All mycobacteria encode a group II sigma factor, σB, closely related to the group I principal housekeeping sigma factor, σA. Group II sigma factors are widely believed to play specialized roles in the general stress response and stationary-phase transition in the bacteria that encode them. Contrary to this widely accepted view, we show an additional housekeeping function of σB that complements the function of σA in logarithmically growing cells. These findings implicate a novel and dynamic partnership between σA and σB in maintaining the expression of housekeeping genes in mycobacteria and can perhaps be extended to other bacterial species that possess multiple group II sigma factors.
KEYWORDS: ChIP-Seq, Mycobacterium, rifampin, sigB, sigma factor
ABSTRACT
Mycobacterial σB belongs to the group II family of sigma factors, which are widely considered to transcribe genes required for stationary-phase survival and the response to stress. Here we explored the mechanism underlying the observed hypersensitivity of ΔsigB deletion mutants of Mycobacteriumsmegmatis, M. abscessus, and M. tuberculosis to rifampin (RIF) and uncovered an additional constitutive role of σB during exponential growth of mycobacteria that complements the function of the primary sigma factor, σA. Using chromatin immunoprecipitation sequencing (ChIP-Seq), we show that during exponential phase, σB binds to over 200 promoter regions, including those driving expression of essential housekeeping genes, like the rRNA gene. ChIP-Seq of ectopically expressed σA-FLAG demonstrated that at least 61 promoter sites are recognized by both σA and σB. These results together suggest that RNA polymerase holoenzymes containing either σA or σB transcribe housekeeping genes in exponentially growing mycobacteria. The RIF sensitivity of the ΔsigB mutant possibly reflects a decrease in the effective housekeeping holoenzyme pool, which results in susceptibility of the mutant to lower doses of RIF. Consistent with this model, overexpression of σA restores the RIF tolerance of the ΔsigB mutant to that of the wild type, concomitantly ruling out a specialized role of σB in RIF tolerance. Although the properties of mycobacterial σB parallel those of Escherichiacoli σ38 in its ability to transcribe a subset of housekeeping genes, σB presents a clear departure from the E. coli paradigm, wherein the cellular levels of σ38 are tightly controlled during exponential growth, such that the transcription of housekeeping genes is initiated exclusively by a holoenzyme containing σ70 (E.σ70).
INTRODUCTION
Transcription in bacteria is carried out by a multisubunit RNA polymerase (RNAP) that associates with an interchangeable sigma subunit and directs the transcription machinery to specific promoter regions (1–4). All bacteria encode an essential principal sigma factor and a variable number of alternative sigma factors. Sigma factors are classified into four groups based on the presence of conserved domains 1 to 4. Group I sigma factors are required for transcription of housekeeping genes and are essential (5–7). Group II sigma factors are closely related to those in group I, lack domain 1.1, but are nonessential. Group III sigma factors contain domains 2 to 4, whereas the group IV sigma factors contain only domains 2 and 4. Mycobacteria encode one sigma factor belonging to each of groups I to III and a variable number of group IV sigma factors: 10 in Mycobacteriumtuberculosis, 16 in M. abscessus, and 25 in M. smegmatis (8). Group IV sigma factors have been studied extensively and are involved in heat shock, cold shock, hypoxia, carbon starvation, surface and oxidative stresses, and virulence (8–10). The mycobacterial σA, a group I sigma factor, is essential and highly similar to the primary sigma factors from other bacteria, suggesting that it is the principal sigma factor in mycobacteria (6, 11). σA mRNA levels are constant under different growth conditions, though the levels of the σA protein have been seen to decrease during stationary phase (6, 7). The mycobacterial σB, a group II sigma factor, lacks domain 1.1 and shows an ∼64% sequence identity with σA; in fact, residues important for recognition of −10 and −35 promoter elements are identical between mycobacterial σA and σB (12, 13). Although it is not essential for survival, σB is >90% conserved across mycobacterial species. A deletion in sigB results in sensitivity to heat, oxidative, and surface stress in vitro and an increased sensitivity to p-aminosalicylic acid, sulfamethoxazole, and ethambutol but does not impact the survival of M. tuberculosis in macrophages or mouse lungs (10, 12, 14–19). Two attempts to characterize the σB regulon yielded contradictory results. The global transcription profile of a strain overexpressing σB identified 72 σB-dependent genes, while the global transcription profile of a ΔsigB strain compared to that of wild-type (WT) bacteria identified only 8 σB-dependent genes during exponential growth (12, 14). This disparity can be resolved by determining the binding sites of σB; although a comprehensive map of transcription regulators, including sigma factors, has been determined in M. tuberculosis using chromatin immunoprecipitation sequencing (ChIP-Seq), this does not include SigB binding sites (20, 21). Exposure to diamide and SDS stress resulted in the downregulation of 40 and 72 genes, respectively, in the ΔsigB strain compared to their expression in wild-type bacteria (14). Furthermore, the transcription of σB was shown to occur from two promoters: one recognized by the stress-inducible sigma factors σE, σH, and σL and the other recognized by σF (22–25). These observations together led to the general notion that σB has little role in exponential growth; rather, it is required solely for the mycobacterial response to stress.
RNA polymerase is a target for the broad-spectrum antibiotic rifampin (RIF), which comprises a frontline therapy against M. tuberculosis infection. RIF exerts its effect by binding to the β subunit of RNA polymerase in a region comprising the DNA/RNA channel and sterically blocks the extrusion of elongating RNA when the transcript exceeds 2 to 3 nucleotides (nt) in length (26). High levels of clinically acquired RIF resistance involve rpoB mutations in four distinct sequence clusters (clusters N, I, II, and III), the majority of which map to cluster I (27–32). In contrast to acquired resistance, the fast-growing mycobacteria, such as M. smegmatis and M. abscessus, are naturally RIF resistant, albeit to various extents. This intrinsic rifampin resistance has been attributed to the presence of a rifampin ADP-ribosyltransferase (Arr), which inactivates the drug by ribosylation (33–35). The association of RNAP with accessory proteins, such as certain sigma factors and RbpA, has also been shown to influence its susceptibility to RIF. Wegrzyn et al. showed that the Escherichiacoli σ70-RNAP is considerably more sensitive than σ32-RNAP in vitro and in vivo and that a deletion of Bacillussubtilis sigB, the orthologue of mycobacterial sigF, renders the bacteria more sensitive to RIF (43). RbpA, an RNAP binding protein conserved in actinomycetes, has been shown to prevent RIF inhibition in vitro (37, 38). While RbpA is essential in mycobacteria, a deletion of RbpA in Streptomyces coelicolor results in RIF sensitivity and a slow-growth phenotype (37, 38). It is unlikely that RbpA is involved in the degradation or efflux of RIF but, rather, modifies RNAP. RbpA interacts exclusively with group I and II sigma factors in Streptomyces and mycobacteria and stabilizes the formation of open promoter complexes, thereby enhancing the transcription efficiency of holoenzymes containing σA and σB. The mechanism by which RbpA confers RIF tolerance is unknown but has been shown to not involve an occlusion of the RIF binding site in RNAP, and its effect is presumably indirect (39, 40).
In the current work, we explore the underlying mechanism of RIF sensitivity of ΔsigB mutants of M. smegmatis, M. abscessus, and M. tuberculosis and demonstrate that the RIF sensitivity of ΔsigB strains is likely not attributable to the lack of transcription of σB-dependent RIF resistance genes. The study has uncovered that, contrary to previous models, σB is transcriptionally active during the exponential phase of growth of M. smegmatis and actively transcribes several σA-dependent housekeeping genes. Our results therefore demonstrate an active role for σB in the exponential phase of mycobacterial growth, in addition to its role as a stress response sigma factor.
RESULTS
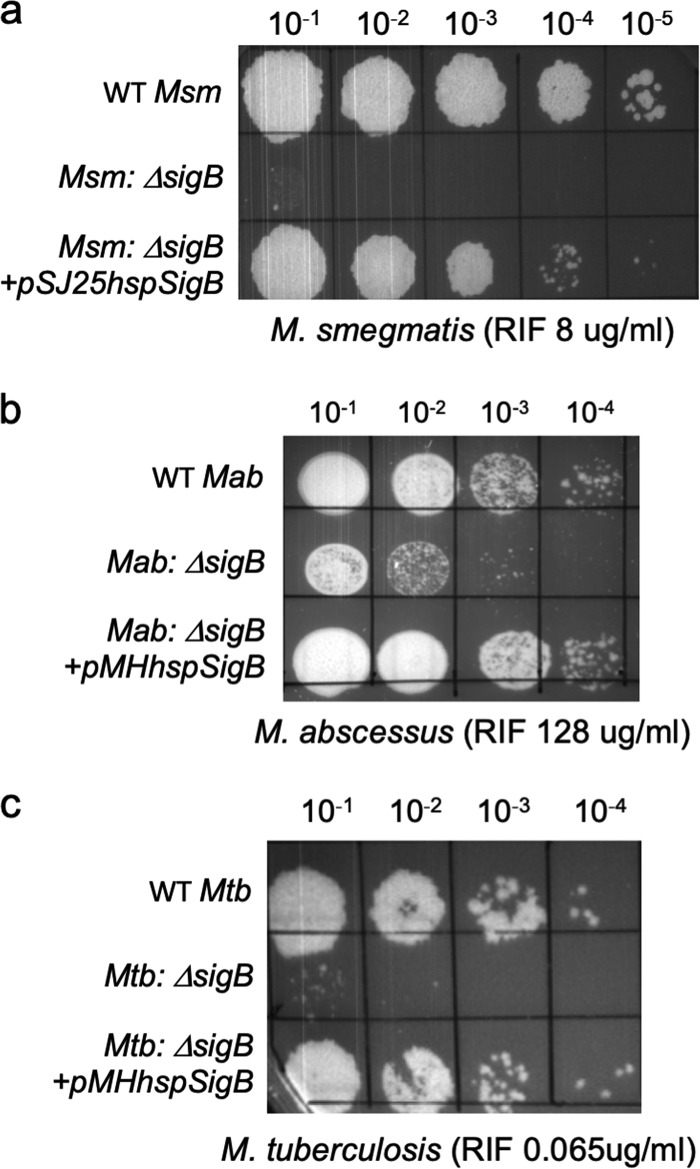
Deletion of sigB results in RIF hypersensitivity in M. smegmatis, M. tuberculosis, and M. abscessus.
