Abstract
Introduction: Prostate cancer (PCa) is the most common cancer among men and the second cause of cancer death among men. For early detection and differentiating PCa from benign prostate hyperplasia (BPH) tissue biopsy has been used for decades. However, circulating cell-free DNA (ccfDNA) testing is a noninvasive, fast, easily repeatable, and sensitive liquid biopsy for cancer detection. Hence, we aimed to investigate the value of the ccfDNA concentration and integrity index in peripheral blood of a population of Iranian prostatic patients for early diagnosis of the disease.
Materials and methods: 100 subjects including 30 PCa, 40 BPH, and 30 healthy individuals were selected. ccfDNA was extracted from fresh blood plasma, and its total concentration and the integrity index were estimated by amplification of ALU115 and ALU247 repeat elements using quantitative real-time PCR.
Results: In the PCa group, the ccfDNA concentration and its integrity were significantly higher than that of the BPH and healthy groups (P-value <0.001 and P-value <0.001). The ccfDNA concentration and its integrity were higher in BPH compared to the healthy group, although it was not statistically significant (P-value =0.836 and P-value =0.053, respectively).
Conclusion: A significant relation between ccfDNA concentration, its integrity, and PCa suggests that the liquid biopsy can be used as a noninvasive early diagnostic biomarker. Determination of a cutoff or a diagnostic range value of the measured parameters for healthy, BPH, and PCa subjects in more samples of Iranian population results in timely, correct, and early detection, which results in better treatment outcomes. Moreover, this method may reduce overdiagnosis and overtreatment procedures.
Keywords: prostate cancer, biopsy, ccfDNA, integrity
Introduction
Prostate cancer (PCa), the first common cancer and the second cause of cancer death among men with multifocal and heterogeneous tumors,1 is a complex polygenic disorder that makes it difficult for targeted molecular diagnosis of the disease
The incidence rate of the disease was reported to be one million in 2006, with a 40% increase regarding aging and population growth in 20162 and an increased prevalence in Iran in the last decade.3,4 Noninvasive methods like the PSA level measurement, rectal exam, and scanning for prostate examinations5 are not sensitive and informative enough for early detection of high score PCa from the low score and those from BPH status.6–8 Although diagnostic imaging like sonography and computed tomography (CT) scan may detect metastasis and prostate enlargement, the definite PCa diagnosis is still through tissue biopsy. Since most of the biopsy patients tend to have BPH and do not need PCa treatments, tissue sampling is not a prerequisite for them and for almost 70% of PCa cases. Moreover, repetitive biopsy for treatment follow-up may not always be possible or be tolerated by at-risk patients. It is estimated that <30% of prostate tumors may cause metastasis to other adjacent tissues and may result in death.9 Most cancer patients experience bone metastases at early diagnosis, which makes the additional sampling difficult, painful, and costly.10 Therefore, there is an urgent need for novel markers that either outperform the conventional biomarkers or are to be used in parallel to them to increase the sensitivity and the specificity of diagnostic tests. The new markers could be examined by noninvasive methods like the measurement of circulating cell-free DNA (ccfDNA) in plasma/serum as a liquid biopsy which could replace tissue biopsy and facilitate the evaluation of focal tumors.11 Studies have been conducted on the effect of the magnitude of the blood plasma ccfDNA fragments originating from tumor tissues as well as qualitative studies like mutation and epigenetic studies.12 It is known that ccfDNA has the same genetic and epigenetic characteristics as the tumor DNA in cancerous patients.13,14 The main source of free DNA in the circulation is apoptotic cell’s DNA fragments of approximately 185–200 bp length.12 In cancer patients, the plasma DNA fragments are created by necrosis, while in other conditions, appearance of the genomic DNA fragments is a reason of autophagy and catastrophic mitosis or mitochondrial disorders due to incomplete and random digestion.12 One of the methods for evaluating ccfDNA concentration is beneficially using repeated ALU repeat elements (247 bp and 115 bp) through quantitative analysis by real-time PCR. For ccfDNA integrity assessment as an index, the ratio of the long DNA fragment concentration to a shorter one is calculated.15–18 Direct real-time PCR (q-RT-PCR) is a quantitative method for amplification of as little as 0.01 pg of DNA with high linearity.19 Moreover, the ccfDNA values can be used as noninvasive, fast, repeatable, and sensitive biomarkers for molecular detection, prognosis, and treatment follow-up in a variety of cancers including PCa.13,14 The purpose of the current study was to assess the plasma level of ccfDNA concentration and its integrity ratio as new tools for early PCa diagnosis and/or screening. The results of this investigation could probably replace other existing methods such as biopsy or surgery before any treatment such as hormone therapy and radiotherapy in the future.
