The genital tract pathogen Chlamydia trachomatis is frequently detected in the gastrointestinal tract, but the host immunity that regulates chlamydial colonization in the gut remains unclear. In a Chlamydia muridarum-C57 mouse model, chlamydial organisms are cleared from the genital tract in ∼4 weeks, but the genital organisms can spread to the gastrointestinal tract.
KEYWORDS: Chlamydia, gamma interferon, Th1 immunity, genital tract immunity, small intestine immunity
ABSTRACT
The genital tract pathogen Chlamydia trachomatis is frequently detected in the gastrointestinal tract, but the host immunity that regulates chlamydial colonization in the gut remains unclear. In a Chlamydia muridarum-C57 mouse model, chlamydial organisms are cleared from the genital tract in ∼4 weeks, but the genital organisms can spread to the gastrointestinal tract. We found that the gastrointestinal chlamydial organisms were cleared from the small intestine by day 28, paralleling their infection course in the genital tract, but persisted in the large intestine for long periods. Mice deficient in α/β T cells or CD4+ T cells but not CD8+ T cells showed chlamydial persistence in the small intestine, indicating a critical role for CD4+ T cells in clearing Chlamydia from the small intestine. The CD4+ T cell-dependent clearance is likely mediated by gamma interferon (IFN-γ), since mice deficient in IFN-γ but not interleukin 22 (IL-22) signaling pathways rescued chlamydial colonization in the small intestine. Furthermore, exogenous IFN-γ was sufficient for clearing Chlamydia from the small intestine but not the large intestine. Mice deficient in developing Chlamydia-specific Th1 immunity showed chlamydial persistence in the small intestine. Finally, IFN-γ-producing CD4+ but not CD8+ T cells from immunized donor mice were sufficient for eliminating Chlamydia from the small intestine but not the large intestine of recipient mice. Thus, we have demonstrated a critical role for Th1 immunity in clearing Chlamydia from the small intestine but not the large intestine, indicating that chlamydial colonization in different regions of the gastrointestinal tract is regulated by distinct immune mechanisms.
INTRODUCTION
Chlamydia trachomatis is a major sexually transmitted bacterial pathogen which may cause pathologies in the upper genital tract (1–4). The Chlamydia muridarum mouse model has been used for investigating C. trachomatis pathogenesis due to its ability to induce long-lasting tubal fibrosis/hydrosalpinx in mice following an intravaginal inoculation (5–10). Using this mouse model, it has been demonstrated that a Th1-dominant immunity is required for controlling chlamydial infection in the genital tract (11). In most cases, wild-type C57 mice significantly reduce chlamydial organism burden in the genital tract on day 21 and clear chlamydial infection by day 28 following an intravaginal inoculation. Mice deficient in CD4+ T cells or major histocompatibility complex (MHC) class II antigen presentation showed significantly prolonged C. muridarum infection in the genital tract, while mice deficient in CD8+ T cells or MHC class I antigen presentation cleared C. muridarum from the genital tract as robustly as wild-type mice (6). Consistently, mice deficient in interleukin 12 (IL-12) (12) or gamma interferon (IFN-γ) (13), which are compromised in developing Th1 adaptive immunity, showed chlamydial persistence and spreading.
C. trachomatis is also frequently detected in the human gastrointestinal (GI) tract (14–17). Although its medical significance remains unclear, it has been proposed that GI tract C. trachomatis may serve as a reservoir for promoting C. trachomatis pathogenicity in the genital tract (18, 19). C. muridarum has been shown to readily spread to the GI tract via the blood circulatory system (20), since mice wearing Elizabethan collars and singly housed in netted cages to prevent coprophagy still showed spreading of genital C. muridarum to the GI tract. Furthermore, hematogenous C. muridarum can establish long-lasting colonization in the mouse GI tract (21). The spreading of genital Chlamydia to the GI tract has been proposed to induce a 2nd hit to promote chlamydial pathogenicity in the upper genital tract (22). Orally delivered C. muridarum is known to colonize the mouse GI tract for long periods (18, 23–26). Although it is unclear whether genital C. trachomatis can also spread to the human GI tract, human sexual behavior should be sufficient for orally delivering C. trachomatis into the GI tract. Regardless of how C. trachomatis is delivered to the GI tract, it is likely that C. trachomatis is able to productively colonize the human GI tract, since live C. trachomatis organisms have been repeatedly detected on human rectal swabs (15, 16, 27–31). It will be interesting to reveal how C. trachomatis tumbles through the human GI tract. Investigating how mouse GI tract immune mechanisms regulate C. muridarum colonization may provide useful information for guiding clinical studies.
On the other hand, if a naive mouse is first exposed to C. muridarum in the GI tract, the GI Chlamydia can act as an oral vaccine to induce protective immunity against subsequent infection with Chlamydia in the genital tract (32) or airway (33). It will be interesting to evaluate whether women whose rectal swabs are positive for C. trachomatis experience reduced rates of repeated infections in their genital tracts. Thus, revealing the host immune mechanisms responsible for regulating chlamydial colonization in the GI tract may also provide useful information for developing an oral Chlamydia vaccine.
In the current study, we have used a C. muridarum mouse model to investigate mechanisms of C. muridarum colonization in the GI tract. In C57 mice, chlamydial organisms were cleared from the small intestine by day 28 after infection but persisted in the large intestine for long periods. Mice deficient in T cell receptor alpha (TCRα) or CD4+ T cells but not CD8+ T cells showed chlamydial persistence in the small intestine, indicating a critical role for CD4+ T cells in clearing Chlamydia. CD4+ T cell-dependent clearance is likely mediated by IFN-γ, since mice deficient in IFN-γ but not IL-22 signaling showed rescue of chlamydial colonization in the small intestine. Furthermore, supplementing TCRα-deficient mice with exogenous IFN-γ or IFN-γ-producing CD4+ but not CD8+ T cells from immunized donor mice was sufficient for eliminating Chlamydia from the small intestine but not the large intestine. Thus, we have demonstrated that Th1 immunity is both necessary and sufficient for clearing Chlamydia from the small intestine. However, the same Th1 immunity had no impact on chlamydial colonization in the large intestine, suggesting that chlamydial colonization in different regions of the mouse gastrointestinal tract may be regulated by different immune mechanisms. The mechanistic information not only will promote our understanding of how orally delivered C. trachomatis interacts with human gastrointestinal tract but also may be useful in developing an oral chlamydial vaccine.
RESULTS
Chlamydia is cleared from the small intestine but persists in the large intestine for a long period.