To understand the role of sigma factors in mycobacterial drug tolerance, we constructed isogenic deletions in 14 out of 28 randomly selected sigma factor genes in M. smegmatis using recombineering and assayed the sensitivity of the deletion strains to a variety of antibiotics (41). Deletion of the primary-like sigma factor sigB resulted in hypersensitivity to RIF (Fig. 1a). We then explored if the phenotype of the ΔsigB mutant could be recapitulated in the pathogenic mycobacteria M. tuberculosis and M. abscessus. sigB deletion mutations were constructed in the attenuated M. tuberculosis strain mc27000 and the M. abscessus ATCC 19977 strain using recombineering. The ΔsigB strains of M. tuberculosis and M. abscessus were found to be hypersensitive to RIF compared to their corresponding wild types (Fig. 1b and c), suggesting that the σB-mediated basal RIF tolerance may be conferred by a conserved mechanism. Growth of the ΔsigB mutant of M. smegmatis in Middlebrook 7H10 agar lacking antibiotics was unaffected but was reduced in 7H9 broth compared to that of WT bacteria (see Fig. S1a and b in the supplemental material). The M. tuberculosis ΔsigB mutant displayed a slow-growth phenotype on Middlebrook 7H10 agar lacking antibiotics and has previously been shown to exhibit slow growth in liquid media (Fig. S1d) (18).
FIG 1.
Deletion of σB confers RIF sensitivity in M. smegmatis (Msm), M. abscessus (Mab), and M. tuberculosis (Mtb). (a to c) Tenfold serial dilutions of M. smegmatis mc2155, M. abscessus ATCC 19977, M. tuberculosis mc27000, and their respective ΔsigB and complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC or OADC containing the indicated concentrations of RIF. Deletion of sigB results in RIF sensitivity in all three strains. The mutant phenotype can be complemented by constitutive expression of the respective sigB gene.
Growth of M. smegmatis in media lacking RIF. (a) Tenfold serial dilutions of M. smegmatis mc2155, the ΔsigB mutant, and complemented strain were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC or OADC lacking RIF. (b) The growth of M. smegmatis mc2155 and the ΔsigB mutant in Middlebrook 7H9-ADC-Tween 20 was monitored over a period of 40 h. Deletion of sigB results in slow growth in liquid media but no apparent growth change on 7H10 agar. (c) Tenfold serial dilutions of M. abscessus ATCC 19977, M. tuberculosis mc2700, and their corresponding ΔsigB mutants and complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 OADC medium lacking RIF. Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
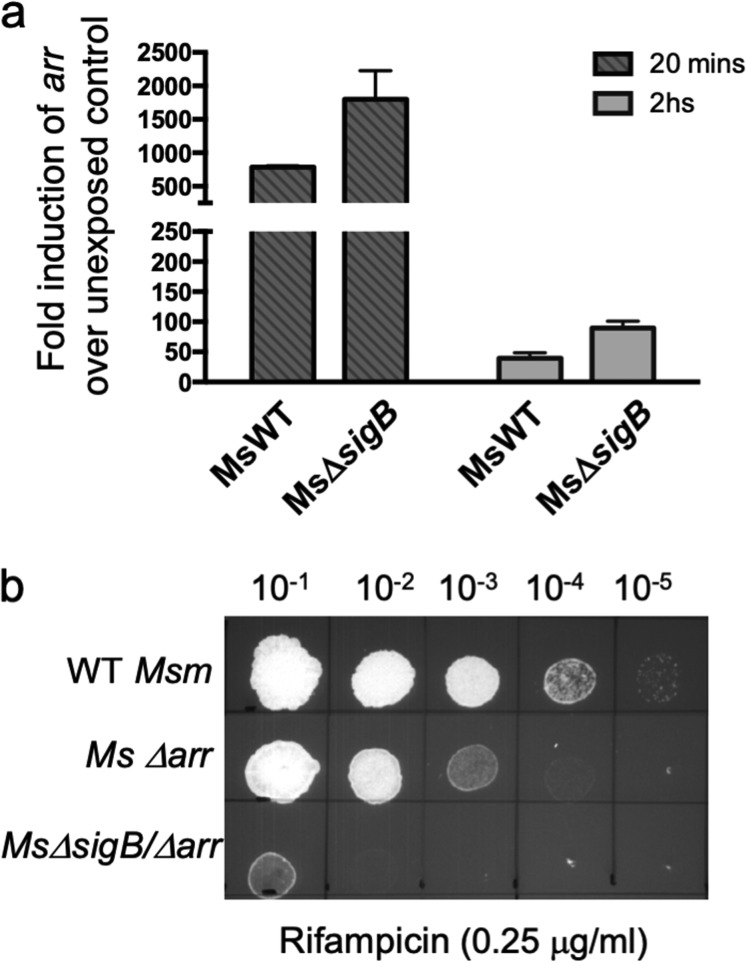
SigB-mediated resistance to RIF is independent of Arr.
Intrinsic tolerance to RIF in mycobacteria has been attributed to the ribosylation of RIF by ADP-ribosyltransferases (Arr), encoded by the fast-growing mycobacteria (42). We first investigated the most likely scenario that σB is required for the transcription of arr, either directly or indirectly, such that a deletion in sigB abrogates arr expression, resulting in RIF sensitivity. We therefore determined the relative abundance of the arr transcript in WT M. smegmatis and the ΔsigB mutant of M. smegmatis (the MsΔsigB mutant) upon exposure to RIF by quantitative PCR (qPCR) analysis. Figure 2a shows that the level of arr induction upon RIF exposure did not decrease in the ΔsigB mutant strain, as would be expected if its transcription were solely dependent on SigB. Instead, arr transcript levels increased ∼6-fold in a ΔsigB strain and may reflect a compensatory response. Although this does not rule out the possibility that σB is required for the transcription of arr, it suggests the presence of redundant pathways for arr expression. Nevertheless, this demonstrates that the RIF sensitivity of the ΔsigB strain cannot be attributed to a compromised transcription of arr. However, it is possible that the cellular level of the Arr protein is indirectly influenced by the absence of σB. If this were the case, we would anticipate that the RIF sensitivities of the Δarr mutant and the Δarr ΔsigB double mutant would be indistinguishable. However, we observed that the RIF sensitivity of the double mutant was significantly higher than that of each of the single mutants, suggesting that their effect is additive and mediated through independent pathways (Fig. 2b; Table 1). Moreover, a ΔsigB strain of M. tuberculosis, which naturally lacks arr, is also hypersensitive to RIF and provides additional support for the suggestion that the σB-mediated resistance to RIF is independent of ADP-ribosyltransferases.
FIG 2.
σB-mediated resistance to RIF is independent of ADP-ribosyltransferase (Arr) and putative effector genes. (a) Wild-type M. smegmatis (MsWT) and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for either 20 min or 2 h, and the amount of M. smegmatis arr transcripts was determined by qPCR and plotted as the fold induction over the level of expression for an unexposed control. Data represent the mean ± SD (n = 3). sigA was used as an endogenous control. (b) Tenfold serial dilutions of WT strain M. smegmatis mc2155 and the MsΔarr and MsΔsigB MsΔarr strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC containing the indicated concentration of RIF.
TABLE 1.
MIC of RIF for the M. smegmatis WT, ΔsigB, Δarr, and ΔsigB Δarr strainsa
| Strain | MIC of RIF (μg/ml) |
|---|---|
| WT mc2155 | 10 |
| mc2155 ΔsigB | 2.5 |
| mc2155 Δarr | 0.25 |
| mc2155 ΔsigB Δarr | 0.0625 |
The survival of the M. smegmatis mc2155 wild-type, ΔsigB, Δarr, and ΔsigB Δarr strains was determined in a 2-fold dilution series of RIF in Middlebrook 7H9 medium. The minimum concentration of antibiotic required to inhibit 99% of the growth is shown.
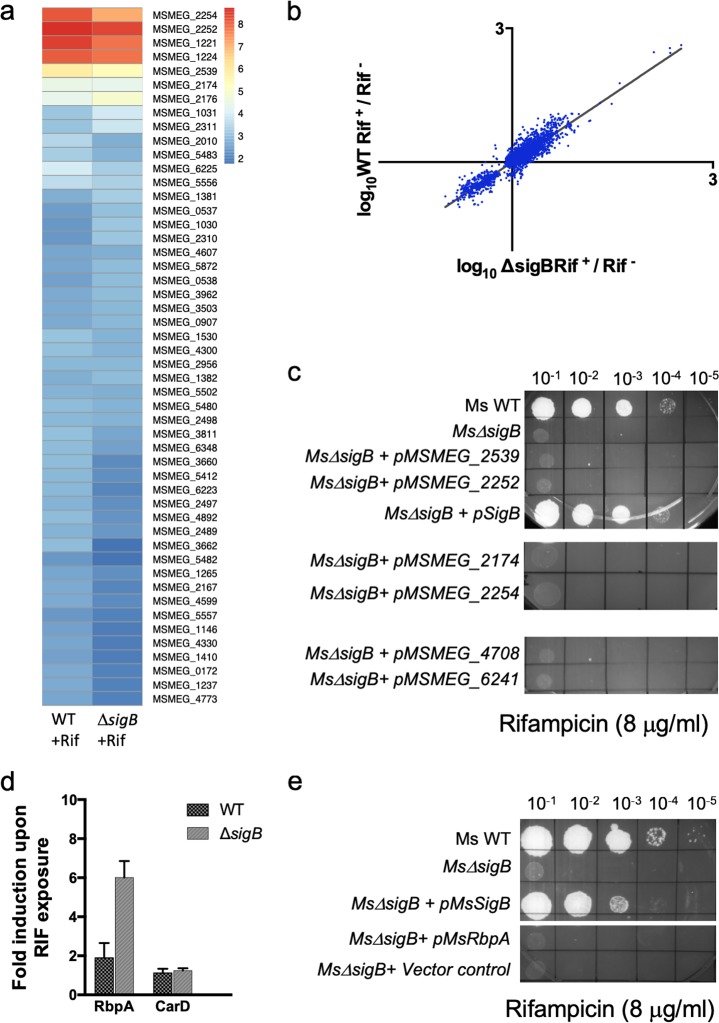
The RIF sensitivity of the ΔsigB mutant is independent of the transcription of known and putative RIF resistance effectors.
