This investigation compared the microbiomes colonizing teeth during the initiation, progression, and resolution of periodontitis in nonhuman primates (Macaca mulatta) at different ages. Subgingival plaque samples were collected at baseline; 0.5, 1, and 3 months following ligature-induced periodontitis; and following naturally occurring disease resolution at 5 months.
KEYWORDS: microbiome, nonhuman primates, periodontitis
ABSTRACT
This investigation compared the microbiomes colonizing teeth during the initiation, progression, and resolution of periodontitis in nonhuman primates (Macaca mulatta) at different ages. Subgingival plaque samples were collected at baseline; 0.5, 1, and 3 months following ligature-induced periodontitis; and following naturally occurring disease resolution at 5 months. Samples were analyzed using 16S amplicon sequencing to identify bacterial profiles across age groups: young (<3 years of age), adolescent (3 to 7 years), adult (12 to 15 years), and aged (17 to 23 years). α-Diversity of the microbiomes was greater in the adult/aged samples than in the young/adolescent samples. β-Diversity of the samples demonstrated clear age group differences, albeit individual variation in microbiomes between animals within the age categories was noted. Phylum distributions differed between the young/adolescent animals and the adult/aged animals at each of the time points, showing an enrichment of the phyla Spirochetes, Fusobacteria, and Bacteroidetes associated with periodontitis. Major differences in the top 50 operational taxonomic units (OTUs) were noted in the young and adolescent microbiomes during initiation and progression postligation compared to the adult and aged animals. The proportions of a large number of species in the top 50 OTUs were lower at baseline and in resolved disease microbiomes in the young samples, while profiles in adolescent animals were more consistent with the disease microbiomes. Microbiome profiles for resolution for adults and aged animals appeared more resilient and generally maintained a pattern similar to that of disease. Use of the model can expand our understanding of the crucial interactions of the oral microbiome and host responses in periodontitis.
INTRODUCTION
Common risk factors enhancing periodontitis include poor plaque control, smoking, diabetes, genetic factors, age, and various systemic chronic inflammatory diseases (1–4). However, the fundamental interactions of the microbial components of the microbiome and the host response to this challenge that are affected by these risk factors remain rather nebulous. Historically, cross-sectional human studies have documented substantial differences in the microbial ecology of healthy and periodontitis sites in almost all studies, using a range of technologies and identifying the proposed pathogenic “red complex” species Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola (5–7) as hallmarks of disease. Nevertheless, limitations of the human disease process do not allow definitive interpretation of how early in an active disease process critical microbial changes occur or when these types of purported pathogens emerge in response to local environmental alterations and help to drive the processes resulting in dysbiosis and progressing disease (8–11).
More recently, with a greater emphasis on the importance of truly understanding both the microbial components and their functional activities in microbiomes in health and disease, investigations using next-generation (NexGen) sequencing for genomics and transcriptomics have become more detailed in characterizing the “entirety” of the microbiome to define changes that occur from health to disease (8). These types of studies have detailed exceptional heterogeneity in the microbiomes in existing health and disease sites (12–16) and provided some insights into the nature of the microbiomes that occur in stable and progressing sites (9), the influence of intrinsic/extrinsic factors on the oral microbiome features (e.g., smoking and diabetes) (17, 18), and changing functions with the microbial community that might be predicted to enhance its pathogenic potential (19, 20). While some of these investigations have identified changes in the autochthonous oral microbial ecology in mice triggered by recognized oral periodontopathogens, with the consequent induction of oral inflammation and alveolar bone loss (21–23), there are differences in the oral microbiome in mice from that in humans, and a human model inducing periodontitis to monitor for the details of the microbial changes over time is not ethical.
Thus, this study used the ligature-induced periodontitis nonhuman primate model, whose oral microbiome is more similar to that of humans (24, 25), to provide new details on the kinetics of changes in the subgingival microbiome with the initiation, progression, and resolution of periodontal lesions. The investigation was also able to compare and contrast these changes across the life span and potentially identify unique characteristics of the microbiome in younger individuals who are less susceptible to periodontitis, as demonstrated in both humans and these nonhuman primates (26–28).