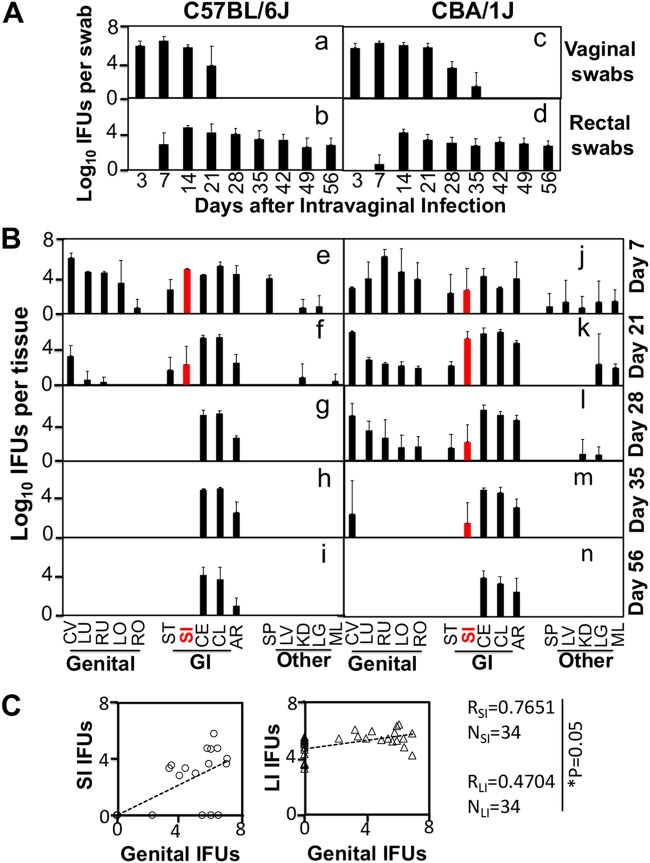
We have previously shown that following an intravaginal inoculation, C. muridarum can spread to and colonize in the GI tract for long periods (20), although the genital C. muridarum is cleared within 4 to 5 weeks in most mouse strains (10). We compared C. muridarum organism distributions in tissues of both C57BL/6J and CBA/1J mice following intravaginal inoculation (Fig. 1). In C57BL/6J mice, C. muridarum live organism burdens in both the genital tract and small intestine were significantly reduced on day 21 and completely cleared by day 28. In CBA/1J mice, C. muridarum burdens in both the genital tract and small intestine were significantly reduced on day 28 and completely cleared by day 56. However, the C. muridarum live organism burdens in the large intestine of both mouse strains remained steady throughout the experiment. These observations suggest that different segments of the GI tract may have evolved distinct mechanisms for controlling chlamydial colonization. The small intestine may use a mechanism similar to that used by the genital tract for controlling chlamydial colonization, while the large intestine is unable to restrict chlamydial colonization at all. Indeed, inclusion-forming units (IFU) recovered from genital tissues correlated significantly better with those from small intestine tissues than large intestine tissues (Fig. 1C). It will be interesting to determine the immune mechanism(s) responsible for the differential clearance of Chlamydia from the GI tract.
FIG 1.
Chlamydia muridarum colonization in the small intestine but not large intestine parallels its infection course in the genital tract. (A) C57BL/6J (n = 5) or CBA/1J (n = 6) mice were intravaginally infected with C. muridarum at 2 × 105 inclusion-forming units (IFU) per mouse. At various time points postinfection as indicated on the x axis, both vaginal (a and c) and rectal (b and d) swab specimens were taken for titration of live organisms. The recovered live organisms were expressed as log10 IFU per swab, as displayed on the y axis. Note that vaginal swabs were cleared of IFU on days 28 for C57 and 42 for CBA mice, but rectal swab IFU were present throughout the experiments in both strains. (B) Parallel groups of mice were sacrificed for monitoring live organisms in various tissues on days 7 (e and j), 21 (f and k), 28 (g and l), 35 (h and m), and 56 (i and n) after infection as indicated on the right. The recovered live chlamydial organisms were expressed as log10 IFU per tissue. The assayed organs/tissues include genital tract tissues, i.e., cervix-vagina (CV), left uterine horn (LU), right uterine horn (RU), left oviduct/ovary (LO), and right oviduct/ovary (RO), gastrointestinal (GI) tract tissues, i.e., stomach (ST), small intestine (SI), cecum (CE), colon (CL), and anorectum (AR), and other tissues, such as spleen (SP), liver (LV), kidney (KD), lung (LG), and mesenteric lymph node (ML). n = 3 to 5 for each time point, and the data came from three independent experiments. Note that IFU from SI (highlighted in red) parallel IFU from the genital tract in both strains of mice. (C) Total IFU from all genital tissues of a given mouse harvested at a given time point as shown in panel B (genital IFU on the x axis) were correlated with IFU from small intestine tissues (SI IFU on the y axis, left panel) and large intestine tissues (LI IFU on the y axis, right panel) using Spearman’s correlation analysis. Note that chlamydial IFU burdens in the genital tract correlate with IFU recovered from the small intestine (RSI = 0.7651) significantly better than those from the large intestine (RLI = 0.4704). NSI or NLI, number of small intestine or large intestine tissue specimens, respectively.
CD4+ but not CD8+ T cells are responsible for clearing Chlamydia from the small intestine.
When mice deficient in T cell receptor alpha gene (TCRα knockout [KO]) were intragastrically inoculated with C. muridarum, the chlamydial organisms remained in the small intestine throughout the experiment (Fig. 2), suggesting that TCRα/β-bearing T lymphocytes are essential for clearing the small intestine of Chlamydia. To further define the subsets of T cells required for clearance, we compared mice with and without deficiency in CD4+ or CD8+ T cells for the ability to clear chlamydial colonization in the small intestine on day 28 after intragastric inoculation (Fig. 3). Mice deficient in CD4+ T cells showed a robust colonization by Chlamydia in all tissue segments of the small intestine, while both wild-type and CD8+ T cell-deficient mice completely cleared Chlamydia from these tissue segments. Furthermore, depletion with an anti-CD4 antibody made the CD8 KO mice susceptible to chlamydial colonization in the small intestine. These observations together demonstrated that CD4+ T cells but not CD8+ T cells are essential for clearing chlamydial colonization from the small intestine. However, regardless of the gene deficiency and/or antibody depletion, chlamydial colonization in the large intestine of the same mice remained steady, suggesting that the large intestine chlamydial organisms are regulated by different immune mechanisms.
FIG 2.