We next tested if σB regulates the expression of genes besides arr that mitigate the effect of RIF. We analyzed the transcription profile of wild-type mc2155 and the ΔsigB mutant upon exposure to sublethal doses of RIF (4 μg/ml) using RNA sequencing (RNA-seq). σB-dependent genes that confer RIF resistance would be detectable as those that are RIF inducible in the wild type but not in the ΔsigB mutant. An exposure time of 20 min was found to be most appropriate to enable detection of the gene expression changes that immediately follow RIF exposure. Exposure of wild-type bacteria to RIF caused a >4-fold induction of 101 genes with a q value of <0.001 (Data Set S1), of which the top 50 are represented in Fig. 3a, left. The most highly induced genes were MSMEG_2252 (homologue of rifampin monooxygenase [Rox]), MSMEG_2539 (thiopurine methyltransferase), MSMEG_2174 (helicase), MSMEG_2254 (oxalate decarboxylase), MSMEG_1221 (ADP-ribosyltransferases [Arr]), and MSMEG_1224 (Arr). Surprisingly, however, genes that were RIF inducible in wild-type bacteria showed comparable levels of induction in the ΔsigB mutant strain (Fig. 3a, right; Fig. 3b; Fig. S2; Data Set S1). Consistent with this observation, the RIF tolerance of the ΔsigB strain could not be restored to wild-type levels by overexpression of MSMEG_2252, MSMEG_2254, MSMEG_2539, or MSMEG_2174 (Fig. 3c). Interestingly, although a deletion in MSMEG_2174 increased RIF susceptibility, its expression was unchanged in the ΔsigB mutant, indicating that the phenotype is sigB independent (Fig. S3).
FIG 3.
Transcriptomic changes accompanying RIF exposure in the wild-type and ΔsigB mutant M. smegmatis strains. (a) Wild-type M. smegmatis and the ΔsigB mutant were exposed to 4 μg/ml of RIF for 20 min and analyzed using RNA-seq. Unexposed samples of both strains were used as controls. Two biological replicates of each sample were used. Genes induced >4-fold with a q value of <0.001 were analyzed further, and the 50 most induced genes are represented as a heat map. (Left) WT; (right) ΔsigB mutant. (b) RIF-induced changes in gene expression in the WT versus ΔsigB mutant are shown using genes with a q value of <0.1. (c) Complemented strains were created by integrating MSMEG_2539, MSMEG_2252, MSMEG_2254, and MSMEG_2174 (genes highly upregulated in the presence of RIF) and MSMEG_4708 and MSMEG_6241(two SigB-dependent genes identified by RNA-seq) at the Bxb1 attB site of mc2155 ΔsigB. Tenfold serial dilutions of M. smegmatis mc2155, the MsΔsigB mutant, and the complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC plates containing the indicated concentration of RIF. Overexpression of the genes listed above did not restore the RIF-sensitive phenotype of mc2155 ΔsigB. (d) Wild-type M. smegmatis and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for 30 min, and the amounts of the rbpA and carD transcripts were determined by qPCR and plotted as the fold induction over the level of expression for an unexposed control. Data represent the mean ± SD (n = 3). sigA was used as an endogenous control. (e) A strain complemented with rbpA was created by integrating MSMEG_3858 at the Bxb1 attB site of mc2155 ΔsigB. Tenfold serial dilutions of M. smegmatis mc2155, the MsΔsigB mutant, and the rbpA complemented strain were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC plates containing the indicated concentration of RIF.
Principal-component analysis (PCA) plot generated from RNA-seq data for M. smegmatis mc2155 and the ΔsigB mutant, unexposed controls, and strains exposed to RIF (4 μg/ml for 20 min) Replicate samples are indicated by the same color. The samples are plotted with respect to their first two principal components, which represent the x and y axes, respectively, and account for 87% and 7% of the variance, respectively. The PCA plot was generated with the DESeq2 package in R. Download FIG S2, PDF file, 0.5 MB (481.1KB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Isogenic deletions in MSMEG_2252, MSMEG_2539, and MSMEG_2174 were created by recombineering, and the strains were assayed for RIF sensitivity. The strain with the MSMEG_2174 deletion displayed increased RIF sensitivity compared to WT bacteria; the RIF sensitivities of the strain with MSMEG_2252 and MSMEG_2539 deletions were unchanged. Download FIG S3, PDF file, 1.6 MB (1.7MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptome of the M. smegmatis wild type and the ΔsigB mutant exposed to 4 μg/ml RIF for 20 min: SD-1 RNA-seq of RIF-induced WT versus SigB.xlsx. Download Data Set S1, XLSX file, 0.02 MB (24.8KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also considered the possibility that σB-dependent RIF resistance effectors are constitutively expressed. We therefore compared the transcription profiles of the wild-type and ΔsigB strains of M. smegmatis mc2155 grown to mid-log phase. RNA-seq analysis showed that 13 genes were significantly (q value < 0.01) underexpressed by >3-fold in the mutant (Table S1 and Data Set S2), which is consistent with previously published results (14). We evaluated the role of two of the most highly affected genes that were underexpressed in the ΔsigB mutant strain: MSMEG_4708, which encodes a methyltransferase, and MSMEG_6241, which encodes an AAATPase. Overexpression of either of these genes did not complement the RIF sensitivity of the ΔsigB mutant strain (Fig. 3c).
List of genes downregulated >3-fold in an mc2155 ΔsigB strain compared to their expression in wild-type bacteria, determined using RNA-seq. Complete data are included in Data Set S2 in the supplemental material. Download Table S1, DOCX file, 0.04 MB (44.4KB, docx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sigB regulons of logarithmically grown M. smegmatis: SD-2 RNA-seq of WT versus SigB.xlsx. Download Data Set S2, XLSX file, 0.02 MB (19.9KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lastly, we evaluated the role of RbpA, an RNA polymerase-associated protein that has been shown to affect the RIF sensitivity of S. coelicolor and is RIF inducible in mycobacteria (37, 38). Figure 3d shows that RbpA transcript levels increased ∼2-fold in wild-type bacteria and ∼6-fold in the ΔsigB mutant upon RIF exposure, consistent with the results of the RNA-seq experiments. Moreover, overexpression of RbpA failed to restore the RIF tolerance of the ΔsigB mutant to that of the wild type (Fig. 3e). Together, these observations suggest that the RIF sensitivity of the ΔsigB mutant cannot be attributed to the lack of RbpA, a known effector of RIF resistance.
σA- and σB-containing holoenzymes are indistinguishable in their RIF susceptibility.
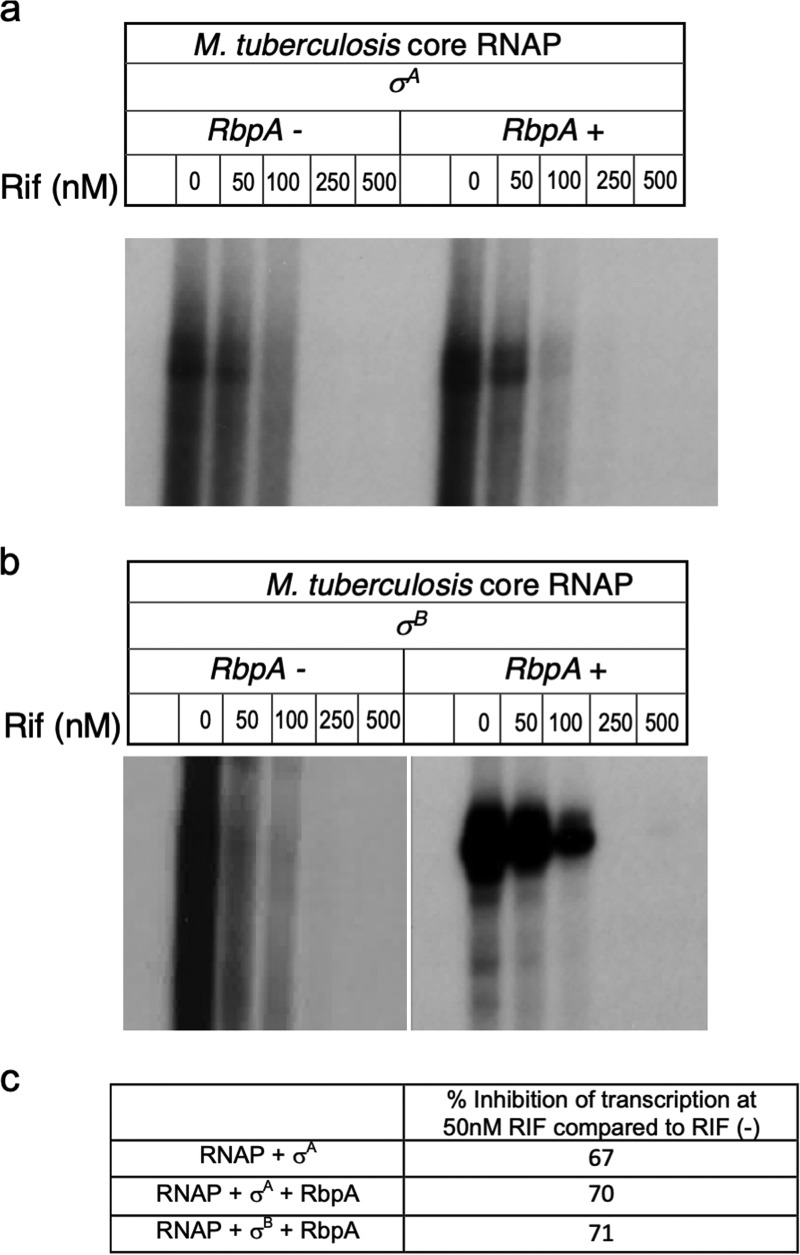
Taken together, the data likely rule out the possibility that the RIF-sensitive phenotype of ΔsigB is due to the lack of expression of either novel or previously described effectors of RIF resistance. We speculated that the observed RIF sensitivity could be a reflection of the interaction of σB with RNAP, the target of RIF. The sensitivity of RNAP to RIF has previously been demonstrated to depend on its association with particular species of sigma factors; holoenzymes associated with primary sigma factors are more sensitive than those associated with alternate sigma factors (38, 43). In addition, σB has been shown to recognize several σA-dependent promoters in vitro (13). Based on these two lines of evidence, we propose that the RIF sensitivity of the ΔsigB mutant can be explained by one of two scenarios: (i) a holoenzyme containing σB (E.σB) is more resistant to RIF than a holoenzyme containing σA (E.σA) and is recruited to housekeeping promoters in the presence of RIF when transcription by E.σA is compromised, or (ii) E.σA and E.σB are equally sensitive to RIF but are both involved in the transcription of housekeeping genes in exponentially growing bacteria. The toxicity of RIF would become pronounced when one of the sigma factors is missing, especially if the expression of neither sigA nor sigB is inducible. Since σA is essential, this phenotype is apparent only in a ΔsigB mutant.