Materials and methods
Study population
In the present study, we selected 100 subjects including 30 patients with PCa, 40 BPH cases, and 30 normal individuals who were referred to Labbafi Nejad and Shohadaye Tajrish hospitals of Shahid Beheshti University of Medical Sciences, from September 1, 2016, to March 20, 2017. The BPH group was selected based on rectal exam, prostate volume (>30 mL), and urologist’s opinion. The PCa group was also selected based on high PSA, biopsy, and pathology response. Blood samples from PCa and BPH patients were taken and studied after status confirmation by pathology. Healthy people were included in the study based on their urination symptoms, abdominal pain, and PSA measurements. Their ultrasonography and urinary culture were considered safe if they had no surgical treatment before because of the prostate condition. Groups were matched for age.
All participants provided written informed consent, and this study was conducted in accordance with the Declaration of Helsinki.
Blood collection and DNA isolation
From each individual, 3 mL fresh venous blood was collected in K2EDTA-containing tubes and processed within 2–4 hrs to separate the plasma. For this purpose, all samples were centrifuged in 3,000 rpm for 15 mins at 4°C and stored at −20°C until ccfDNA extraction. Using NucleoSpin plasma XS (NS) Kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany), 250 μL of plasma was subjected to the extraction20 and the isolated purified ccfDNA was suspended in TE buffer with a final volume of 20 μL and stored at −20°C for further analysis.
Real-time PCR
According to the previous works,21 following the quantification of the extracted ccfDNA by spectrophotometer at 260 nm, the ccfDNA samples were also quantified by RT-PCR, where ALU repeat elements of 115 bp and 247 base pairs were amplified by specific primers. One set of primers for amplification of ALU115 and a second primer set for ALU247 amplicons were adapted as it was previously reported.22–25 Sequences of the primers were as follows:
ALU115 forward primer was 5ʹCCTGAGGTCAGGAGTTCGAG3ʹ and the reverse primer was 5ʹCCCGAGTAGCTGGGATTACA3ʹ;
ALU247 forward primer was 5ʹ GTGGCTCACGCCTGTTAATC 3ʹ and the reverse primer was 5ʹ CAGGCTGGAGTGCAGTGG3ʹ.
Quantitative RT-PCR was performed on Rotor Gene 6000 (Corbett, Mortlake, NSW, Australia). Each reaction consisted of 11 μL Real Q Plus 2x Master Mix Green (Amplicon “No Rox”, Denmark), 2 μL of DNA sample, 10 pmol/L of each forward and reverse primers, and H2O to a total volume of 25 μL.
The thermal program for ALU115 started by an initial denaturation at 95°C for 10 mins, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 64°C for 15 s, and extension at 72°C for 20 s ending with a final replication at 72°C for 5 mins. Thermal program for ALU247 was as follows: An initial denaturation at 95°C for 10 mins, 40 cycles repetition of denaturation at 95°C for 10 s, 15-s annealing at 67°C, 20-s extension at 72°C which was followed by an extra extension step at 72°C for 5 mins. A negative control was included in every run. The Ct values were used for calculating the ccfDNA concentration by absolute quantification through the standard curve. The ratio of ALU247 to ALU115 was considered as the integrity value as was previously described.15,18
Statistical analysis
Statistical analysis was performed by SPSS 13.0, and the p-value <0.05 was considered as statistically significant. The differences between plasma ccfDNA levels of PCa, BPH, and healthy groups were analyzed by unpaired t-test, and Chi-square. Correlations between clinicopathological parameters and plasma ccfDNA levels or the integrity index were performed by one-way ANOVA test and post hoc.
Results
Patient characteristics
The mean age was 63.7±8.0 (48–79 years) in PCa patients, 60.8±8.1 (37–75 years) in BPH group, and 60.6±7.6 (48–76 years) in healthy individuals. There was no significant difference in age between the undergone study groups (P=0.23) (Table 1).
Table 1.