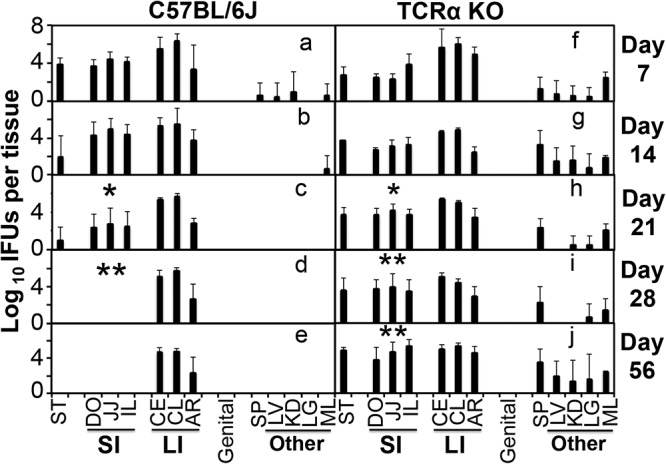
Chlamydia muridarum colonization in the gastrointestinal tract of mice deficient in α/β T cells. Mice without (a to e) or with deficiency in the T cell receptor alpha gene (TCRα KO [f to j]) were intragastrically inoculated with Chlamydia muridarum at 2 × 105 IFU per mouse. On days 7, 14, 21, 28, and 56 after infection, as indicated on the right side of each panel, groups of 3 to 5 mice were sacrificed for monitoring live chlamydial organisms in various tissues, including stomach, segments of small intestine such as duodenum (DO), jejunum (JJ), and ileum (IL), large intestine tissues such as cecum, colon, and anorectum, genital tract tissue, and other tissues, such as spleen, liver, kidney, lung, and mesenteric lymph node, as indicated on the x axis. The number of live organisms recovered from each tissue sample was expressed as log10 IFU. Note that C57BL/6J mice showed significantly reduced IFU in small intestine by day 21 and cleared IFU by day 28, while TCRα KO mice maintained significant levels of small intestine IFU throughout the experiment. The large intestine IFU remained steadily high in both strains. *, P < 0.05; **, P < 0.01 (Wilcoxon, panel c versus panel b, panel d versus panel b, panel h versus panel c, panel i versus panel d, and panel j versus panel e). Data were acquired from 3 independent experiments.
FIG 3.
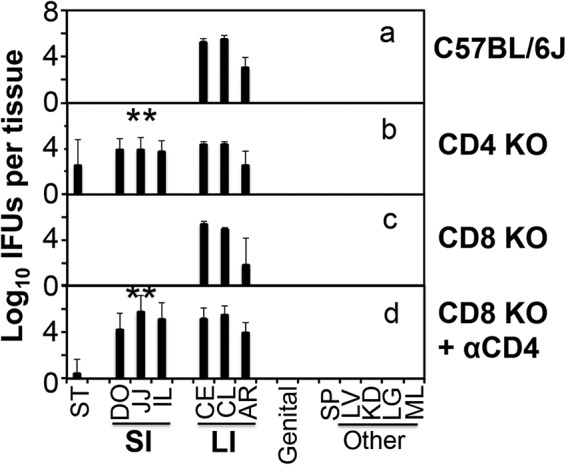
Chlamydia muridarum colonization in the gastrointestinal tract of mice deficient in CD4+ or CD8+ T cells. Mice without (a; n = 4) or with deficiency in CD4+ T cells (CD4 KO) (b; n = 3) or CD8+ T cells (CD8 KO) (c and d; n = 4 for each panel) were intragastrically inoculated with Chlamydia muridarum at 2 × 105 IFU per mouse. Some CD8+ KO mice were also treated with an anti-CD4 antibody (d). On day 28 after infection, all mice were sacrificed for monitoring live chlamydial organisms in various tissues as described in the Fig. 2 legend and shown on the x axis. The number of live organisms recovered from each tissue sample was expressed as log10 IFU. Note that mice deficient in CD4+ but not CD8+ T cells were defective in clearing chlamydial colonization in the small intestine and that depletion with an anti-CD4 antibody made the CD8 KO mice susceptible to chlamydial colonization in the small intestine. **, P < 0.01 (Wilcoxon, panel b versus panel a and panel d versus panel a or c). Data were acquired from 3 independent experiments.
IFN-γ-mediated immunity is sufficient for clearing Chlamydia from the small intestine but not the large intestine.
We next compared mice with and without deficiencies in IFN-γ receptor (IFN-γR) or IL-22 for the ability to clear Chlamydia from GI tissues (Fig. 4). The wild-type C57 mice completely cleared chlamydial organisms from the small intestine on day 28 after an intragastrical inoculation with C. muridarum. However, mice deficient in IFN-γR were defective in clearing chlamydial organisms from the small intestine and elsewhere, demonstrating that chlamydial clearance is dependent on the IFN-γ signaling pathway. However, mice deficient in IL-22 cleared Chlamydia from the small intestine as efficiently as the wild-type mice, indicating that the IL-22 signaling pathway is not required. Furthermore, when exogenous IFN-γ was provided to TCRα-deficient mice, chlamydial organisms were cleared from the small intestine but not large intestine (Fig. 5), demonstrating that IFN-γ is sufficient for eliminating Chlamydia from the small but not large intestine.
FIG 4.
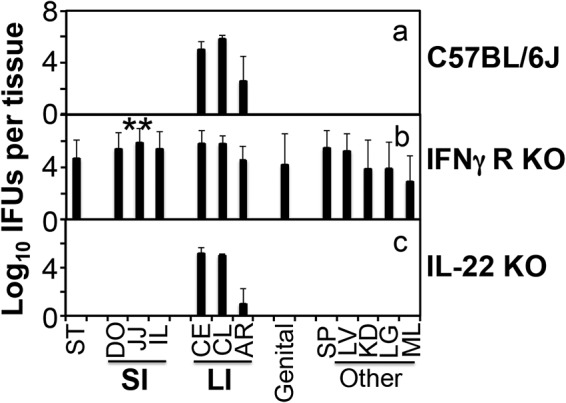
Chlamydia muridarum colonization in the gastrointestinal tract of mice deficient in IFN-γ receptor or IL-22. Mice without (a; n = 4) or with deficiency in IFN-γ receptor (b; n = 8) or IL-22 (c; n = 4) were intragastrically inoculated with C. muridarum at 2 × 105 IFU per mouse. On day 28 after infection, all mice were sacrificed for monitoring live chlamydial organisms in various tissues as described in the Fig. 2 legend and shown on the x axis. The number of live organisms recovered from each tissue sample was expressed as log10 IFU. Note that mice deficient in IFN-γR but not IL-22 were defective in clearing Chlamydia. **, P < 0.01 (Wilcoxon, panel b versus panel a or c). Data were acquired from 3 independent experiments.
FIG 5.
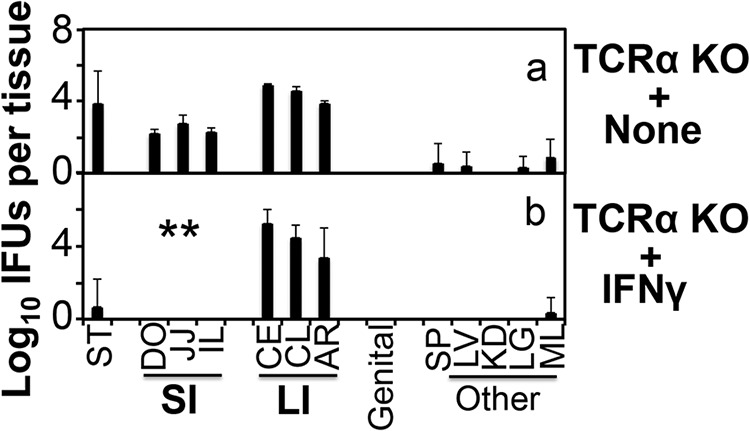
Inhibition of Chlamydia muridarum colonization in the small intestine of TCRα KO mice by exogenous IFN-γ. TCRα KO mice not receiving (panel a, n = 4) or receiving injection of exogenous IFN-γ (b; n = 5) were intragastrically inoculated with C. muridarum at 2 × 105 IFU per mouse. On day 28 after infection, all mice were sacrificed for monitoring live chlamydial organisms as described in the Fig. 2 legend and shown on the x axis. The number of live organisms recovered from each tissue sample was expressed as log10 IFU. Note that exogenous IFN-γ cleared IFU from the small intestine but not large intestine of TCRα KO mice. **, P < 0.01 (Wilcoxon, panel b versus panel a). Data were acquired from 2 independent experiments.