We determined the RIF sensitivity of σA-RNAP and σB-RNAP by assaying their activity at the sigA promoter (sigAP) in multiple-round in vitro transcription assays. Assays were performed both in the presence and in the absence of RbpA, since RbpA has been shown to assist with open complex formation by σA and σB, as well as offer protection against RIF inhibition (13, 38–40, 44, 45). Figure 4a and b show that RbpA greatly (>100-fold) enhanced transcription by E.σB, but its effect on E.σA was modest (∼2-fold) and is consistent with previously published results (13, 40). In the presence of RbpA, although the overall yield of the transcript was ∼100-fold higher when using E.σB, the inhibition of transcription at each RIF concentration was comparable when using either E.σA or E.σB (∼70% inhibition was seen with both holoenzymes at 50 nM RIF) (Fig. 4c). This suggests that E.σA and E.σB are equally RIF sensitive and that the association of σB with RNAP does not offer any additional protection against RIF.
FIG 4.
σA and σB containing holoenzymes are equally RIF susceptible. (a and b) Multiple-round in vitro transcription assays were performed on the sigA promoter using 200 nM σA-RNAP/σB-RNAP. RbpA (600 nM) was added where indicated. RIF was added to the indicated concentrations for 30 min at 37°C. Transcription was initiated by addition of 2 μl of an NTP mix (1.5 mM ATP, GTP, and CTP and 0.5 mM UTP) plus 2 μCi of [α-32P]UTP. The reaction mixtures were incubated at 37°C for 30 min, and the reactions were terminated by the addition of 5 mM EDTA and 100 μg/ml tRNA. Samples were ethanol precipitated and separated using denaturing PAGE (6% urea polyacrylamide gel). (c) The products were visualized using a Typhoon imager (GE Healthcare) and quantitated using ImageQuant software. Inhibition of RNAP activity at 50 nM RIF is expressed as a ratio of the activity in the presence and absence of RIF.
σB actively transcribes housekeeping genes in exponentially growing M. smegmatis.
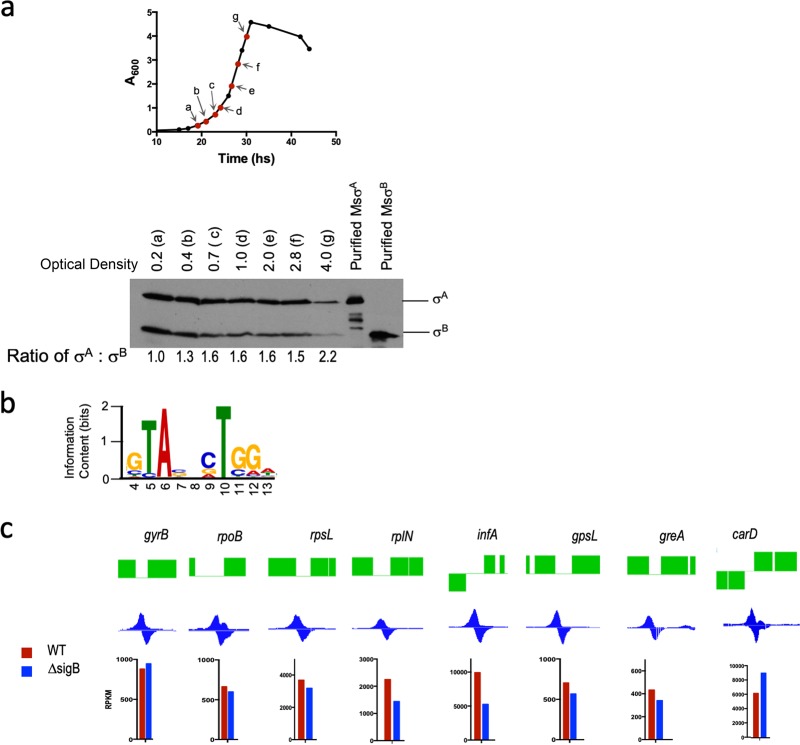
We next explored the alternate scenario that E.σA and E.σB are both involved in the transcription of housekeeping genes in exponentially growing bacteria. This hypothesis is contrary to the currently accepted notion that σB is required only during transition to stationary phase and in response to environmental stress (7, 8, 12, 14). However, RNA-seq of exponentially growing M. smegmatis showed comparable levels of sigA and sigB transcripts (Data Set S2) (10). We therefore determined the relative levels of the σA and σB proteins at various stages of M. smegmatis growth by Western blot analysis using an anti-σ70 antibody that recognizes an epitope in domain 3.1 common to mycobacterial σA and σB and E. coli σ70 (46) (Fig. S4a). Figure 5a shows that σB is consistently present in exponentially growing M. smegmatis; in fact, σB protein levels were comparable to those of σA during early logarithmic phase of growth.
FIG 5.
σB is transcriptionally active in exponentially growing M. smegmatis. (a) (Top) Growth kinetics of wild-type M. smegmatis indicating the growth phase and samples used for Western blotting. (Bottom) Relative levels of the σA and σB proteins at the indicated optical densities determined by Western blotting using an anti-σ70 monoclonal antibody. Samples were normalized by wet weight and protein concentration to ensure equivalent loading at each OD. Purified σA and σB proteins were used as controls. The ratio of σA/σB was quantitated using ImageJ software and is shown below. Equivalent amounts of protein were loaded in each lane of the Coomassie-stained gel (see Fig. S4b in the supplemental material). (b) Sequence logo of enriched motif in σB-FLAG-bound sites identified using the MEME Suite of tools (MEME E value = 7.0e−003). (c) The ChIP-Seq peaks of σB bound to promoters of key housekeeping genes visualized with SignalMap software are shown. The transcript levels of the corresponding genes (RPKM values) in the wild type (red) and the ΔsigB strain (blue) are plotted.
(a) Sequence conservation of mycobacterial σA and σB and E. coli σ70. The conserved epitope recognized by the 2G10 antibody is shown. (b) Coomassie-stained gel serving as a loading control for the assay whose results are presented in Fig. 5a. Equivalent amounts of protein were loaded in each lane. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In order to determine if σB is transcriptionally active in exponentially growing bacteria, we analyzed the genomewide binding profile of σB in M. smegmatis using chromatin immunoprecipitation sequencing (ChIP-Seq). The sigB gene was C-terminally tagged with the 3×-FLAG epitope at its native chromosomal location, grown to mid-exponential phase (optical density at 600 nm [OD600] = 0.5), and DNA-nucleoprotein complexes were immunoprecipitated with anti-FLAG monoclonal antibodies. Wild-type strain mc2155 lacking a 3×-FLAG fusion was used as a control. Sequenced library reads were mapped to the reference genome using the Bowtie 2 algorithm, and peaks were called using a previously published Python script, Peakcaller (47), and viewed with SignalMap software (Fig. S5a). We identified 327 genomewide peaks of σB covering 306 genomic regions (peaks within 100 bp of each other were merged), of which 266 peaks were intergenic and 40 mapped within genes (Data Set S3). Transcription start sites (TSSs) for annotated genes in the vicinity of 210 out of the 266 intergenic σB ChIP-Seq peaks have been previously published; ∼85% of these peaks were located within 11 nt of a TSS and are therefore highly likely to be σB dependent (Fig. S5b; Data Set S3) (48). σB binding sites were found to be associated with essential housekeeping genes encoding ribosomal proteins, rpoB, carD, gyrA, and, most prominently, the genes for rRNA. Using the MEME Suite of tools, we could identify a highly enriched motif in 101 σB ChIP-Seq regions (Fig. 5b) (49). The central core of this motif resembled the −10 consensus sequence 5′-TANNNT-3′ proposed for housekeeping promoters and could be detected in 191 out of 210 intergenic σB ChIP-Seq peaks found in close proximity to known TSSs (Data Set S3) (8, 50, 51); these included experimentally determined −10 sequences published previously, such as those for ideR, rpsL, rrnP, and sigA (52–54). Curiously, however, this motif differed considerably from the −10 consensus 5′-NNGNNG-3′ previously published for M. tuberculosis sigB, which could have been a consequence of the methods employed (12). The 5′-NNGNNG-3′ motif was derived using sequence analysis of 5′ untranslated regions of genes that were identified to be σB dependent using microarrays upon overexpression of σB. The identified genes presumably represent a combination of σB-dependent genes that are transcribed during exponential phase and in response to stress, as well as several additional nonspecific genes that are known to be identified during global transcriptomic analyses using overexpressed proteins (55). The σB binding sites identified in this study were determined using σB that was FLAG tagged in its native chromosomal location and were derived from sites that are recognized by σB only during logarithmic phase. Determination of σB binding sites under various environmental stresses is likely to identify promoter motifs that differ from the 5′-TANNNT-3′ motif identified here.