RESULTS
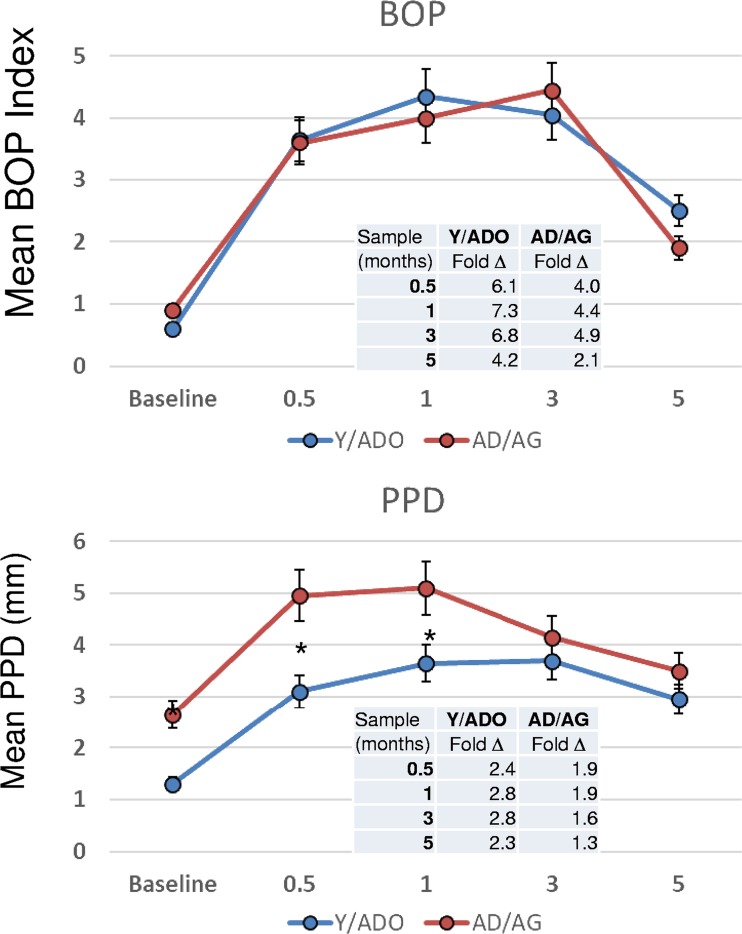
Clinical findings from the study are shown in Fig. 1. The results demonstrated that the younger animals exhibited an extent and kinetics of inflammation, as bleeding on probing (BOP), similar to those of the older animals. Also, while a significantly lower magnitude of tissue destruction was observed in the younger group, as reflected in the probing pocket depths (PPDs), the rates of change in PPD over the initiation and early progression of disease (i.e., baseline to 1 month) were similar (y = 2.94x + 2.25 [R2 = 0.5987] for the young/adolescent [Y/ADO] groups; y = 2.36x + 3.05 [R2 = 0.4466] for the adult/aged [AD/AG] groups). Thus, the variations of PPD at baseline in health between the groups might explain the potential difference in the oral microbiomes, even with a clinically healthy periodontium, and support that aging could contribute to a dysbiotic oral microbiome even in the absence of clinical changes in the periodontium.
FIG 1.
Clinical measures of bleeding on probing (BOP) and mouth mean probing pocket depth (PPD) in nonhuman primates. The animals were organized into young/adolescent (Y/ADO) and adult/aged (AD/AG) groups. The points denote group means for Y/ADO animals (n = 18) and AD/AG animals (n = 18), and the vertical brackets depict 1 standard deviation (SD). The asterisks denote a significant difference between groups at a time point (baseline and 0.5, 1, 3, and 5 months) at a P value of <0.05.