Characteristics of the studied groups
| Number of patients | ccfDNA (ALU115) median (range), (ng/μL) | P-value | Integrity (ALU247/ALU115) | P-value | |
|---|---|---|---|---|---|
| PCa | 30 | 51.03±10.15a | <0.001 | 0.35±0.050 | <0.001 |
| Age (years) | |||||
| 63.73±8.09 | – | – | 0.231 | – | – |
| tPSA (ng/mL) | |||||
| <4 | 6 | 68.30±41.36a | 0.024 | 0.33±0.05 | 0.737 |
| >4 | 24 | 46.71±8.09a | 0.35±0.05 | ||
| Gleason score | |||||
| <6 | 15 | 52.48±35.51a | 0.723 | 0.34±0.05 | 0.724 |
| 7 | 8 | 38.60±8.25a | 0.36±0.05 | ||
| 8–10 | 7 | 62.11±12.11a | 0.36±0.05 | ||
| Metastatic | |||||
| Yes | 10 | 98.68±24.81 | <0.001 | 0.36±0.06 | 0.016 |
| No | 20 | 27.20±3.69 | 0.34±0.04 | ||
| BPH | 40 | 13.50±4.40 | 0.836* | 0.16±0.049 | 0.053 |
| Age (years) | |||||
| 60.80±8.14 | – | – | 0.231 | – | – |
| tPSA (ng/mL) | |||||
| <4 | 28 | 12.07±3.56 | 0.083 | 0.15±0.04 | 0.091 |
| >4 | 12 | 16.83±4.49 | 0.18±0.05 | ||
| Healthy subjects | 30 | 9.28±1.51 | 0.13±0.02 | ||
| Age (years) | |||||
| 60.63±7.69 | – | – | 0.231 | – | – |
| tPSA (ng/mL) | |||||
| <4 | 30 | – | – | – | – |
| >4 | – | ||||
Notes: aSE; *correlation between BPH group and healthy group.
Abbreviations: BPH, benign prostate hypertrophy; tPSA, total prostate-specific antigen; ccfDNA, circulation cell-free DNA; PCa, prostate cancer.
Real-time PCR
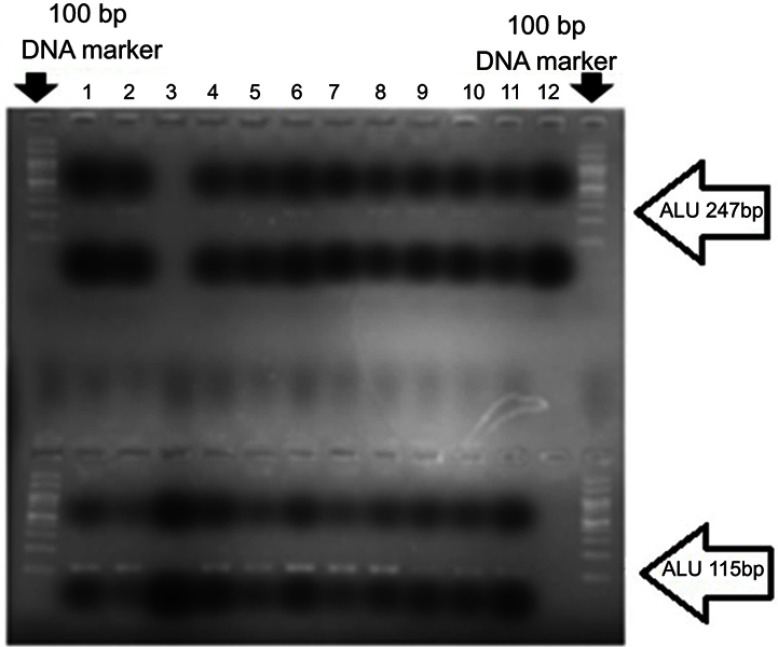
The amplified products with melting temperature at ~82°C were analyzed for the ALU sequences of 247 bp and 115 bp on 2% agarose gel electrophoresis (Figure 1). Amplification efficiency and accuracy were confirmed by drawing a standard curve in acceptable −3.3 concentration gradient and R≥0.98, R2≥0.98 for each of the ALU sequences. The concentration of ccfDNA in patients with PCa was 51.03±10.15 ng/mL, significantly higher than that of patients with BPH (13.5±4.40 ng/mL) and of healthy controls (9.28±1.51 ng/mL, P<0.001). The concentration level of ALU115 in patients’ ccfDNA with metastatic PCa (98.68±24.81 ng/mL) showed to be significantly higher than that of patients with nonmetastatic PCa (27.20±3.69 ng/mL, P<0.001).