Antigen-specific Th1 immunity is sufficient for clearing Chlamydia from the small intestine but not the large intestine.
The observations that CD4+ T cells and IFN-γ were both essential for clearing Chlamydia from mouse small intestine suggest a critical role for a Th1-dominant adaptive immunity in the clearance. We next compared mice with and without deficiency in IL-12p35 for the ability to clear Chlamydia (Fig. 6). Mice deficient in IL-12p35 are significantly defective in developing Th1 immunity against C. muridarum infection in the genital tract (12). We indeed found that IL-12p35-deficient mice were susceptible to chlamydial colonization in the small intestine on day 28 after infection, when Chlamydia was completely cleared from the small intestine of wild-type C57 mice. Furthermore, OT2 mice (with TCR on all CD4+ T cells engineered to recognizing an ovalbumin epitope only and unable to develop chlamydial-antigen-specific CD4+ T cells) also failed to clear Chlamydia in the small intestine, indicating that chlamydial-antigen-specific CD4+ T cells are required for clearance.
FIG 6.
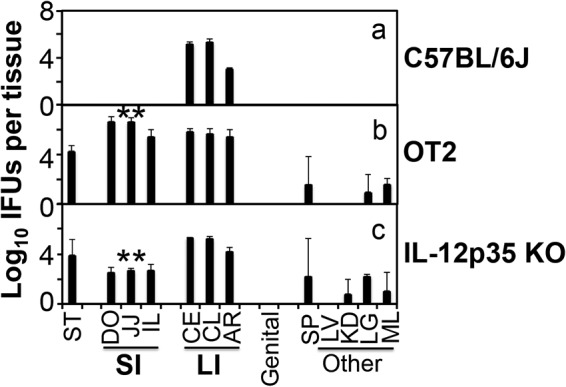
Chlamydia muridarum colonization in the small intestine of mice deficient in developing Th1-dominant adaptive immunity. Mice without (a; n = 3) or with TCR on all CD4+ T cells engineered to recognize an ovalbumin epitope only (OT2) (b; n = 5) or deficient in IL-12p35 subunit gene (c; n = 5) were intragastrically inoculated with C. muridarum at 2 × 105 IFU per mouse. On day 28 after infection, all mice were sacrificed for monitoring live chlamydial organisms as described in the Fig. 2 legend and shown on the x axis. The number of live organisms recovered from each tissue sample was expressed as log10 IFU. Note that both the transgenic OT2 and IL-12p35 KO mice were susceptible to chlamydial colonization in the small intestine. **, P < 0.01 (Wilcoxon, panel b or c versus panel a). Data were acquired from 2 independent experiments.
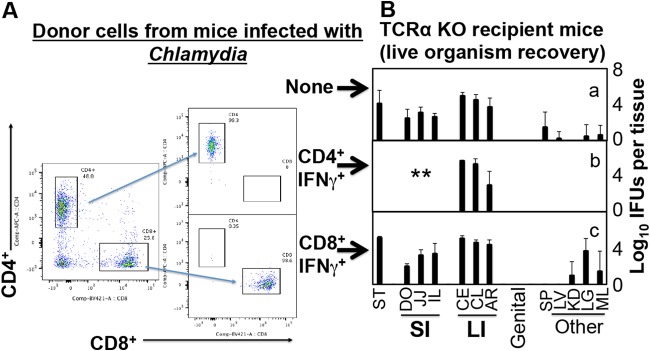
Finally, we used an adoptive transfer approach for testing whether Chlamydia-induced IFN-γ-producing CD4+ T cells are sufficient for controlling chlamydial infection in the small intestine (Fig. 7). To make donor cells, CD4+ or CD8+ T cells that express IFN-γ (designated CD4+ IFN-γ+ or CD8+ IFN-γ+) were sorted from IFN-γ reporter mice (that express enhanced yellow fluorescent protein [eYFP] under the control of the endogenous IFN-γ promoter as a bicistronic IFN-γ–IRES–eYFP mRNA) after the reporter mice were infected with C. muridarum for 10 days. The TCRα-deficient mice were used as recipient mice. The recipient mice that received CD4+ IFN-γ+ donor cells cleared Chlamydia from the small intestine, while those receiving the same amount of CD8+ IFN-γ+ donor cells failed to do so. The above observations together demonstrate that Chlamydia-specific IFN-γ-producing CD4+ (or Th1) but not CD8+ T cells are sufficient for clearing chlamydial colonization in the small intestine. Interestingly, the same Th1 cells did not affect chlamydial colonization in the large intestine, suggesting that the Chlamydia-specific Th1 cells were not able to gain access to, or were suppressed from, Chlamydia-infected cells in the large intestine.
FIG 7.
Adoptive transfer of IFN-γ-producing T cells to enhance resistance to Chlamydia muridarum colonization in the small intestine. Green fluorescent protein-positive (GFP+) CD4+ (CD4+ IFN-γ+) or GFP+ CD8+ (CD8+ IFN-γ+) T cells were purified from donor mice that express GFP under the control of the IFN-γ promoter on day 10 after the donor mice were intravaginally infected with C. muridarum. After flow cytometry sorting, the donor cells reached purity of >98% and were transferred into TCRα KO recipient mice that were intragastrically infected with C. muridarum. On day 28 after intragastric infection, recipient mice were sacrificed for monitoring live organisms as described in the Fig. 2 legend and shown on the x axis. Note that recipient TCRα KO mice receiving CD4+ IFN-γ+ but not CD8+ IFN-γ+ T cells cleared Chlamydia from the small intestine. **, P < 0.01 (Wilcoxon, panel b or c versus panel a). Data were from 2 independent experiments with 4 or 5 mice in each group.
DISCUSSION
Chlamydia has been frequently detected in the GI tracts of both humans (14–17) and animals (20, 23–25). C. trachomatis in the GI tract has been proposed to serve as a reservoir for promoting chlamydial pathogenicity in women’s genital tracts (18, 19), while C. muridarum has been shown to readily spread to the GI tract (20), which may indirectly promote chlamydial pathogenicity in mouse genital tracts (22). However, when a naive mouse is first exposed to C. muridarum in the GI tract, the GI tract C. muridarum may act as an oral vaccine to induce transmucosal immunity (32, 33). Thus, it is important to understand how GI Chlamydia is regulated by host immune mechanisms.