(a) Genomewide binding profile of σB visualized with SignalMap software. (b) Histogram showing the frequency distribution of distances between SigB ChIP-Seq peak centers and TSSs. Download FIG S5, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ChIP-Seq of σB: SD-3 ChIP-Seq binding sites.xlsx. Download Data Set S3, XLSX file, 0.1 MB (70.2KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A comparison of the σB ChIP-Seq data with the RNA-seq data for the ΔsigB strain revealed that none of the genes whose promoters were bound by σB were significantly downregulated in the ΔsigB mutant (Fig. 5c; Data Set S3). This supports the idea that promoters that are recognized by σB during exponential growth must also be recognized by an additional sigma factor, likely σA. A previously published study of σA binding sites in exponentially growing M. smegmatis was performed using E. coli anti-σ70 antibody, which recognizes both mycobacterial σA and σB; this data set therefore represents a combination of σA and σB binding sites. A comparison of the sites bound by σ70 with those bound by σB-FLAG showed the presence of at least 541 sites that were bound by σA alone; these included essential genes, such as those encoding subunits of DNA polymerase III, initiation factor 2 (IF-2), peptide release factor 2, RecO, FtsZ, rpsS, etc., and thereby explains the essentiality of σA (46, 56). The 306 σB binding sites (identified here using anti-FLAG antibody) comprise a subset of total sites identified by E. coli σ70 and likely represent sites that are recognized either by σB alone or by both σA and σB. In order to distinguish between these possibilities, we overexpressed a FLAG-tagged σA using the constitutive hsp60 promoter from a chromosomally integrated location. However, ChIP-Seq using this strain was highly inefficient. Repeated attempts to add a FLAG tag at the C-terminal end of sigA at its native chromosomal location were also unsuccessful, suggesting that the presence of a FLAG tag may compromise the function of σA. Despite the inefficiency of ChIP-Seq using σA-FLAG, we could identify 94 σA binding sites that were common with σB binding sites (and that were also recognized by E. coli anti-σ70 antibody) and likely represent high-affinity binding sites of σA. Out of the 94 σA binding sites, 61 were associated with known TSSs, and these therefore constitute the minimum number of promoters that are recognized by both σA and σB, including promoters for essential genes such as those for rRNA, tRNAs, ribosomal proteins, sigA, and rpoB (Data Set S3).
Overexpression of σA restores the RIF tolerance of MsΔsigB to that of the wild type.
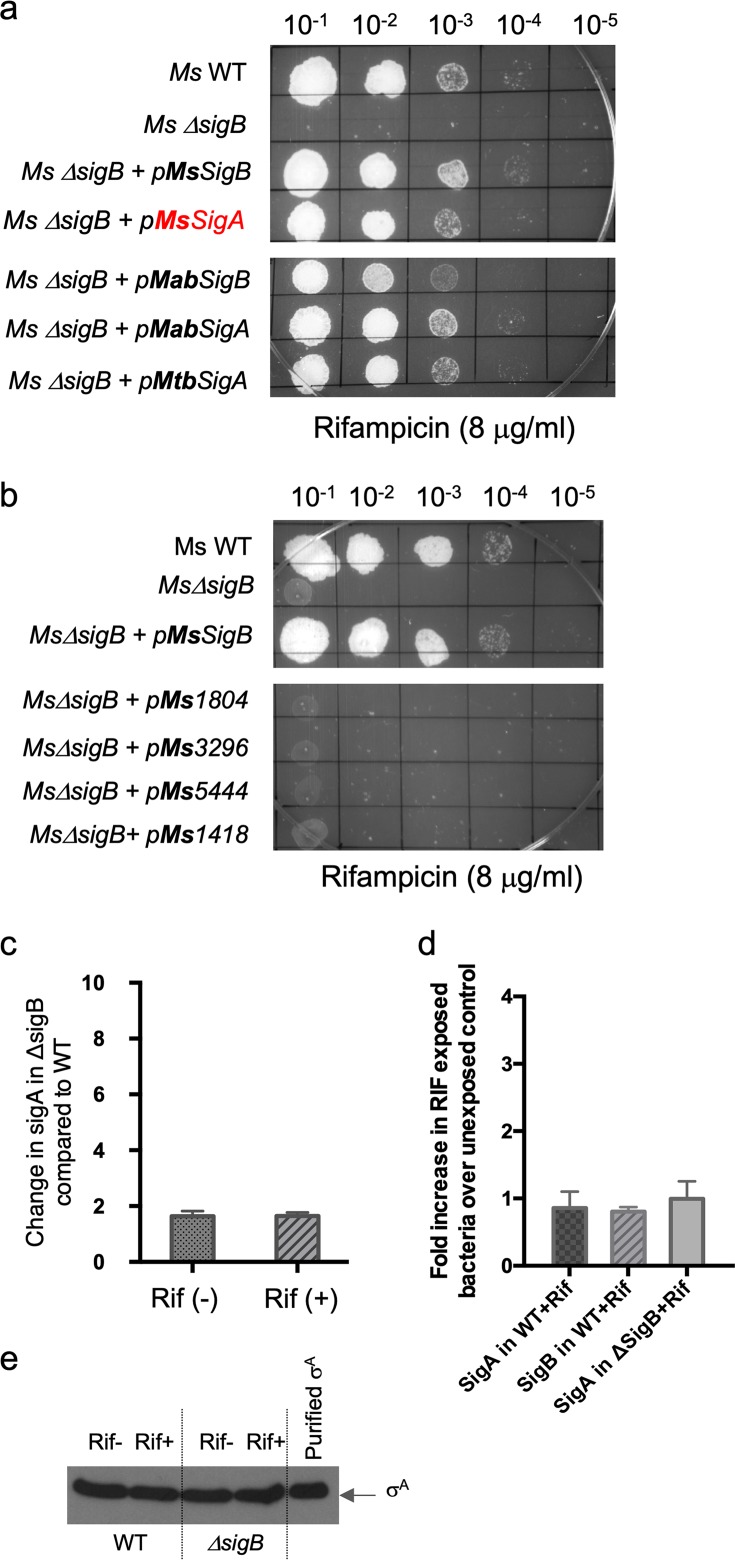
We reasoned that if transcription of housekeeping genes is initiated by both E.σA and E.σB, the absence of σB could be compensated for by increasing the copy number of σA in a way that mitigates the deleterious effect of RIF. Figure 6a and Table 2 show that the constitutive overexpression of M. smegmatis sigA from a chromosomally integrated location restored the RIF sensitivity of the MsΔsigB mutant. Further, we observed that overexpression of either sigA or sigB from M. abscessus and M. tuberculosis could complement the phenotype of the MsΔsigB mutant (Fig. 6a). However, this effect was restricted to the group I and II sigma factors; sigF (group III) and the extracytoplasmic function (ECF) sigma factors were unable to restore the RIF sensitivity of the MsΔsigB mutant (Fig. 6b), presumably because of their inability to initiate transcription at housekeeping promoters. An alternate explanation is that a deletion of sigB reduces the levels of σA, which can be complemented by overexpression of σA. We therefore followed the expression of the sigA transcript as well as protein levels in the MsΔsigB mutant in the presence and absence of exposure to RIF. Figure 6c and d show that sigA transcript and protein levels did not decrease in the MsΔsigB bacteria compared to wild-type bacteria; furthermore, sigA expression was also not RIF inducible in either the wild type or the MsΔsigB mutant (Fig. 6e).
FIG 6.
Overexpression of σA restores the RIF sensitivity of the MsΔsigB mutant. (a to c) Complemented strains were created by integrating Ms_SigA, Mtb_SigA, Mab_SigA, Ms_SigB, Mtb_SigB, Mab_SigB, Ms_Ms1804, Ms_Ms3296, Ms_Ms5444, and MSMEG_1418 at the Bxb1 attB site of mc2155 ΔsigB. Tenfold serial dilutions of M. smegmatis mc2155, the mc2155 ΔsigB mutant, and the complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC plates containing the indicated concentrations of RIF. The RIF sensitivity of mc2155 ΔsigB could be complemented by the constitutive expression of sigA and sigB from all mycobacterial strains but not by ECF sigma factors. (b) Wild-type M. smegmatis and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for 30 min, and the amount of the M. smegmatis sigA transcript was determined by qPCR and plotted as the fold induction of sigA levels in the MsΔsigB strain over the level of expression in the wild-type strain. The data represent the mean ± SD (n = 3). MSMEG_4936 was used as an endogenous control, as its levels were unchanged under various conditions in RNA-seq experiments. (d) Wild-type M. smegmatis and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for 30 min, and the amounts of the M. smegmatis sigA and sigB transcripts were determined by qPCR and plotted as the fold induction upon RIF exposure over the level of expression for an unexposed control. The data represent the mean ± SD (n = 3). MSMEG_4936 was used as an endogenous control. (e) Wild-type M. smegmatis and the MsΔsigB strain were grown to an A600 of 0.7 and exposed to 4 μg/ml RIF for 30 min. The levels of σA protein were determined by Western blotting using an anti-σ70 monoclonal antibody. Samples were normalized by wet weight and protein concentration to ensure equivalent loading of each sample. Purified σA was used as a control.
TABLE 2.
MIC of RIF for M. smegmatis wild-type, ΔsigB, and ΔsigB strains overexpressing either M. smegmatis sigB or sigAa
| Strain | MIC of RIF (μg/ml) |
|---|---|
| WT mc2155 | 10 |
| mc2155 ΔsigB | 2.5 |
| mc2155 ΔsigB sigBOE | >10 |
| mc2155 ΔsigB sigAOE | >10 |
The survival of M. smegmatis wild-type strain mc2155, mc2155 ΔsigB, and mc2155 ΔsigB overexpressing M. smegmatis sigB and sigA (mc2155 ΔsigB sigBOE and mc2155 ΔsigB sigAOE, respectively) was determined in a 2-fold dilution series of RIF in Middlebrook 7H9 medium. The minimum concentration of antibiotic required to inhibit 99% of the growth is shown.