Figure 1.
A number of cancerous samples were examined after amplification by real-time PCR. The products were mixed with loading buffer and loaded on 2% agarose gel. Lanes 1–12 are examined samples. The sequences of ALU247 (above) and ALU115 (at the bottom) are clearly visible. The first and the last wells show the 100 bp DNA marker.
The DNA integrity (ALU247/ALU115 ratio) in patients with PCa (0.35±0.05 ng/mL) was also significantly higher than patients with BPH (0.16±0.04 ng/mL) and of healthy individuals (0.13±0.02 ng/mL, P<0.001). Also, the DNA integrity in metastatic patients (0.36±0.06 ng/mL) was significantly higher than patients with nonmetastatic PCa (0.34±0.04 ng/mL, P<0.001). Moreover, the concentration level of ccfDNA and its integrity in BPH patients demonstrated no significant increase compared to the healthy individuals (P=0.83, P=0.05, respectively). In patients with PCa, no statistically significant association was found between total ccfDNA level and Gleason score (P=0.72) and prostate volume (P=0.20). 20% of PCa and 70% of BPH groups had normal serum PSA level (≤4 ng/mL). A significant association was obtained between the increased level of ccfDNA and normal level of total serum PSA in the PCa group (P=0.02). Furthermore, in patients with PCa, there was no significant association neither between the DNA integrity and Gleason score (P=0.72) nor between the DNA integrity and total serum PSA (P=0.73).
Remarkably, a significant association was found between the DNA integrity and the prostate volume in the studied groups (P=0.001).
Discussion
The involvement of approximately 40–70% of men with BPH,26 a more common condition associated with aging men, compared with 16% prevalence PCa is considerable.27 Therefore, new screening biomarkers are needed to prevent unnecessary frequent biopsies and useless treatment of slow-growing and nonmetastatic cancers9,28–30 to overcome losing early diagnosis of subjects with normal PSA, subjects without clinical symptoms and people with BPH who will be affected with PCa.
Our results were consistent with the previous studies17,21,30 where ccfDNA total and its integrity index in our patients with PCa were significantly higher than those of BPH and healthy group. The reason for using ALU repeats to measure ccfDNA short-fragment concentrations and their integrity is the detection of low concentration DNA pieces in plasma by an easy assay with direct q-RT-PCR. Regardless of the differences in the stages of the disease and methods which could affect the value achievement, the difference in the ranges of the screening can vary. The span of ccfDNA values depends on the ethnicity and the genetic background.31 Little differences have been evidenced in different populations when the same method was utilized31 But, when different methods like DNA DipStick and RT-PCR were used to evaluate ccfDNA concentration in newly diagnosed Greek PCa patients, the values were considered significantly different as 236.58±257.8 ng/mL and 20.2±18.7 ng/mL, respectively.16 Therefore, for a fully reliable assessment, a common method with high sensitivity is required to study patients with different conditions.
Fawzy et al found no statistically significant relationship between ALU115 ccfDNA integrity, Gleason scores, and stages of the disease, as in our study, and PSA (contrary to our study).21 Also, we found a significant relationship between ALU115 ccfDNA (P<0.001) and integrity (P=0.05) in metastatic and nonmetastatic patients like them.
In Feng’s study, the level of ccfDNA (ALU115) and its integrity in 96 Chinese patients with PCa was higher than those of patients with BPH as in our study.17 Also, Feng et al found a significant association between ccfDNA and Gleason score unlike our results, but could not detect any association between DNA integrity and Gleason score, like us. They also revealed that ccfDNA had early diagnosis ability to differentiate PCa from BPH patients with increased PSA (≥4 ng/mL), although a probability of false-negative report exists in the normal range of total PSA. Unexpectedly and contrary to previous studies, our results showed a significant correlation between the level of ccfDNA and low PSA in PCa individuals (P=0.024, Table 1). As we indicated, the ccfDNA concentration in 20% of PCa patients with normal PSA was higher than other patients in the PCa group. We did not exclude patients with normal PSA from our final conclusion unlike Feng’s. Our trial showed that individuals with normal PSA may have increased ccfDNA concentrations and possibly are susceptible to develop cancer. Therefore, the measurement of PSA alongside with the ccfDNA could reduce the false-negative rate and help in early diagnosis and screening of a group of potential patients instead of missing them from the detection. We conclude that normal PSA should not be excluded from the evaluation because early detection and differentiation of biopsy candidate PCa and BPHs is the purpose of the study.32 Reports indicate that seldom small-cell prostate carcinoma show normal PSA.33,34 In our assay, two of six patients with normal PSA who were candidates for BPH surgery (OP/TURP) were diagnosed with PCa after operation and through pathology results.
