding of microbial metabolism depends on integrating meta-omics technologies with more targeted approaches. Only through such combinations will it be possible to gain both a broad and a deep understanding of how the uncultured majority live.
Reflecting this, many of the greatest metabolic discoveries in the last few years have depended on synergies between meta-omic and targeted approaches. For example, following the discovery of the first complete ammonia-oxidizing (comammox) bacteria through genome-resolved metagenomics (19, 20), the first isolate of such an organism was cultured (Nitrospira inopinata); this provided a more sophisticated understanding of their ecophysiology, revealing that comammox bacteria are high-affinity ammonia oxidizers adapted for an oligotrophic lifestyle (21). Innovations are also possible independent of culturing. For example, metagenomic approaches led to the discovery of novel archaeal lineages harboring divergent methyl-CoM reductase genes (22); the subsequent experimental detection of the unprecedented intermediate butyl-coenzyme M in a butane-oxidizing enrichment led to the realization that some such enzymes mediate unprecedented pathways of anaerobic alkane activation (23). In addition, protein biochemistry has also been used on rare occasions to reveal information about otherwise intractable processes. In a particularly impressive example, Krüger et al. confirmed how anaerobic methane oxidation occurs by directly purifying the key enzyme responsible from a methane-oxidizing microbial mat (24); this supported a weight of evidence from metagenomic, biogeochemical, and imaging studies that anaerobic methanotrophs (ANME) function through performing a reverse methanogenesis pathway.
In our experience, integrative approaches are rewarding because of the different perspectives they provide and the synergies that can be achieved by working across disciplines. As outlined in Fig. 1, our own research program is becoming highly integrated. We benefit from diverse expertise, both within our group and through various collaborations, to develop information about survival-related processes at the protein, cellular, and ecosystem levels. We then integrate these findings to gain a comprehensive understanding of the basis and significance of different metabolic processes. For example, through such approaches, we are currently developing understanding of how atmospheric H2 oxidation is mediated at both the molecular level and the ecosystem scale. These approaches are largely inspired by our experiences in medical microbiology, where integrative approaches are commonplace. For example, similar approaches have transformed understanding of how pathogens such as Mycobacterium tuberculosis generate energy during latent infection of host tissues; a gold standard approach is to identify novel metabolic enzymes through systems biology approaches, determine their function and mechanism through biochemical and genetic approaches, and validate their importance through phenotypic studies both in vitro and in vivo (25). There are also some areas of environmental microbiology where strong enzyme-to-ecosystem links have also been developed, for example, in the area of microbial methane cycling (26), but these are presently exceptions rather than the rule.
FIG 1.
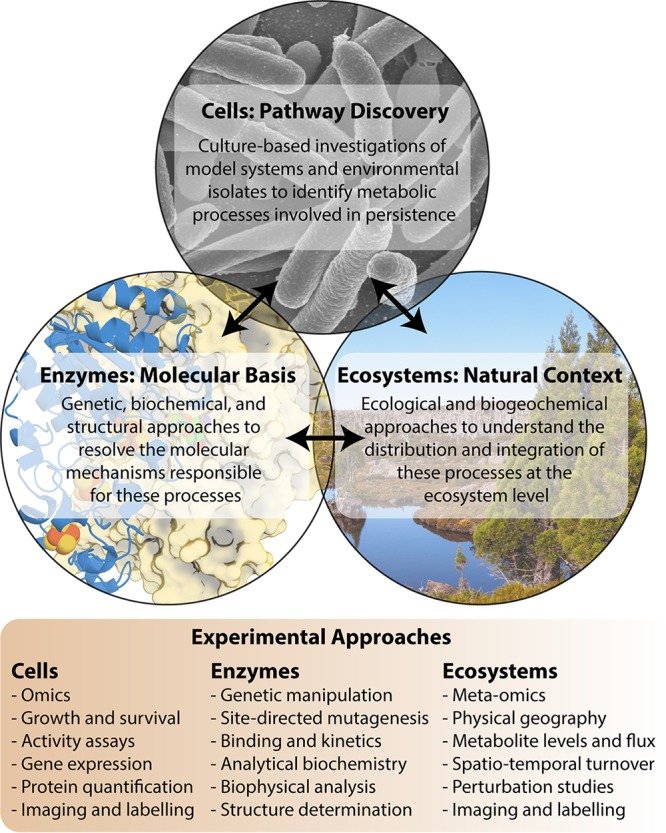
Overview of how synthesizing information across enzymatic, cellular, and ecosystem scales enables the discovery and characterization of metabolic processes involved in microbial persistence.
Looking ahead, several ongoing methodological developments have the potential to transform knowledge of how the microbial majority live. First, a range of labeling and imaging approaches enable researchers to precisely determine which microorganisms mediate certain biogeochemical processes. For example, highly resolved mass spectrometry techniques such as nanoscale secondary-ion mass spectrometry (NanoSIMS) allow detection of specific metabolic activities at the single-cell level. In parallel, the development of high-throughput culturing approaches (also known as culturomics) facilitates the bacteriological and genomic description of new isolates at unprecedented scale. In our view, these approaches ought to be combined with in-depth approaches to gain a sophisticated understanding of the ecophysiology of organisms, for example, through profiling gene expression and enzymatic activity of cells in their dormant states. We also predict that genetic and biochemical approaches will become more broadly available for a range of systems. Currently, genetic tools are unavailable for many ecologically dominant and biogeochemically important phyla, including Acidobacteria, Thaumarchaeota, Nitrospirae, and Verrucomicrobia; however, recent developments in CRISPR interference technologies may start to change this (27). Finally, from a biochemistry standpoint, the rise of cryo-electron microscopy has allowed for protein structure determination at high resolution, using extremely small volumes of purified sample. This has allowed for structure determination of low-copy-number proteins expressed in their native organism, ushering in a new chapter of structural biology (28). We argue that, by leveraging both current and emerging technologies, it will be possible for researchers to gain a broad and deep understanding of the dormant majority.