DISCUSSION
All mycobacterial species contain a highly conserved group II sigma factor, the product of the sigB gene. Global transcriptomic analyses have failed to identify sizable numbers of σB-dependent genes during exponential growth of mycobacteria. Moreover, sigB mRNA levels increase upon entry into stationary phase and in response to heat shock and surface and oxidative stresses (5, 7, 10, 14). These observations have led to the inference that σB is specialized for transcription during transition to stationary phase and in the global stress response but does not play an active role in transcription during the logarithmic phase of growth. Herein we present a series of observations which together illuminate a role for σB during exponential growth, in addition to it being a stress response sigma factor.
Using ChIP-Seq analysis under exponential growth conditions, we demonstrated that σB binds to over 200 promoter regions, several of which control the transcription of essential housekeeping genes, such as the rRNA gene, carD, rpoB, etc. This finding is consistent with that of RNA-seq analysis, which showed that sigB mRNA is as abundant as sigA mRNA during exponential phase of M. smegmatis growth (see Data Set S2 in the supplemental material) and that the σB protein is consistently present in all stages of growth (Fig. 5a). A limited ChIP-Seq data set for ectopically overexpressed FLAG-tagged σA confirmed at least 61 promoter sites that were recognized by both σA and σB, including those that control the vital cellular functions of ribosome biogenesis and transcription. The most plausible explanation for σB occupancy at such crucial sites is that it is engaged in active transcription of these genes, or else its occupancy would interfere with the σA-dependent transcription initiation from these sites. These results imply that E.σA and E.σB together transcribe a subset of housekeeping genes during exponential growth of M. smegmatis. While further data are required to determine the relative occupancy of E.σA and E.σB at a given promoter, we predict that this varies with the promoter site, its association with accessory proteins, as well as with changing growth and environmental conditions. An example of such a scenario has previously been demonstrated in vitro by Hu et al., in which E.σA and E.σB are transcriptionally active at the sigAP but open complex formation on this promoter is seen only with E.σB in the presence of RbpA, suggesting a higher affinity of E.σB at the sigA promoter (13).
It is noteworthy that the transcript levels of sigA as well as σA protein levels are not responsive to the lack of σB in a ΔsigB mutant strain (Fig. 6c and e); sigA is also not RIF inducible in either the wild type or the ΔsigB mutant (Fig. 6d). The absence of σB in a ΔsigB mutant strain must therefore result in a decrease in the concentration of holoenzyme that is available for transcription of a subset of housekeeping genes. We would therefore predict a decrease in transcription of some housekeeping genes in the ΔsigB mutant strain; however, this was not reflected in our RNA-seq experiments. Since the most prominent binding of σB is observed at rRNA gene promoters (rrnP), it is plausible that the deletion of sigB largely impacts transcription from rRNA promoters, changes in which cannot be captured by the RNA-seq approach. Moreover, the rRNA gene promoter has been shown to be particularly susceptible to inhibition by RIF (40). It is therefore tempting to suggest that σB-RNAP plays a crucial role in maintaining the levels of rRNA and that the slow-growth phenotype of ΔsigB in liquid media could be a reflection of the decrease in rRNA levels. Addition of RIF to a ΔsigB strain could potentially reduce the transcription at rrnP further, resulting in growth arrest and the observed sensitivity to RIF. Although we cannot completely rule out the possibility that a σB-dependent gene affects the translation of a RIF effector transcript, we favor an explanation that the addition of RIF targeting a decreased pool of holoenzyme capable of transcribing housekeeping genes contributes to its increased lethality. Restoration of RIF tolerance in the ΔsigB mutant by overexpression of σA further rules out the possibility of a specialized role of σB in RIF tolerance. The RIF-sensitive phenotype of ΔsigB mutants of M. abscessus, M. tuberculosis, and M. smegmatis and the cross-species functional complementarity between σA and σB among these species suggest that the role of σB in the transcription of housekeeping genes during exponential growth is likely to be conserved in all mycobacterial species.
The RNAP-associated protein RbpA has previously been shown to protect against RIF inhibition in S. coelicolor. Although RbpA is incapable of protecting against RIF inhibition at mycobacterial rrnP as well as sigAP, we and others have noted that the overall levels of transcription by E.σA at these promoters are higher in the presence of RbpA; moreover, the transcription efficiency of E.σB at these promoters greatly exceeds that of E.σA but is strictly RbpA dependent (Fig. 4) (40). Consistent with this observation, a CHIP-Seq analysis of RbpA showed that >75% of σB-bound sites are also bound by RbpA (K. Hurst-Hess, R. Biswas, and P. Ghosh, unpublished results). Curiously, expression of RbpA increases ∼2-fold in wild-type bacteria and ∼6-fold in ΔsigB mutant bacteria upon RIF exposure (Fig. 3d). We speculate that in the presence of RIF, the growth of bacteria can be stimulated by RbpA by increasing the transcription efficiency of E.σA and E.σB and the increase in RbpA transcription in ΔsigB reflects an attempt to compensate for the lack of transcription by E.σB.
The behavior of mycobacterial σB is reminiscent of that of rpoS (σ38), a group II sigma factor of E. coli induced during stress and stationary phase with a high degree of similarity to the primary sigma factor of E. coli, σ70. σ70 and σ38 display extensive overlap between their target promoters, and σ38 has been shown to take over several housekeeping duties of σ70 during stationary phase (57–59). Despite the overlap in promoter recognition, E. coli σ70 and σ38 have distinct but complementary roles in vivo, and σ38 transcribes its regulon only under relevant physiological conditions (59, 60). This is achieved in part by tightly regulating the cellular concentration of σ38 at the levels of transcription, translation, and proteolysis such that the σ38 protein is nearly undetectable during exponential growth but increases during entry into stationary phase (58, 61–63). Additionally, the promoter specificity of E.σ38 is modulated to allow transcription of housekeeping genes under appropriate conditions; the precise mechanism by which this is achieved is unclear but may be mediated in part by cis-acting promoter features as well as trans-acting proteins, such as Crl, an activator that stimulates E.σ38 activity at certain promoters, and global regulators like H-NS and IHF (64–68). The association of mycobacterial σB with RbpA has similarly been shown to allow the recognition of σA-specific promoters by σB (13). Although the roles of Crl and RbpA appear to be similar, they have been shown to act via distinct mechanisms: Crl increases the affinity of σ38 to core RNAP, whereas RbpA stimulates open complex formation without stabilizing the holoenzyme. Mycobacterial σB, nevertheless, presents a clear departure from the E. coli paradigm: the cellular levels of σB are not controlled during exponential growth, and σB instead actively participates in the cotranscription of housekeeping genes. Variation in the relative levels of these sigma factors may play a key role in the global regulation of gene expression. We speculate that the presence of σB may offer an advantage in the survival of mycobacteria under conditions where either the function of σA is compromised or the bacteria could benefit from the increased transcription of housekeeping genes. Since the expression of σA itself is noninducible, any demand for increasing the housekeeping gene expression could be achieved by inducing σB expression. Moreover, we speculate that this mechanism of regulation of gene expression could be more widely utilized, especially in the closely related Streptomyces coelicolor and cyanobacteria that encode multiple group II sigma factors (69).
MATERIALS AND METHODS
Media and strains.
Mycobacterium smegmatis was grown at 37°C in Middlebrook 7H9 (Difco) supplemented with 10% albumin-dextrose-catalase (ADC) and 0.05% Tween 20. Mycobacterium abscessus ATCC 19977 was grown at 37°C in Middlebrook 7H9 (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) and 0.05% Tween 20. Mycobacterium tuberculosis mc27000, an attenuated strain of Mycobacterium tuberculosis H37Rv which carries deletions in the RD1 and panCD loci, both of which are critical for the virulence of M. tuberculosis, was grown at 37°C in Middlebrook 7H9 (Difco) supplemented with 10% OADC and 0.05% Tween 20 (70). Antibiotics were added as required to the amounts indicated below. Gene replacement mutants were constructed using recombineering as described previously (41). The recombineering construct was generated by cloning in the multiple-cloning sites flanking the apramycin cassette of pYUB854. Mutant clones were checked using the Fcheck and Rcheck primers flanking the deletion site. The sigB-FLAG-tagged strain was confirmed using sequencing as well as by Western blot analysis with anti-FLAG antibody.
Antibiotic sensitivity assays.
Wild-type and mutant strains of M. smegmatis, M. abscessus, and M. tuberculosis were grown to an A600 of 0.6 to 0.7. Cells were tested for their susceptibility to RIF by spotting a 10-fold serial dilution on Middlebrook 7H10 (Difco) plates containing the concentration of RIF indicated above and below. Antibiotic susceptibility in liquid media was assayed by inoculating the desired strain in a 2-fold dilution series of each antibiotic at an initial A600 of 0.0004. The cultures were incubated at 37°C, and the A600 was measured after 48 h for M. smegmatis.
RNA preparation, qPCR, and RNA-seq analysis.
Wild-type M. smegmatis mc2155 as well as the ΔsigB deletion strain were grown to exponential phase (OD600 = 0.4) in Middlebrook 7H9-ADC, exposed to 4 μg/ml of RIF for various periods of time (0 to 90 min), and evaluated for the lethality of RIF. Total RNA was prepared from wild-type and mutant strains exposed to RIF (4 μg/ml) for 20 min using a Qiagen RNA preparation kit, followed by DNase I treatment. Unexposed samples were used as controls. Approximately 5-μg total RNA samples were treated by the Ribo-Zero rRNA removal procedure (Illumina) to enrich for mRNA. Approximately 500 ng of RNA was used for library preparation using a ScriptSeq (v2) RNA-seq kit and high-throughput sequencing on an Illumina NextSeq platform. The sequence data were analyzed using the reference-based analysis and default parameters in the Rockhopper (v2.03) program, in which the data are normalized by upper quartile normalization and transcript abundance is reported as the number of reads per kilobase per million (RPKM). Differential gene expression was tested for each transcript, and q values, which control the false-discovery rate, were then reported (71, 72). RNA-seq experiments were performed three independent times, using two biological replicates each time.