Although live C. muridarum organisms have been shown to last for long periods on rectal swabs, the organisms are cleared from the small intestine as fast as they are eliminated from the genital tract. In the current study, we have demonstrated for the first time that similar to the C. muridarum organisms in the mouse genital tract (11), the organisms in the mouse small intestine are eliminated by Th1-dominant immunity. First, mice deficient in TCRα or CD4+ T cells showed chlamydial persistence in the small intestine. Second, mice deficient in CD8+ T cells cleared Chlamydia from the small intestine as efficiently as wild-type mice, but depletion of CD4+ T cells rescued chlamydial colonization in the small intestine of the CD8+ T cell-deficient mice. Third, mice deficient in IFN-γ but not IL-22 signaling rescued chlamydial colonization in the small intestine. Fourth, supplementing TCRα-deficient mice with exogenous IFN-γ was sufficient for clearing Chlamydia from the small intestine. Fifth, mice deficient in developing Chlamydia-specific CD4+ T cell immunity or Th1 immunity permitted chlamydial persistence in the small intestine. Finally, IFN-γ-producing CD4+ but not CD8+ T cells from immunized donor mice were sufficient for eliminating Chlamydia from the small intestine of recipient mice.
Despite the critical role of CD4+ T cell-produced IFN-γ in clearing C. muridarum from both the mouse genital tract (13, 34, 35) and small intestine (current study), the precise mechanism for how IFN-γ clears C. muridarum remains unknown. Indoleamine dioxygenase (IDO) and nitric oxide (NO) synthase are among the most prominent and well-studied IFN-γ-inducible effectors. IDO works in human cells, while NO works in mouse cells (36). However, blockage of NO synthesis only partially rescued chlamydial growth, suggesting that additional mechanisms remain to be determined. IFN-inducible guanylate binding proteins (GBPs) are known to mediate cell-autonomous host resistance to bacterial pathogens (37). However, C. muridarum is resistant to GBP-mediated restrictions on bacterial growth. GBPs neither bind to C. muridarum inclusions nor restrict C. muridarum growth, although GBPs are still able to promote inflammasome activation in C. muridarum-infected cells (38). It is possible that IFN-γ may restrict C. muridarum infection by inducing host cell apoptosis since Chlamydia is selected to inhibit host cell apoptosis (39, 40). Here we have shown that exogenous IFN-γ is sufficient for eliminating C. muridarum from small intestine, which has provided an ideal animal model for further revealing the mechanisms. It is worth noting that besides CD4+ T cells, B cells may also play an important role in anti-Chlamydia immunity (34). IFN-γ may also cooperate with other immune components, such as antibodies and neutrophils, for enhancing chlamydial clearance (41, 42).
Once arriving in the large intestine, C. muridarum lasts for long periods. The question is why Th1 immunity that is able to clear C. muridarum in the small intestine spares C. muridarum in the large intestine. There are regional differences in the development and composition of the adaptive immune landscape of the intestine (43). The Th1 immunity identified in the small intestine may not necessarily work in the large intestine. It appears that C. muridarum organisms in different segments of the mouse GI tract are regulated by distinct immune mechanisms. A lack of T cells or CD4+ T cells and Th1 immunity did not significantly increase C. muridarum colonization in the large intestine. More importantly, supplementation with exogenous IFN-γ or IFN-γ-producing CD4+ T cells from Chlamydia-immunized donor mice failed to significantly reduce C. muridarum colonization in the large intestine. Since the same immunological conditions significantly inhibited C. muridarum colonization in the small intestine of the same mice, we hypothesize that the failure of Th1 immunity to impact the large intestine Chlamydia may be due to lack of accessibility to Chlamydia-infected cells in the large intestine by the Th1 cells. The large intestine is known to host the highest concentration of microbiota species, and high levels of regulatory T cells (Tregs) have been detected in the large intestine (44–46). Chlamydia-infected epithelial cells in the large intestine may be protected from the Th1 immunity by Tregs, which will be investigated in our future studies.
The information acquired from mouse model study may be useful in guiding clinical studies on C. trachomatis colonization in the gut. C. trachomatis is frequently detected on rectal swabs (15, 16, 27–31), suggesting that C. trachomatis is introduced into the human GI tract. It will be worth testing whether C. trachomatis is also efficiently cleared from human small intestine but persists in the large intestine for a long period. More importantly, we have recently shown that oral delivery of C. muridarum is nonpathogenic while inducing transmucosal immunity against subsequent challenge infection in the mouse genital tract (32). Investigating the mechanisms of chlamydial colonization in the GI tract would provide mechanistic information useful in developing an oral chlamydial vaccine. The current study has revealed that robust Th1-dominant immunity is induced by the orally delivered Chlamydia, which efficiently cleared the chlamydial organisms from the small intestine within 4 weeks, demonstrating the capacity of orally delivered chlamydial organisms to induce protective immunity against genital tract infection. It is well established that a Th1-dominant response is protective against chlamydial infection in the genital tract (34). The question is whether the gastrointestinal C. trachomatis can induce protective immunity in human genital tracts, which is testable by evaluating whether humans whose rectal swabs are positive for C. trachomatis can develop Chlamydia-specific Th1 immunity and display lower rates of repeated infections. Thus, mouse studies can provide mechanistic information for guiding clinical investigations.
We are aware of the distinction between C. trachomatis and C. muridarum. C. trachomatis is naturally transmitted among humans sexually, while C. muridarum may be naturally transmitted among mice via the oral-fecal route (instead of sexually). This is because the C. muridarum plasmid or plasmid-encoded proteins or some chromosomal proteins have been shown to be more important for C. muridarum to colonize the GI tract than to infect the genital tract (47, 48). In addition, the C. muridarum genome still maintains three copies of a full-length cytotoxin gene with significant homology to genes for large clostridial cytotoxins (49). C. muridarum may have acquired the cytotoxin gene from other enteric bacteria, and maintaining the full-length cytotoxins may be necessary for C. muridarum to compete against other enteric bacteria in the GI tract. However, C. trachomatis has been transmitted sexually among humans, forcing C. trachomatis to adapt to the human genital mucosa and allowing the cytotoxin genes to be lost or significantly shortened in C. trachomatis (49). Nevertheless, C. trachomatis has been frequently detected in the GI tract in humans but without any significant association with GI pathologies (14–17, 50). C. trachomatis may have experienced selection pressures from both the genital and GI tracts. It will be worth testing whether C. trachomatis is regulated by different immune mechanisms in different regions of the human GI tract.
MATERIALS AND METHODS
Chlamydial organism growth.
Chlamydia muridarum used in the current study was clone G13.32.1 (51), which was derived from strain Nigg3 (GenBank accession number CP009760.1). The C. muridarum organisms were propagated in HeLa cells (human cervical carcinoma epithelial cells; ATCC catalog number CCL2.1) and purified as elementary bodies (EBs) as reported previously (20, 39). Aliquots of purified EBs were stored at −80°C until use. The storage buffer (SPG) consisted of 220 mM sucrose, 12.5 mM phosphate, and 4 mM l-glutamic acid (pH 7.5).
Mouse infection.
Mouse experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (52). The protocol was approved by the Committee on the Ethics of Laboratory Animal Experiments of the University of Texas Health Science Center at San Antonio.