Chun et al observed the relationship between ccfDNA concentrations with increasing prostate volume.32 We did not find such a significant association, but instead, we found a significant association between the integrity index and prostate volume elevation (P-value =0.001) (Table 2). However, both the measured ccfDNA concentration and its integrity values were significantly higher in metastatic condition (P-value <0.001) while they had no correlation with Gleason’s score. The link between ccfDNA level and metastasis could be used for invasive stages. Therefore, as more circulating tumor cells (CTCs), micrometastatic cells, and diffuse malignant cells are disseminated, the more ccfDNA is produced.
Table 2.
Relation between patient’s characteristics with ccfDNA (ALU115) and integrity (ALU247/ALU115) in increased prostate volume and normal prostate volume group
| Number of patients | ccfDNA (ALU115) Median(range) (ng/μL) | P-value | P-value | Integrity (ALU247/ALU115) | P-value | P-value | |
|---|---|---|---|---|---|---|---|
| PCa | 0.001 | ||||||
| Prostate volume | |||||||
| <35 g (25 cc) | 5 | 49.09 | 0.956 | 0.33 | 0.368 | ||
| 35g (25 cc)–50g (70 cc) | 13 | 50.59 | 0.34 | ||||
| ≫50 g (70 cc) | 12 | 57.12 | 0.37 | ||||
| BPH | |||||||
| Prostate volume | |||||||
| <35 g (25 cc) | 6 | 12.15 | 0.161 | 0.207 | 0.14 | 0.363 | |
| 35 g (25 cc)–50 g (70 cc) | 20 | 13.06 | 0.16 | ||||
| ≫50 g (70 cc) | 14 | 15.60 | 0.17 | ||||
| Healthy subject | |||||||
| Prostate volume | |||||||
| <35 g (25 cc) | 27 | – | – | – | – | ||
| 35 g (25 cc)–50 g (70 cc) | 3 | ||||||
| ≫50 g (70 cc) | 0 |
Abbreviations: PCa, prostate cancer; BPH, benign prostate hypertrophy; ccfDNA, circulation cell-free DNA.
In order to achieve the definite and applicable range of ccfDNA level for PCa diagnosis in different cancer statues, a larger sample size is necessary to be examined and utilized as a standard level to differentiate healthy from BPH and cancer-affected men.
Moreover, a possible cutoff point for plasma ccfDNA concentration can be introduced to differentiate BPH and nonmalignant states from healthy conditions for Iranian population screening purposes. Also, introducing a uniform method for the disease diagnosis is of importance regardless of the minor differences due to the population differences or hygienic conditions. While this research demonstrated that the aberrant levels of ccfDNA encompass the quantitative abnormalities, it registered that DNA integrity is a qualitative change of PCa disease.12
The present study lacked large number of samples and separation of metastatic from nonmetastatic understudy groups which should be reconsidered in the future studies.
Conclusions
Total ccfDNA concentration in PCa patients’ plasma is significantly higher than those of healthy and BPH groups. Also, ccfDNA and its integrity are higher in metastatic PCa than in localized PCa. Therefore, plasma ccfDNA levels and its integrity can be used to differentiate PCa from BPH in prostatic patients and in healthy conditions for early detection. Plasma ccfDNA and its integrity can act as a facile discriminating marker instead of unnecessary prostate biopsies. The examination of ccfDNA by measuring the concentration of ALU sequences in the process of screening and tracking PCa in Iranian male population apparently plays an important role in early detection of the disease.
Suggestions
Using a similar investigation with a larger population will show the validity of our data in the future. Based on the found relationship between ccfDNA with PSA, we recommend the ccfDNA testing for BPH surgical candidates.