M. smegmatis wild-type and ΔsigB deletion strains were exposed to RIF (4 μg/ml) for the required times. Total RNA was prepared using a Qiagen RNA preparation kit, followed by DNase I treatment. Primers for quantitative reverse transcription-PCR (qRT-PCR) were generated using Primer Quest software (Integrated DNA Technologies). cDNA was generated using random hexamers and Maxima reverse transcriptase (Fisher Scientific), and qRT-PCR was performed using the Maxima SYBR green qPCR master mix (Fisher Scientific) and the following primer pair for MSMEG_1221: 5′-CCTGTGGTTCGCGGAAA-3′/5′-CCCTGCTCAAGAATCTCACC-3′. An Applied Biosystems 7300 real-time PCR system was used with cycling conditions of 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Chromatin immunoprecipitation sequencing and data analysis.
ChIP-Seq was performed as previously described with minor modifications (73). mc2155 sigB-FLAG was grown at 37°C in Middlebrook 7H9 broth (Becton, Dickinson) supplemented with ADS (albumin [50 g liter−1], dextrose [20 g liter−1], NaCl [8.1 g liter−1]), 0.2% glycerol, and 0.05% Tween 80 to an OD600 of 0.4. This was followed by cross-linking with 1% formaldehyde for 30 min with constant agitation and quenching with 250 mM glycine. The cells were pelleted, washed with Tris-buffered saline buffer, and resuspended in buffer 1 containing a protease inhibitor cocktail (73). Cells were lysed on a Covaris S220 Focused ultrasonicator for 30 min (amplitude = 20%, intensity = 5, number of cycles/burst = 200), immunoprecipitated with anti-FLAG monoclonal antibody M2 (Sigma) for 12 h at 4°C, and further processed as described previously (73). Each CHIP-Seq experiment was performed three independent times using two replicates of culture each time.
Genomic DNA libraries enriched for σB binding were sequenced on the Illumina platform (Wadsworth Center, Sequencing Core Facility). The reads were aligned to the reference genome using the Bowtie2 and SAMtools algorithms (74). Regions of enrichment were identified using a custom Python script as described previously (47). Briefly, for each replicate data set in the pair, an appropriate threshold, T1 or T2, was determined for the plus and minus strands. Values for T1 and T2 were considered to be between 1 and 1,000. For each combination of values for T1 and T2, the number of genome positions with values greater than or equal to the value for T1 in the first replicate and with values greater than or equal to the value for T2 in the second replicate was determined. The false-discovery rate was estimated using the null hypothesis that no regions are enriched. The combination of thresholds yielding the highest number of true-positive positions with an estimated false-discovery rate of less than 0.01 was selected. Once T1 and T2 were chosen, a region was identified as a peak if both replicates showed enrichment above the corresponding thresholds for each strand. For a peak to be called, there must be a peak on the plus strand within a threshold distance of a peak on the minus strand. Peaks obtained with the Peakcaller program were verified using the MACS2 algorithm and viewed with SignalMap (v2.0.05) software (Roche NimbleGen). Relative enrichment is reported as the fold-over-threshold (FAT) score. The enriched regions were analyzed using MEME Suite (v5.0.3) tools and the default parameters (49).
Protein overexpression and purification.
M. tuberculosis σA, σB, and RbpA were cloned in pET21a with a C-terminal His tag, transformed into E. coli BL21(DE3)pLysS, grown to an A600 of 0.4, and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C. The cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 5% glycerol, and the clarified lysate was loaded on an Ni-nitrilotriacetic acid (NTA) column (Qiagen). Nonspecifically bound proteins were removed by washing with lysis buffer containing 40 mM, 35 mM, and 20 mM imidazole, and the protein was eluted with 150 mM imidazole. For purification of M. tuberculosis RNA polymerase, BL21(DE3)pLysS was cotransformed with pETDuet-Mtbββ′ and pRsfDuet-Mtbαω, grown at 30°C to an A600 of 0.4, and induced with 0.4 mM IPTG at 16°C for a period of 18 h. The cells were lysed by sonication and passed through an Ni-NTA column (Qiagen) that had been equilibrated with 50 mM Tris, 300 mM NaCl, and 5% glycerol (lysis buffer). The column was washed with lysis buffer and 40 mM imidazole and eluted with lysis buffer and 150 mM imidazole. Fractions containing RNAP were loaded on a heparin-Sepharose matrix (GE Healthcare) that had been equilibrated with 50 mM Tris, 300 mM NaCl, and 5% glycerol and eluted with a buffer containing 1 M NaCl.
In vitro transcription assays.
Multiple-round in vitro transcription was performed as previously described (40). In short, 200 nM M. tuberculosis RNAP was assembled with 600 nM the desired sigma factor in a volume of 10 μl for 10 min at 37°C. RbpA (600 nM) was added during assembly to the indicated samples, followed by a further incubation for 5 min. sigAP DNA (20 nM) was added to the mixtures, and the mixtures were incubated for 10 min at 37°C. RIF was added to the concentrations indicated below for 30 min at 37°C. Transcription was initiated by addition of 2 μl of a nucleoside triphosphate (NTP) mix (1.5 mM ATP, GTP, and CTP and 0.5 mM UTP) plus 2 μCi of [α-32P]UTP. The reaction mixtures were incubated at 37°C for 30 min, and the reactions were terminated by the addition of 5 mM EDTA plus 100 μg/ml tRNA. Samples were ethanol precipitated, resuspended in stop buffer (80% [vol/vol] formamide, 10 mM EDTA, 0.01% xylene cyanol, 0.01% bromophenol blue), and separated using denaturing PAGE (18% urea polyacrylamide gel). The products were visualized using a Typhoon imager (GE Healthcare) and quantitated using ImageQuant software.
Western blot analysis.
M. smegmatis was grown in Middlebrook 7H9 supplemented with ADS and Tween 20. Aliquots were removed at different stages of growth (A600 = 0.2, 0.4, 0.7, 1.0, 2.0, 2.8, and 4.0), pelleted, and washed with phosphate-buffered saline (PBS) buffer. Pellets were normalized by weight, resuspended in the required volumes of PBS, and lysed by sonication. The lysate was clarified by centrifugation, the protein concentration was determined at the A260, and equal quantities of protein from different growth stages were separated using 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with anti-σ70 monoclonal antibody 2G10. Purified σA and σB were used as controls.
ACKNOWLEDGMENTS
We thank The Wadsworth Center’s Applied Genomics Technology Core for sequencing of RNA-seq libraries, the Media Core for the preparation of media and buffers, and the Wadsworth Bioinformatics Core. pETDuet-Mtbββ′ was a kind gift from Jayant Mukhopadhyay, Bose Institute, Kolkata, India. We thank Joe Wade and Anil Ojha for discussions and critical reading of the manuscript.
P.G. is supported by a Cystic Fibrosis Foundation grant and the Wadsworth Center.
Footnotes
Citation Hurst-Hess K, Biswas R, Yang Y, Rudra P, Lasek-Nesselquist E, Ghosh P. 2019. Mycobacterial SigA and SigB cotranscribe essential housekeeping genes during exponential growth. mBio 10:e00273-19. https://doi.org/10.1128/mBio.00273-19.