C. muridarum EBs were used to inoculate 6- to 7-week-old female mice (Jackson Laboratories, Inc., Bar Harbor, ME) intravaginally or intragastrically as described previously (10, 20, 26, 48). The following mice were used in the current study. The two wild-type strains were CBA1/J (stock number 000656) and C57BL/6J (0006640). Gene knockout (KO) strains were CD4 KO (002663 [B6.129S2-Cd4tm1Mak/J]), CD8 KO (002665 [B6.129S2-Cd8atm1Mak/J]), IFN-γR KO (003288 [B6.129S7-Ifngr1tm1Agt/J]), IL-22 KO (027524 [C57BL/6-Il22tm1.1(icre)Stck/J]), IL-12p35 KO (002692 [B6.129S1-Il12atm1Jm/J]), and TCRα KO (002116 [B6.129S2-Tcratm1Mom/J]). The transgenic mouse strains were OT2 {004194 [B6.Cg-Tg(TcraTcrb)425Cbn/J]} and IFN-γ reporter (017581 [B6.129S4-Ifngtm3.1Lky/J]). For intravaginal inoculation, stock EBs diluted in 10 μl of SPG that contained 2 × 105 IFU or the desired IFU as indicated for individual experiments were delivered to the ectocervix area using a 20-μl micropipette tip. Five days prior to inoculation, each mouse was injected subcutaneously with 2.5 mg of Depo-Provera (Pharmacia Upjohn, Kalamazoo, MI) suspended in sterile phosphate-buffered saline (PBS). For intragastric inoculation, EBs diluted in 100 μl of SPG that contained 2 × 105 IFU or the desired number of IFU as indicated for individual experiments were delivered to the stomach using a straight-balled end needle designed for mouse oral gavage (N-PK 020; Braintree Scientific Inc., Braintree, MA). Following the initial inoculation, both vaginal and rectal swabs were taken periodically or organs/tissues were harvested (after mice were sacrificed) for titration of viable organisms as described previously (20, 21, 26). In some experiments, mice were treated with an anti-CD4 antibody, while in others, exogenous molecules or donor T cells were provided as described below. At designated time points after inoculation, mice were sacrificed for titration of viable organisms as described below.
Antibody depletion, providing exogenous molecules or cells (adoptive transfer).
In some experiments, mice were treated with anti-CD4 antibody (clone number GK1.5, rat IgG2b, κ, purified as described previously [41] or purchased from Bio X Cell, West Lebanon, NH). The antibody treatment began on day 5 after intragastric inoculation and then was continued twice weekly throughout experiments. Each injection was administered intraperitoneally with 400 μg of IgG in 400 μl of PBS as described previously (41, 53).
For injecting exogenous IFN-γ into mice, IFN-γ (catalog no. 575308; Biolegend, Inc., San Diego, CA) was diluted in sterile PBS and injected intraperitoneally at 16 μg/injection in a 200-μl volume. The injection started on day 7 after chlamydial infection and continued once every 2 days throughout the experiments.
For adoptive-transfer experiments, both the spleen and mesenteric lymph nodes were harvested from IFN-γ reporter mice on day 10 after chlamydial infection. Single cell suspensions were labeled with the following antibodies: anti-CD4 (conjugated with allophycocyanin [APC], clone number GK1.5, rat IgG2b, κ, catalog number 100404; Biolegend, Inc., San Diego, CA), anti-CD8 (conjugated with Efluor 450, clone number eBioH35-17.2, catalog number 12-0083-81; Thermo Fisher Scientific, Inc., Waltham, MA), and anti-CD3 (conjugated with phycoerythrin [PE], clone number 17A2, rat IgG2b, κ, catalog number 100206; Biolegend). Donor cells were then purified using flow cytometry sorting (BD FACSAria II, catalog number 642886; BD Biosciences San Jose, CA), gating CD3 and YFP doubly positive cells and then sorting for CD4+ or CD8+ cells. The sorted donors were designated IFN-γ+ CD4+ or IFN-γ+ CD8+ T cells. The flow cytometry sorting ensured >98% purity for both donor cell populations prior to adoptive transfer. For adoptive transfer, each recipient mouse received 2 × 106 corresponding donor T cells three times on days 7, 14, and 21 after the recipient mice were infected with C. muridarum. Retro-orbital injection was used to deliver the donor cells.
Titrating live chlamydial organisms recovered from swabs and tissue homogenates.
To quantitate live chlamydial organisms on vaginal or rectal swabs, each swab was soaked in 0.5 ml of SPG and vortexed with glass beads, and the chlamydial organisms released into the supernatants were titrated on HeLa cell monolayers in duplicate. The infected cultures were processed for immunofluorescence assay as described previously (54, 55) and below. Inclusions were counted in five random fields per coverslip under a fluorescence microscope. For coverslips with less than one IFU per field, entire coverslips were counted. Coverslips showing obvious cytotoxicity of HeLa cells were excluded. The total number of IFU per swab was calculated based on the mean IFU per view, the ratio of the view area to that of the well, dilution factor, and inoculation volumes. Where possible, a mean IFU/swab was derived from the serially diluted and duplicate samples for any given swab. The total number of IFU/swab was converted into log10, which was used to calculate the mean and standard deviation across mice of the same group at each time point.
For quantitating live organisms from mouse organs and tissue segments, each organ or tissue segment was transferred to a tube containing 0.5 to 5 ml of SPG depending the sizes of the organs. Each GI tract was cut into different segments/portions as indicated for individual experiments, including stomach (ST), small intestine (SI), cecum (CE), colon (CL), and anorectum (AR). In some experiments, SI was further divided into duodenum, jejunum, and ileum. Each genital tract was divided into segments, including vagina/cervix (CV), left uterine horn (LU), right uterine horn (RU), left oviduct/ovary (LO), and right oviduct/ovary (RO). Other tissues, including spleen (SP), liver (LV), kidney (KD), lung (LG), and mesenteric lymph node (ML), were also measured. The organs and tissue segments were homogenized in cold SPG using a 2-ml tissue grinder (K885300-0002; Fisher Scientific, Pittsburgh, PA) or an automatic homogenizer (TH115; Omni International, Kennesaw, GA). The homogenates were briefly sonicated and spun at 3000 rpm for 5 min to pellet remaining large debris. The supernatants were titrated for live C. muridarum organisms on HeLa cells as described above. The results were expressed as log10 IFU per organ or tissue segment.
Immunofluorescence assay.
The immunofluorescence assay used for titration of live organisms was carried out as described previously (39, 56). A rabbit antibody (designated R1604, raised with purified C. muridarum EBs) was used as a primary antibody to label all C. muridarum organisms in HeLa cells, which were visualized with a goat anti-rabbit IgG conjugated with Cy2 (green, catalog number 111-225-144; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). The DNA dye Hoechst 3328 (blue; Sigma-Aldrich, St. Louis, MO) was used to visualize nuclei. The doubly labeled samples were used for counting C. muridarum organisms under a fluorescence microscope (AX70; Olympus) equipped with a charge-coupled-device (CCD) camera (Hamamatsu).