Also, we propose to compare the ccfDNA amount in PCa patients with normal PSA of different stages and PCa controls with variable PSA of different stages for a future study. A patient of any stage may have increased ccfDNA but normal PSA and there is no definitive direct relationship between PSA and PCa progression35 It can be proposed that ccfDNA is associated with PCa without a mediator like PSA. Moreover, in poor-differentiated adenocarcinoma prostate patients with low PSA level where early diagnosis is difficult, the ccfDNA concentration may aid to the diagnosis of the disease.
Acknowledgments
We would like to appreciate Mr Naghi Taghipour, Mr Mehdi Sahraie and Maziar Ganji from Shahid Beheshti University of Medical Sciences who managed the sampling procedure. The present study was supported by the Research Council and was approved by Shahid Beheshti University of Medical Sciences Ethics Committee.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2.Dhillon PK, Mathur P, Nandakumar A, et al. The burden of cancers and their variations across the states of India: the Global Burden of Disease Study 1990–2016. Lancet Oncol. 2018;19:1289–1306. doi: 10.1016/S1470-2045(18)30447-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosseini SY, Moharramzadeh M, Ghadian AR, Hooshyar H, Lashay AR, Safarinejad MR. Population‐based screening for prostate cancer by measuring total serum prostate‐specific antigen in Iran. Int J Urol. 2007;14(5):406–411. doi: 10.1111/j.1442-2042.2006.01729.x [DOI] [PubMed] [Google Scholar]
- 4.Safarinejad M. Population-based screening for prostate cancer by measuring free and total serum prostate-specific antigen in Iran. Ann Oncol. 2006;17(7):1166–1171. doi: 10.1093/annonc/mdl087 [DOI] [PubMed] [Google Scholar]
- 5.Braunwald E, Fauci AS, Kasper DL, Hauser SL, Longo DL, Jameson JL. CD Harrison’s Principles of Internal Medicine. New York: McGraw-Hill; 2008. [Google Scholar]
- 6.Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324(17):1156–1161. doi: 10.1056/NEJM199104253241702 [DOI] [PubMed] [Google Scholar]
- 7.Klotz L, Zhang L, Lam A, Nam R, Mamedov A, Loblaw A. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol. 2009;28(1):126–131. doi: 10.1200/JCO.2009.24.2180 [DOI] [PubMed] [Google Scholar]
- 8.Andriole GL, Crawford ED, Grubb III RL, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–1319. doi: 10.1056/NEJMoa0810696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ploussard G, Epstein JI, Montironi R, et al. The contemporary concept of significant versus insignificant prostate cancer. Eur Urol. 2011;60(2):291–303. doi: 10.1016/j.eururo.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 10.McKay RR, Zukotynski KA, Werner L, et al. Imaging, procedural and clinical variables associated with tumor yield on bone biopsy in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2014;17(4):325–331. doi: 10.1038/pcan.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Z-H, Xu H-X, Zhang S-R, et al. The significance of liquid biopsy in pancreatic cancer. J Cancer. 2018;9(18):3417–3426. doi: 10.7150/jca.24591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 13.Francis G, Stein S. Circulating cell-free tumour DNA in the management of cancer. Int J Mol Sci. 2015;16(12):14122–14142. doi: 10.3390/ijms160614122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stötzer OJ, Lehner J, Fersching-Gierlich D, Nagel D, Holdenrieder S. Diagnostic relevance of plasma DNA and DNA integrity for breast cancer. Tumor Biol. 2014;35(2):1183–1191. doi: 10.1007/s13277-013-1158-4 [DOI] [PubMed] [Google Scholar]
- 15.Allen D, Butt A, Cahill D, Wheeler M, Popert R, Swaminathan R. Role of cell‐free plasma DNA as a diagnostic marker for prostate cancer. Ann N Y Acad Sci. 2004;1022(1):76–80. doi: 10.1196/annals.1318.013 [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulou E, Davilas E, Sotiriou V, et al. Cell-free DNA and RNA in plasma as a new molecular marker for prostate cancer. Oncol Res Feat Preclin Clin Cancer Ther. 2004;14(9):439–445. doi: 10.3727/0965040041791473 [DOI] [PubMed] [Google Scholar]