A general analysis of the samples for α-diversity is provided in Table S1 in the supplemental material. The α-diversity of the samples demonstrated by analyses of the number of operational taxonomic units (OTUs), the Shannon index, the inverse Simpson index, the Chao1 estimator, and the abundance-based coverage estimator (ACE) demonstrated a greater diversity in the adult and aged samples than in the young and adolescent samples. Figure S1 in the supplemental material displays a principal-coordinate analysis (PCoA) describing the β-diversity of the samples. Of note, ∼27% of the variation in the microbiomes of the samples was related to the group source of the samples, supporting important individual variation in microbiome composition between animals even within the age categories.
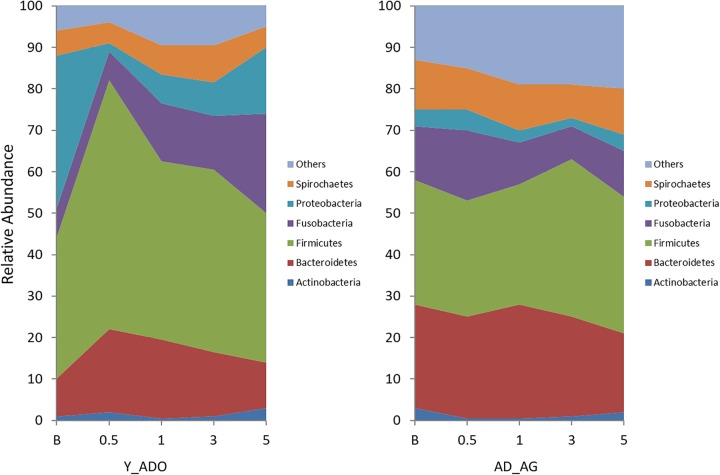
The results in Fig. 2 provide a summary of the phylum distribution in the microbiome of the ligated teeth at baseline; at 0.5, 1, and 3 months of disease; and 5 months following ligature removal. There were clear differences in this distribution between the young/adolescent groups of animals and the adult/aged groups of animals at each of the time points, showing an enrichment of the phyla Spirochetes, Fusobacteria, and Bacteroidetes associated with periodontitis, even before the onset of disease, particularly in the adult/aged animals. Also of interest were the more dramatic changes in the phylum distribution in the younger animals following ligation, which might reflect a less-well-established microbiome that is more greatly influenced by environmental changes (i.e., ligature placement). Finally, it appears that, generally, the phylum distribution in both groups approached baseline profiles at the 5-month sampling period following ligature removal and when clinical parameters returned to near-normal levels, except for the Fusobacteria proportions in the young/adolescent group, which remained substantially elevated. Similarly, Fig. 3A to D depict the signals for the top 50 OTUs compared across baseline samples among the age groups, and Tables S2 to S5 in the supplemental material provide the processed data for the dominant 200 OTUs/species for each age group and across all time points. The graphs display a greater relative abundance of these species within the oral microbiome of the adult and aged animals, albeit as shown in Fig. S2 in the supplemental material, the top 50 OTUs for the young and adolescent animals represented approximately 90% of the overall OTU reads, while in the adult and aged groups, this subset of OTUs provided microbiome coverage that averaged 75% to 80%.
FIG 2.
Area graph demonstrating the group means of the relative abundances of bacterial phylotypes in the Y/ADO and AD/AG groups at each time point.