Statistical analyses.
The time courses of live organism shedding (area under the curve [AUC]) or IFU from individual tissues were compared between two groups using the Wilcoxon rank sum test (an in-house Excel sheet), while category data, including number of mice positive for live organism shedding, were analyzed using Fisher’s exact test (http://vassarstats.net/tab2x2.html). Correlations of chlamydial colonization in different tissues were analyzed by calculating Spearman rank order correlation coefficients (http://vassarstats.net/corr_rank.html). Furthermore, the significance of the difference between two correlation coefficients was also calculated (http://vassarstats.net/rdiff.html).
ACKNOWLEDGMENTS
This study is supported in part by U.S. NIH grants (R01AI047997 and R01AI121989 to G.Z.) and Central South University Funds for Supporting Preliminary Research (2017zzts031 to H.L.).
REFERENCES
- 1.Centers for Disease Control and Prevention. 2017. Sexually transmitted disease surveillance, 2016. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/std/stats16/default.htm. [Google Scholar]
- 2.Budrys NM, Gong S, Rodgers AK, Wang J, Louden C, Shain R, Schenken RS, Zhong G. 2012. Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet Gynecol 119:1009–1016. doi: 10.1097/AOG.0b013e3182519326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers AK, Budrys NM, Gong S, Wang J, Holden A, Schenken RS, Zhong G. 2011. Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertil Steril 96:715–721. doi: 10.1016/j.fertnstert.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodgers AK, Wang J, Zhang Y, Holden A, Berryhill B, Budrys NM, Schenken RS, Zhong G. 2010. Association of tubal factor infertility with elevated antibodies to Chlamydia trachomatis caseinolytic protease P. Am J Obstet Gynecol 203:494.e7–494.e14. doi: 10.1016/j.ajog.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Maza LM, Pal S, Khamesipour A, Peterson EM. 1994. Intravaginal inoculation of mice with the Chlamydia trachomatis mouse pneumonitis biovar results in infertility. Infect Immun 62:2094–2097. [8168974] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison RP, Feilzer K, Tumas DB. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 63:4661–4668. [7591120] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah AA, Schripsema JH, Imtiaz MT, Sigar IM, Kasimos J, Matos PG, Inouye S, Ramsey KH. 2005. Histopathologic changes related to fibrotic oviduct occlusion after genital tract infection of mice with Chlamydia muridarum. Sex Transm Dis 32:49–56. doi: 10.1097/01.olq.0000148299.14513.11. [DOI] [PubMed] [Google Scholar]
- 8.Pal S, Peterson EM, de la Maza LM. 2005. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect Immun 73:8153–8160. doi: 10.1128/IAI.73.12.8153-8160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murthy AK, Li W, Guentzel MN, Zhong G, Arulanandam BP. 2011. Vaccination with the defined chlamydial secreted protein CPAF induces robust protection against female infertility following repeated genital chlamydial challenge. Vaccine 29:2519–2522. doi: 10.1016/j.vaccine.2011.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Zhang H, Zhou Z, Yang Z, Ding Y, Zhou Z, Zhong E, Arulanandam B, Baseman J, Zhong G. 2014. Chlamydial induction of hydrosalpinx in 11 strains of mice reveals multiple host mechanisms for preventing upper genital tract pathology. PLoS One 9:e95076. doi: 10.1371/journal.pone.0095076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison RP, Caldwell HD. 2002. Immunity to murine chlamydial genital infection. Infect Immun 70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Lei L, Zhou Z, He J, Xu S, Lu C, Chen J, Yang Z, Wu G, Yeh IT, Zhong G, Wu Y. 2013. Contribution of interleukin-12 p35 (IL-12p35) and IL-12p40 to protective immunity and pathology in mice infected with Chlamydia muridarum. Infect Immun 81:2962–2971. doi: 10.1128/IAI.00161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry LL, Feilzer K, Caldwell HD. 1997. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol 158:3344–3352. [PubMed] [Google Scholar]
- 14.Peters RPH, Dubbink JH, van der Eem L, Verweij SP, Bos MLA, Ouburg S, Lewis DA, Struthers H, McIntyre JA, Morré SA. 2014. Cross-sectional study of genital, rectal, and pharyngeal Chlamydia and gonorrhea in women in rural South Africa. Sex Transm Dis 41:564–569. doi: 10.1097/OLQ.0000000000000175. [DOI] [PubMed] [Google Scholar]
- 15.Gratrix J, Singh AE, Bergman J, Egan C, McGinnis J, Drews SJ, Read R. 2014. Prevalence and characteristics of rectal chlamydia and gonorrhea cases among men who have sex with men after the introduction of nucleic acid amplification test screening at 2 Canadian sexually transmitted infection clinics. Sex Transm Dis 41:589–591. doi: 10.1097/OLQ.0000000000000176. [DOI] [PubMed] [Google Scholar]
- 16.Gratrix J, Singh AE, Bergman J, Egan C, Plitt SS, McGinnis J, Bell CA, Drews SJ, Read R. 2015. Evidence for increased Chlamydia case finding after the introduction of rectal screening among women attending 2 Canadian sexually transmitted infection clinics. Clin Infect Dis 60:398–404. doi: 10.1093/cid/ciu831. [DOI] [PubMed] [Google Scholar]
- 17.Musil K, Currie M, Sherley M, Martin S. 2016. Rectal chlamydia infection in women at high risk of chlamydia attending Canberra Sexual Health Centre. Int J STD AIDS 27:526–530. doi: 10.1177/0956462415586317. [DOI] [PubMed] [Google Scholar]
- 18.Yeruva L, Spencer N, Bowlin AK, Wang Y, Rank RG. 2013. Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 68:88–95. doi: 10.1111/2049-632X.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bavoil PM, Marques PX, Brotman R, Ravel J. 2017. Does active oral sex contribute to female infertility? J Infect Dis 216:932–935. doi: 10.1093/infdis/jix419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Huang Y, Gong S, Yang Z, Sun X, Schenken R, Zhong G. 2015. In vivo and ex vivo imaging reveals a long-lasting chlamydial infection in the mouse gastrointestinal tract following genital tract inoculation. Infect Immun 83:3568–3577. doi: 10.1128/IAI.00673-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai J, Zhang T, Wang L, Shao L, Zhu C, Zhang Y, Failor C, Schenken R, Baseman J, He C, Zhong G. 2016. Intravenous inoculation with chlamydia muridarum leads to a long-lasting infection restricted to the gastrointestinal tract. Infect Immun 84:2382–2388. doi: 10.1128/IAI.00432-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong G. 2018. Chlamydia spreading from the genital tract to the gastrointestinal tract—a two-hit hypothesis. Trends Microbiol 26:611–623. doi: 10.1016/j.tim.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igietseme JU, Portis JL, Perry LL. 2001. Inflammation and clearance of Chlamydia trachomatis in enteric and nonenteric mucosae. Infect Immun 69:1832–1840. doi: 10.1128/IAI.69.3.1832-1840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry LL, Hughes S. 1999. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect Immun 67:3686–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeruva L, Melnyk S, Spencer N, Bowlin A, Rank RG. 2013. Differential susceptibilities to azithromycin treatment of chlamydial infection in the gastrointestinal tract and cervix. Antimicrob Agents Chemother 57:6290–6294. doi: 10.1128/AAC.