- 17.Feng J, Gang F, Li X, et al. Plasma cell-free DNA and its DNA integrity as biomarker to distinguish prostate cancer from benign prostatic hyperplasia in patients with increased serum prostate-specific antigen. Int Urol Nephrol. 2013;45(4):1023–1028. doi: 10.1007/s11255-013-0491-2 [DOI] [PubMed] [Google Scholar]
- 18.Bodega B, Orlando V. Repetitive elements dynamics in cell identity programming, maintenance and disease. Curr Opin Cell Biol. 2014;31:67–73. doi: 10.1016/j.ceb.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 19.Umetani N, Hiramatsu S, Hoon DS. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci. 2006;1075(1):299–307. doi: 10.1196/annals.1368.040 [DOI] [PubMed] [Google Scholar]
- 20.Khani M, Pouresmaeili F, Mirfakhraie R. Evaluation of extracted circulating cell free DNA concentration by Standard Nucleospin Plasma XS (NS) kit protocol compared to its modified protocol. Urol Nephrol Open Access. 2017;4(4):00137. [Google Scholar]
- 21.Fawzy A, Sweify KM, El-Fayoumy HM, Nofal N. Quantitative analysis of plasma cell-free DNA and its DNA integrity in patients with metastatic prostate cancer using ALU sequence. J Egypt Natl Canc Inst. 2016;28(4):235–242. doi: 10.1016/j.jnci.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 22.Choi J-J, Reich III CF, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005;115(1):55–62. doi: 10.1111/imm.2005.115.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzenbach H, Alix-Panabières C, Müller I, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res. 2009;15(3):1032–1038. doi: 10.1158/1078-0432.CCR-08-1910 [DOI] [PubMed] [Google Scholar]
- 24.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao S-L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Prev Biomarkers. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 25.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N‐ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86(4):774–779. [DOI] [PubMed] [Google Scholar]
- 26.Verhamme K, Dieleman J, Bleumink G, Van der Lei J, Sturkenboom M, Panel TPEE. Incidence and prevalence of lower urinary tract symptoms suggestive of benign prostatic hyperplasia in primary care—the Triumph project. Eur Urol. 2002;42(4):323–328. doi: 10.1016/S0302-2838(02)00354-8 [DOI] [PubMed] [Google Scholar]
- 27.Howlader N. SEER cancer statistics review, 1975–2008. 2011. Available from: http://seercancergov/csr/1975_2008/. Accessed October 11, 2011.
- 28.Swanson GP, Yu C, Kattan MW, Hermans MR. Validation of postoperative nomograms in prostate cancer patients with long-term follow-up. Urology. 2011;78(1):105–109. doi: 10.1016/j.urology.2011.01.061 [DOI] [PubMed] [Google Scholar]
- 29.Freedland SJ. Screening, risk assessment, and the approach to therapy in patients with prostate cancer. Cancer. 2011;117(6):1123–1135. doi: 10.1002/cncr.25477 [DOI] [PubMed] [Google Scholar]
- 30.Chang R, Kirby R, Challacombe B. Is there a link between BPH and prostate cancer? Practitioner. 2012;256(1750):13–17. [PubMed] [Google Scholar]
- 31.Yin C, Luo C, Hu W, Ding X, Yuan C, Wang F. Quantitative and qualitative analysis of circulating cell-free DNA can be used as an adjuvant tool for prostate cancer screening: a meta-analysis. Dis Markers. 2016;2016:1–12. doi: 10.1155/2016/3825819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun FKH, Mueller I, Lange I, et al. Circulating tumour‐associated plasma DNA represents an independent and informative predictor of prostate cancer. BJU Int. 2006;98(3):544–548. doi: 10.1111/j.1464-410X.2006.06352.x [DOI] [PubMed] [Google Scholar]
- 33.Trotz C. Prostate cancer with a normal PSA: small cell carcinoma of the prostate – a rare entity. J Am Board Family Pract. 2003;16(4):343–344. [DOI] [PubMed] [Google Scholar]
- 34.Nishio R, Furuya Y, Nagakawa O, Fuse H. Metastatic prostate cancer with normal level of serum prostate-specific antigen. Int Urol Nephrol. 2003;35(2):189–192. [DOI] [PubMed] [Google Scholar]
- 35.Vickers AJ, Cronin AM, Roobol MJ, et al. The relationship between prostate-specific antigen and prostate cancer risk: the Prostate Biopsy Collaborative Group. Clin Cancer Res. 2010;16(17):4374–4381. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]