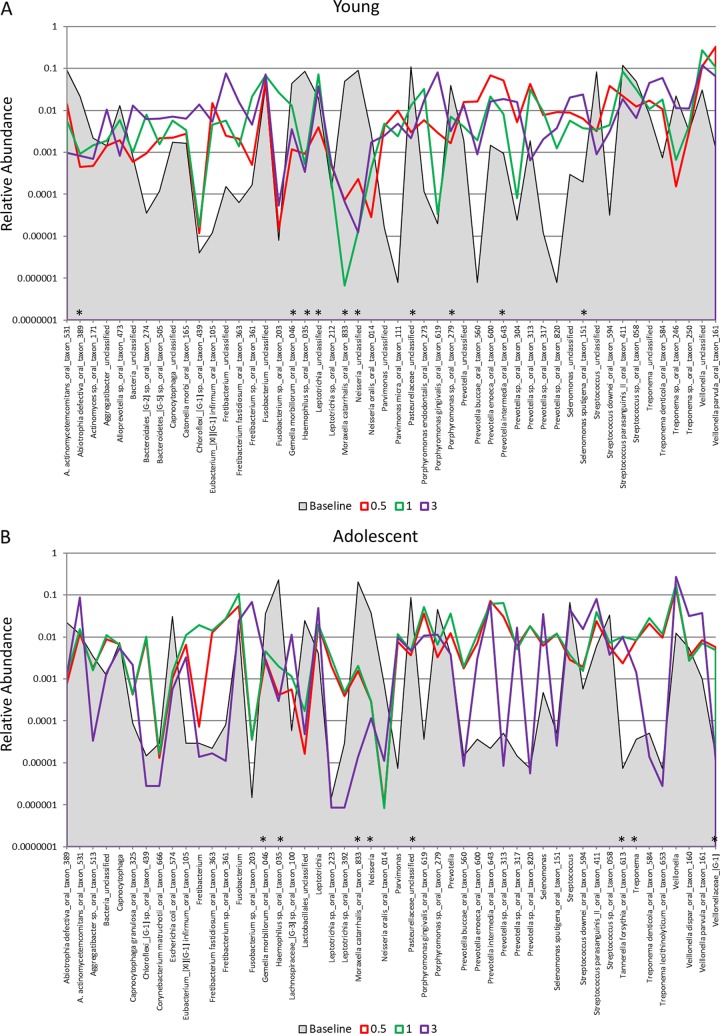
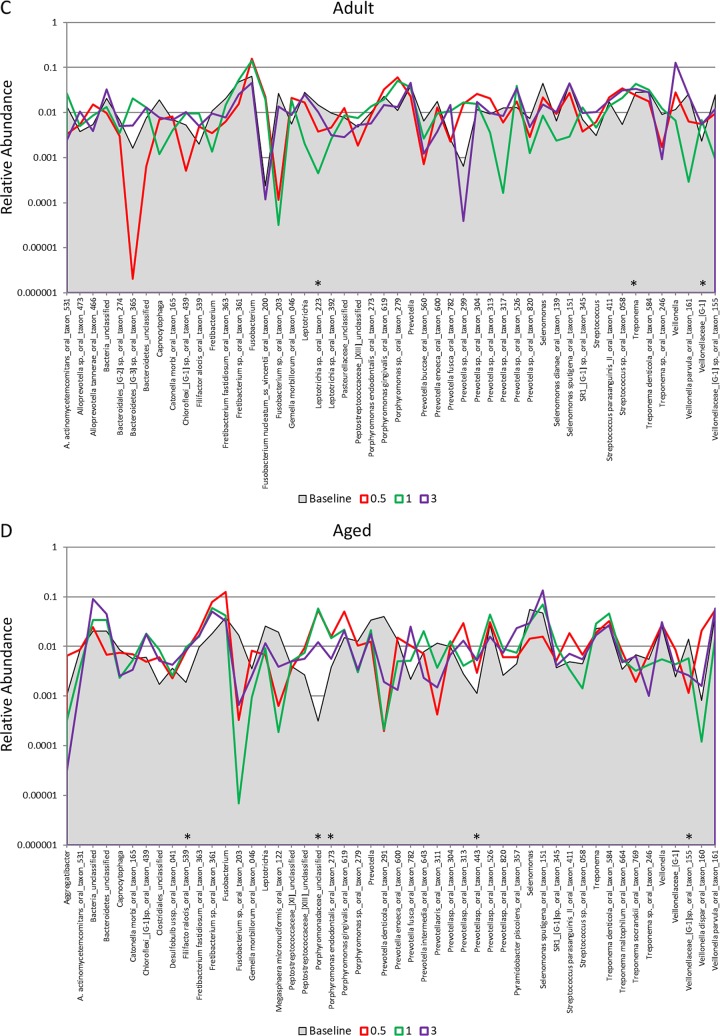
FIG 3.