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Zhang Q, Zhang T, Zhang Y, Zhu C, Sun X, Zhang N, Xue M, Zhong G. 2016. The Chlamydia muridarum organisms fail to auto-inoculate the mouse genital tract after colonization in the gastrointestinal tract for 70 days. PLoS One 11:e0155880. doi: 10.1371/journal.pone.0155880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khosropour CM, Soge OO, Suchland R, Leipertz G, Unutzer A, Pascual R, Hybiske K, Barbee LA, Manhart LE, Dombrowski JC, Golden MR. 2019. Recurrent/intermittent vaginal and rectal chlamydial infection following treatment: a prospective cohort study among female STD clinic patients. J Infect Dis 2019:jiz113. doi: 10.1093/infdis/jiz113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dukers-Muijrers NH, Schachter J, van Liere GA, Wolffs PF, Hoebe CJ. 2015. What is needed to guide testing for anorectal and pharyngeal Chlamydia trachomatis and Neisseria gonorrhoeae in women and men? Evidence and opinion. BMC Infect Dis 15:533. doi: 10.1186/s12879-015-1280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dukers-Muijrers NH, Speksnijder AG, Morre SA, Wolffs PF, van der Sande MA, Brink AA, van den Broek IV, Werner MI, Hoebe CJ. 2013. Detection of anorectal and cervicovaginal Chlamydia trachomatis infections following azithromycin treatment: prospective cohort study with multiple time-sequential measures of rRNA, DNA, quantitative load and symptoms. PLoS One 8:e81236. doi: 10.1371/journal.pone.0081236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Javanbakht M, Gorbach P, Stirland A, Chien M, Kerndt P, Guerry S. 2012. Prevalence and correlates of rectal Chlamydia and gonorrhea among female clients at sexually transmitted disease clinics. Sex Transm Dis 39:917–922. doi: 10.1097/OLQ.0b013e31826ae9a2. [DOI] [PubMed] [Google Scholar]
- 31.Khosropour CM, Dombrowski JC, Barbee LA, Manhart LE, Golden MR. 2014. Comparing azithromycin and doxycycline for the treatment of rectal chlamydial infection: a retrospective cohort study. Sex Transm Dis 41:79–85. doi: 10.1097/OLQ.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Zhu C, Zhang T, Tian Q, Zhang N, Morrison S, Morrison R, Xue M, Zhong G. 2018. Nonpathogenic colonization with chlamydia in the gastrointestinal tract as oral vaccination for inducing transmucosal protection. Infect Immun 86:e00630-17. doi: 10.1128/IAI.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu C, Lin H, Tang L, Chen J, Wu Y, Zhong G. 2018. Oral Chlamydia vaccination induces transmucosal protection in the airway. Vaccine 36:2061–2068. doi: 10.1016/j.vaccine.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Morrison SG, Su H, Caldwell HD, Morrison RP. 2000. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun 68:6979–6987. doi: 10.1128/IAI.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. 2008. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol 180:3375–3382. doi: 10.4049/jimmunol.180.5.3375. [DOI] [PubMed] [Google Scholar]
- 36.Roshick C, Wood H, Caldwell HD, McClarty G. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect Immun 74:225–238. doi: 10.1128/IAI.74.1.225-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeij JP, Frickel EM. 2017. Exposing Toxoplasma gondii hiding inside the vacuole: a role for GBPs, autophagy and host cell death. Curr Opin Microbiol 40:72–80. doi: 10.1016/j.mib.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, Yamamoto M, Nagarajan UM, Miao EA, Coers J. 2015. Guanylate binding proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun 83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, Greenberg AH, Zhong G. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med 187:487–496. [9463399] doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Y, Weininger M, Pirbhai M, Dong F, Zhong G. 2006. Inhibition of staurosporine-induced activation of the proapoptotic multidomain Bcl-2 proteins Bax and Bak by three invasive chlamydial species. J Infect 53:408–414. doi: 10.1016/j.jinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 41.Naglak EK, Morrison SG, Morrison RP. 2016. IFNgamma is required for optimal antibody-mediated immunity against genital chlamydia infection. Infect Immun 84:3232–3242. doi: 10.1128/IAI.00749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naglak EK, Morrison SG, Morrison RP. 2017. Neutrophils are central to antibody-mediated protection against genital chlamydia. Infect Immun 85:e00409-17. doi: 10.1128/IAI.00409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agace WW, McCoy KD. 2017. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46:532–548. doi: 10.1016/j.immuni.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 45.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, Begum-Haque S, Kasper DL, Kasper LH. 2015. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes 6:234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao L, Melero J, Zhang N, Arulanandam B, Baseman J, Liu Q, Zhong G. 2017. The cryptic plasmid is more important for Chlamydia muridarum to colonize the mouse gastrointestinal tract than to infect the genital tract. PLoS One 12:e0177691. doi: 10.1371/journal.pone.0177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao L, Zhang T, Liu Q, Wang J, Zhong G. 2017. Chlamydia muridarum with mutations in chromosomal genes tc0237 and/or tc0668 is deficient in colonizing the mouse gastrointestinal tract. Infect Immun 85:e00321-17. doi: 10.1128/IAI.00321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belland RJ, Scidmore MA, Crane DD, Hogan DM, Whitmire W, McClarty G, Caldwell HD. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci U S A 98:13984–13989. doi: 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig AP, Kong FY, Yeruva L, Hocking JS, Rank RG, Wilson DP, Donovan B. 2015. Is it time to switch to doxycycline from azithromycin for treating genital chlamydial infections in women? Modelling the impact of autoinoculation from the gastrointestinal tract to the genital tract. BMC Infect Dis 15:200. doi: 10.1186/s12879-015-0939-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad TA, Gong S, Yang Z, Matulich P, Keck J, Beltrami N, Chen C, Zhou Z, Dai J, Zhong G. 2016. The chromosome-encoded hypothetical protein TC0668 is an upper genital tract pathogenicity factor of Chlamydia muridarum. Infect Immun 84:467–479. doi: 10.1128/IAI.01171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 53.Morrison SG, Morrison RP. 2001. Resolution of secondary Chlamydia trachomatis genital tract infection in immune mice with depletion of both CD4+ and CD8+ T cells. Infect Immun 69:2643–2649. doi: 10.1128/IAI.69.4.2643-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G. 2014. Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect Immun 82:983–992. doi: 10.1128/IAI.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang L, Zhang H, Lei L, Gong S, Zhou Z, Baseman J, Zhong G. 2013. Oviduct infection and hydrosalpinx in DBA1/j mice is induced by intracervical but not intravaginal inoculation with Chlamydia muridarum. PLoS One 8:e71649. doi: 10.1371/journal.pone.0071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong G, Fan P, Ji H, Dong F, Huang Y. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med 193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]