REFERENCES
- 1.Gruber TM, Gross CA. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 2.Saecker RM, Record MT Jr, Dehaseth PL. 2011. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biol 4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 5.Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I, Proveddi R. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol 186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez M, Doukhan L, Nair G, Smith I. 1998. sigA is an essential gene in Mycobacterium smegmatis. Mol Microbiol 29:617–628. doi: 10.1046/j.1365-2958.1998.00960.x. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Coates AR. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachdeva P, Misra R, Tyagi AK, Singh Y. 2010. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J 277:605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev 30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 10.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol 31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 11.Predich M, Doukhan L, Nair G, Smith I. 1995. Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol Microbiol 15:355–366. doi: 10.1111/j.1365-2958.1995.tb02249.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Karakousis PC, Bishai WR. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol 190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Morichaud Z, Perumal AS, Roquet-Baneres F, Brodolin K. 2014. Mycobacterium RbpA cooperates with the stress-response sigmaB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res 42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. 2009. The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doukhan L, Predich M, Nair G, Dussurget O, Mandic-Mulec I, Cole ST, Smith DR, Smith I. 1995. Genomic organization of the mycobacterial sigma gene cluster. Gene 165:67–70. doi: 10.1016/0378-1119(95)00427-8. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Coates AR. 1999. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J Bacteriol 181:1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee R, Chatterji D. 2005. Evaluation of the role of sigma B in Mycobacterium smegmatis. Biochem Biophys Res Commun 338:964–972. doi: 10.1016/j.bbrc.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 18.Yang SS, Hu YB, Wang XD, Gao YR, Li K, Zhang XE, Chen SY, Zhang TY, Gu J, Deng JY. 2017. Deletion of sigB causes increased sensitivity to para-aminosalicylic acid and sulfamethoxazole in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e00551-17. doi: 10.1128/AAC.00551-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pisu D, Provvedi R, Mata Espinosa D, Barrios Payan J, Boldrin F, Palu G, Hernandez-Pando R, Manganelli R. 2017. The alternative sigma factors SigE and SigB are involved in tolerance and persistence to antitubercular drugs. Antimicrob Agents Chemother 61:e01596-17. doi: 10.1128/AAC.01596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minch KJ, Rustad TR, Peterson EJ, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. 2015. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect Immun 74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 24.Raman S, Song T, Puyang X, Bardarov S, Jacobs WR Jr, Husson RN. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chauhan R, Ravi J, Datta P, Chen T, Schnappinger D, Bassler KE, Balazsi G, Gennaro ML. 2016. Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis. Nat Commun 7:11062. doi: 10.1038/ncomms11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 27.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 28.Lisitsyn NA, Gur'ev SO, Sverdlov ED, Moiseeva EP, Nikiforov VG. 1984. Nucleotide substitutions in the rpoB gene leading to rifampicin resistance of E. coli RNA polymerase. Bioorg Khim 10:127–128. (In Russian.) [PubMed] [Google Scholar]
- 29.Lisitsyn NA, Sverdlov ED, Moiseyeva EP, Danilevskaya ON, Nikiforov VG. 1984. Mutation to rifampicin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol Gen Genet 196:173–174. doi: 10.1007/BF00334112. [DOI] [PubMed] [Google Scholar]
- 30.Ovchinnikov YA, Monastyrskaya GS, Guriev SO, Kalinina NF, Sverdlov ED, Gragerov AI, Bass IA, Kiver IF, Moiseyeva EP, Igumnov VN, Mindlin SZ, Nikiforov VG, Khesin RB. 1983. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol Gen Genet 190:344–348. doi: 10.1007/BF00330662. [DOI] [PubMed] [Google Scholar]
- 31.Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 32.Severinov K, Soushko M, Goldfarb A, Nikiforov V. 1994. RifR mutations in the beginning of the Escherichia coli rpoB gene. Mol Gen Genet 244:120–126. [DOI] [PubMed] [Google Scholar]
- 33.Quan S, Imai T, Mikami Y, Yazawa K, Dabbs ER, Morisaki N, Iwasaki S, Hashimoto Y, Furihata K. 1999. ADP-ribosylation as an intermediate step in inactivation of rifampin by a mycobacterial gene. Antimicrob Agents Chemother 43:181–184. doi: 10.1128/AAC.43.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quan S, Venter H, Dabbs ER. 1997. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother 41:2456–2460. doi: 10.1128/AAC.41.11.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rominski A, Roditscheff A, Selchow P, Bottger EC, Sander P. 2017. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother 72:376–384. doi: 10.1093/jac/dkw466. [DOI] [PubMed] [Google Scholar]
- 36.Reference deleted. [Google Scholar]
- 37.Dey A, Verma AK, Chatterji D. 2010. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology 156:873–883. doi: 10.1099/mic.0.033670-0. [DOI] [PubMed] [Google Scholar]
- 38.Newell KV, Thomas DP, Brekasis D, Paget MS. 2006. The RNA polymerase-binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Mol Microbiol 60:687–696. doi: 10.1111/j.1365-2958.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- 39.Hubin EA, Tabib-Salazar A, Humphrey LJ, Flack JE, Olinares PD, Darst SA, Campbell EA, Paget MS. 2015. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc Natl Acad Sci U S A 112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y, Morichaud Z, Chen S, Leonetti JP, Brodolin K. 2012. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the sigma A-containing RNA polymerase holoenzyme. Nucleic Acids Res 40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 42.Alexander DC, Jones JR, Liu J. 2003. A rifampin-hypersensitive mutant reveals differences between strains of Mycobacterium smegmatis and presence of a novel transposon, IS1623. Antimicrob Agents Chemother 47:3208–3213. doi: 10.1128/AAC.47.10.3208-3213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wegrzyn A, Szalewska-Pałasz A, Błaszczak A, Liberek K, Wegrzyn G. 1998. Differential inhibition of transcription from sigma70- and sigma32-dependent promoters by rifampicin. FEBS Lett 440:172–174. doi: 10.1016/S0014-5793(98)01449-5. [DOI] [PubMed] [Google Scholar]
- 44.Tabib-Salazar A, Liu B, Doughty P, Lewis RA, Ghosh S, Parsy M-L, Simpson PJ, O’Dwyer K, Matthews SJ, Paget MS. 2013. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res 41:5679–5691. doi: 10.1093/nar/gkt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bortoluzzi A, Muskett FW, Waters LC, Addis PW, Rieck B, Munder T, Schleier S, Forti F, Ghisotti D, Carr MD, O'Hare HM. 2013. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J Biol Chem 288:14438–14450. doi: 10.1074/jbc.M113.459883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breyer MJ, Thompson NE, Burgess RR. 1997. Identification of the epitope for a highly cross-reactive monoclonal antibody on the major sigma factor of bacterial RNA polymerase. J Bacteriol 179:1404–1408. doi: 10.1128/jb.179.4.1404-1408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Mei H, Chen F, Tang Q, Yu Z, Cao X, Andongma BT, Chou SH, He J. 2017. Transcriptome landscape of Mycobacterium smegmatis. Front Microbiol 8:2505. doi: 10.3389/fmicb.2017.02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 50.Agarwal N, Tyagi AK. 2006. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res 34:4245–4257. doi: 10.1093/nar/gkl521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez M, Smith I. 2000. Determinants of mycobacterial gene expression, p 111–129. In Hatfull GF, Jacobs WR Jr (ed), Molecular genetics of mycobacteria. ASM Press, Washington, DC. [Google Scholar]
- 52.Kenney TJ, Churchward G. 1996. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol 178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bashyam MD, Kaushal D, Dasgupta SK, Tyagi AK. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol 178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-y-Merchand JA, Colston MJ, Cox RA. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 55.Chiner-Oms Á, González-Candelas F, Comas I. 2018. Gene expression models based on a reference laboratory strain are poor predictors of Mycobacterium tuberculosis complex transcriptional diversity. Sci Rep 8:3813. doi: 10.1038/s41598-018-22237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landick R, Krek A, Glickman MS, Socci ND, Stallings CL. 2014. Genome-wide mapping of the distribution of CarD, RNAP sigma(A), and RNAP beta on the Mycobacterium smegmatis chromosome using chromatin immunoprecipitation sequencing. Genom Data 2:110–113. doi: 10.1016/j.gdata.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ. 2005. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol Cell 20:357–366. doi: 10.1016/j.molcel.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A 90:8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. 2001. Promoter recognition and discrimination by EsigmaS RNA polymerase. Mol Microbiol 42:939–954. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A 90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hengge R. 2009. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol 160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 63.Lange R, Hengge-Aronis R. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev 8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 64.Typas A, Becker G, Hengge R. 2007. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol Microbiol 63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- 65.Arnqvist A, Olsen A, Normark S. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol 13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 66.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol 7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 67.Arnqvist A, Olsen A, Pfeifer J, Russell DG, Normark S. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol 6:2443–2452. [DOI] [PubMed] [Google Scholar]
- 68.Colland F, Barth M, Hengge-Aronis R, Kolb A. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and lrp transcription factors. EMBO J 19:3028–3037. doi: 10.1093/emboj/19.12.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buttner MJ, Lewis CG. 1992. Construction and characterization of Streptomyces coelicolor A3(2) mutants that are multiply deficient in the nonessential hrd-encoded RNA polymerase sigma factors. J Bacteriol 174:5165–5167. doi: 10.1128/jb.174.15.5165-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tjaden B. 2015. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16:1. doi: 10.1186/s13059-014-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaini S, Lyubetskaya A, Gomes A, Peterson M, Tae Park S, Raman S, Schoolnik G, Galagan J. 2014. Transcription factor binding site mapping using ChIP-Seq. Microbiol Spectr 2:MGM2-0035-2013. doi: 10.1128/microbiolspec.MGM2-0035-2013. [DOI] [PubMed] [Google Scholar]
- 74.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of M. smegmatis in media lacking RIF. (a) Tenfold serial dilutions of M. smegmatis mc2155, the ΔsigB mutant, and complemented strain were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 ADC or OADC lacking RIF. (b) The growth of M. smegmatis mc2155 and the ΔsigB mutant in Middlebrook 7H9-ADC-Tween 20 was monitored over a period of 40 h. Deletion of sigB results in slow growth in liquid media but no apparent growth change on 7H10 agar. (c) Tenfold serial dilutions of M. abscessus ATCC 19977, M. tuberculosis mc2700, and their corresponding ΔsigB mutants and complemented strains were grown to an A600 of 0.7 and spotted on Middlebrook 7H10 OADC medium lacking RIF. Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principal-component analysis (PCA) plot generated from RNA-seq data for M. smegmatis mc2155 and the ΔsigB mutant, unexposed controls, and strains exposed to RIF (4 μg/ml for 20 min) Replicate samples are indicated by the same color. The samples are plotted with respect to their first two principal components, which represent the x and y axes, respectively, and account for 87% and 7% of the variance, respectively. The PCA plot was generated with the DESeq2 package in R. Download FIG S2, PDF file, 0.5 MB (481.1KB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Isogenic deletions in MSMEG_2252, MSMEG_2539, and MSMEG_2174 were created by recombineering, and the strains were assayed for RIF sensitivity. The strain with the MSMEG_2174 deletion displayed increased RIF sensitivity compared to WT bacteria; the RIF sensitivities of the strain with MSMEG_2252 and MSMEG_2539 deletions were unchanged. Download FIG S3, PDF file, 1.6 MB (1.7MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptome of the M. smegmatis wild type and the ΔsigB mutant exposed to 4 μg/ml RIF for 20 min: SD-1 RNA-seq of RIF-induced WT versus SigB.xlsx. Download Data Set S1, XLSX file, 0.02 MB (24.8KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of genes downregulated >3-fold in an mc2155 ΔsigB strain compared to their expression in wild-type bacteria, determined using RNA-seq. Complete data are included in Data Set S2 in the supplemental material. Download Table S1, DOCX file, 0.04 MB (44.4KB, docx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sigB regulons of logarithmically grown M. smegmatis: SD-2 RNA-seq of WT versus SigB.xlsx. Download Data Set S2, XLSX file, 0.02 MB (19.9KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) Sequence conservation of mycobacterial σA and σB and E. coli σ70. The conserved epitope recognized by the 2G10 antibody is shown. (b) Coomassie-stained gel serving as a loading control for the assay whose results are presented in Fig. 5a. Equivalent amounts of protein were loaded in each lane. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) Genomewide binding profile of σB visualized with SignalMap software. (b) Histogram showing the frequency distribution of distances between SigB ChIP-Seq peak centers and TSSs. Download FIG S5, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ChIP-Seq of σB: SD-3 ChIP-Seq binding sites.xlsx. Download Data Set S3, XLSX file, 0.1 MB (70.2KB, xlsx) .
Copyright © 2019 Hurst-Hess et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.