Distribution and relative abundance of signals for the top 50 OTUs in baseline samples from young (A), adolescent (B), adult (C), and aged (D) animals and readouts for each OTU/species and for samples collected at 2 weeks (0.5 months) and at 1 and 3 months postligation during initiation and progression of disease. The asterisks denote a significant difference from the baseline with disease (0.5, 1, and 3 months) at a P value of at least <0.05 using Z-score normalization.
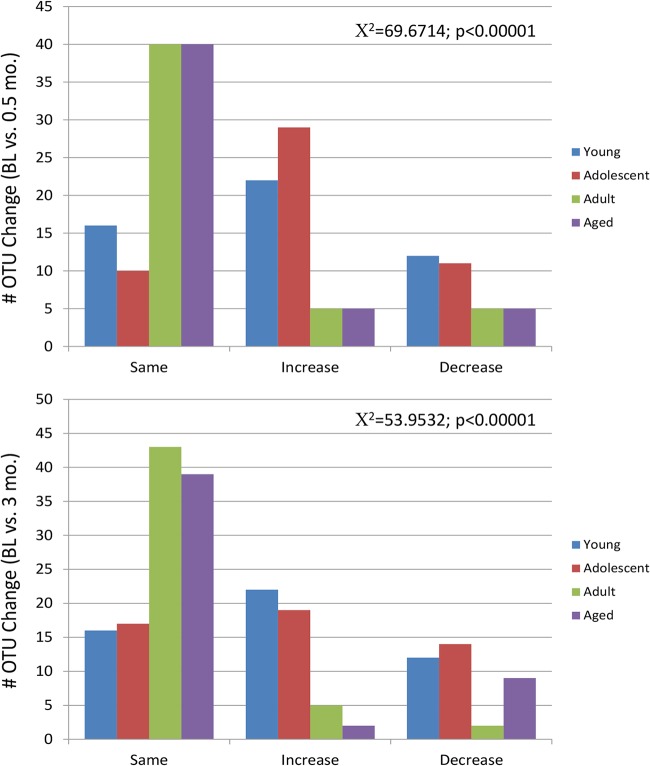
Figure 4 summarizes the changes in the microbiome patterns that occur during the initiation of the disease process from baseline through 2 weeks. The first striking result was the substantial differences in the young and adolescent animals during this initial interval postligation compared to the adult and aged animals. This is also noted in Fig. 5, showing a statistically significantly higher frequency of major microbiome species changes (≥5-fold increased or decreased) within the top 50 OTUs at 2 weeks (0.5 months) in the young and adolescent animals versus adult and aged groups. Also note that within the top 50 OTUs across the age groups, only 4% to 10% of the species were not represented in multiple groups, with these differences generally being in the lower tertile of the overall reads for the top 50 species.
FIG 4.
Distribution of the number of changes in signals for the top 50 OTUs comparing baseline (BL) to disease initiation at 2 weeks (0.5 months) (top) or disease progression at 3 months (bottom), identified as a ≥5-fold or ≤5-fold (increase or decrease) change from the baseline or remaining the same in the 4 groups of animals.