ACKNOWLEDGMENTS
Our research is supported by an ARC DECRA Fellowship (DE170100310; awarded to C.G.), an NHMRC New Investigator Grant (APP5191146; awarded to C.G.), a Sir Henry Wellcome Research Fellowship (106077/Z/14/Z; awarded to R.G.), and a Swiss National Science Foundation Early Postdoc Mobility Fellowship (P2EZP3_178421; awarded to E.C.).
We thank all members of our laboratory for helpful discussions and Gregory Cook, Ralf Conrad, Sergio Morales, Michael Berney, and Philippe Constant for their inspirations.
The authors declare no conflict of interest.
mSystems® vol. 4, no. 3, is a special issue sponsored by Illumina.
REFERENCES
- 1.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 2.Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A 111:4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greening C, Carere CR, Rushton-Green R, Harold LK, Hards K, Taylor MC, Morales SE, Stott MB, Cook GM, Liam K. 2015. Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc Natl Acad Sci U S A 112:10497–10502. doi: 10.1073/pnas.1508385112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. 2014. An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci U S A 111:11479–11484. doi: 10.1073/pnas.1407034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carere CR, Hards K, Houghton KM, Power JF, McDonald B, Collet C, Gapes DJ, Sparling R, Boyd ES, Cook GM, Greening C, Stott MB. 2017. Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J 11:2599–2610. doi: 10.1038/ismej.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daims H, Lücker S, Wagner M. 2016. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol 24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mußmann M, Pjevac P, Krüger K, Dyksma S. 2017. Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J 11:1276. doi: 10.1038/ismej.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori JF, Scott JJ, Hager KW, Moyer CL, Küsel K, Emerson D. 2017. Physiological and ecological implications of an iron- or hydrogen-oxidizing member of the Zetaproteobacteria, Ghiorsea bivora, gen. nov., sp. nov. ISME J 11:2624. doi: 10.1038/ismej.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Constant P, Poissant L, Villemur R. 2008. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J 2:1066–1076. doi: 10.1038/ismej.2008.59. [DOI] [PubMed] [Google Scholar]
- 12.Constant P, Chowdhury SP, Hesse L, Pratscher J, Conrad R. 2011. Genome data mining and soil survey for the novel group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H2-oxidizing bacteria. Appl Environ Microbiol 77:6027–6035. doi: 10.1128/AEM.00673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liot Q, Constant P. 2016. Breathing air to save energy—new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. Microbiologyopen 5:47–59. doi: 10.1002/mbo3.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam ZF, Cordero PRF, Feng J, Chen Y-J, Bay S, Gleadow RM, Jirapanjawat T, Carere CR, Stott MB, Chiri E, Greening C. 2019. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J doi: 10.1038/s41396-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, Montgomery K, Lines T, Beardall J, Van Dorst J, Snape I, Stott MB, Hugenholtz P, Ferrari BC. 2017. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552:400–403. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 17.Kessler AJ, Chen Y-J, Waite DW, Hutchinson T, Koh S, Popa ME, Beardall J, Hugenholtz P, Cook PLM, Greening C. 2019. Bacterial fermentation and respiration processes are uncoupled in anoxic permeable sediments. Nat Microbiol doi: 10.1038/s41564-019-0391. [DOI] [PubMed] [Google Scholar]
- 18.Overmann J, Abt B, Sikorski J. 2017. Present and future of culturing bacteria. Annu Rev Microbiol 71:711–730. doi: 10.1146/annurev-micro-090816-093449. [DOI] [PubMed] [Google Scholar]
- 19.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. 2017. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549:269. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 23.Laso-Pérez R, Wegener G, Knittel K, Widdel F, Harding KJ, Krukenberg V, Meier DV, Richter M, Tegetmeyer HE, Riedel D, Richnow H-H, Adrian L, Reemtsma T, Lechtenfeld OJ, Musat F. 2016. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539:396. doi: 10.1038/nature20152. [DOI] [PubMed] [Google Scholar]
- 24.Kruger M, Meyerdierks A, Glockner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt J, Bocher R, Thauer RK, Shima S. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- 25.Cook GM, Hards K, Dunn E, Heikal A, Nakatani Y, Greening C, Crick DC, Fontes FL, Pethe K, Hasenoehrl E, Berney M. 2017. Oxidative phosphorylation as a target space for tuberculosis: success, caution, and future directions. Microbiol Spectr 5(3) doi: 10.1128/microbiolspec.TBTB2-0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henneberger R, Chiri E, Bodelier PEL, Frenzel P, Lüke C, Schroth MH. 2015. Field‐scale tracking of active methane‐oxidizing communities in a landfill cover soil reveals spatial and seasonal variability. Environ Microbiol 17:1721–1737. doi: 10.1111/1462-2920.12617. [DOI] [PubMed] [Google Scholar]
- 27.Peters JM, Koo B-M, Patino R, Heussler GE, Hearne CC, Qu J, Inclan YF, Hawkins JS, Lu CHS, Silvis MR, Harden MM, Osadnik H, Peters JE, Engel JN, Dutton RJ, Grossman AD, Gross CA, Rosenberg OS. 2019. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol 4:244. doi: 10.1038/s41564-018-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