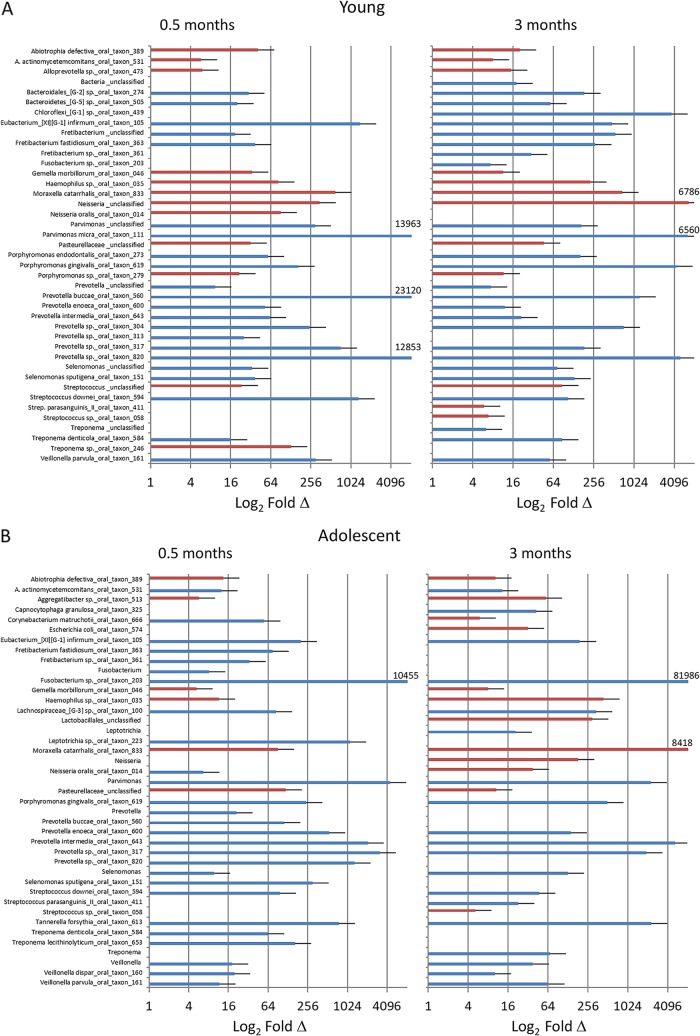
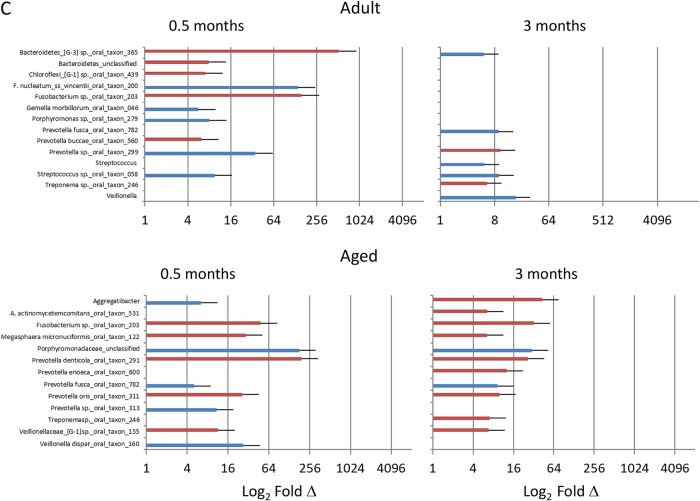
FIG 5.
Identification of OTU/species changes (≥5-fold [blue] or ≤5-fold [red]) in young (A), adolescent (B), and adult/aged (C) samples comparing baseline to disease initiation (0.5 months) and progression (3 months).
Comparison of the microbiomes during ligature-induced periodontitis occurring from 2 weeks through 3 months of disease is shown in Fig. 6A to D. Substantial differences in the levels of the top 50 OTUs were seen in these microbiomes during the disease process. This was particularly noticeable in the young and adolescent samples, with a much higher frequency of species demonstrating level changes of ≥5-fold. Specifically, the frequencies of 34/50 and 38/50 of this subset of species increased or decreased at this level in the young microbiome at 0.5 and 3 months, respectively. Similarly, 41/50 and 35/50 of the top 50 species in the adolescent animals changed from baseline to 0.5 and 3 months, respectively. These findings were compared with those for the microbiome samples from the adult and aged groups, where only 7 to 10 species specifically changed from baseline patterns. The details of the affected species are summarized in Fig. 7 and Table 1, with increases in species of Porphyromonas, Prevotella, Fusobacteria, Treponema, Tannerella, and Fretibacterium being observed in most age groups, consistent with human periodontitis microbiomes (8, 16, 17, 29).
FIG 6.
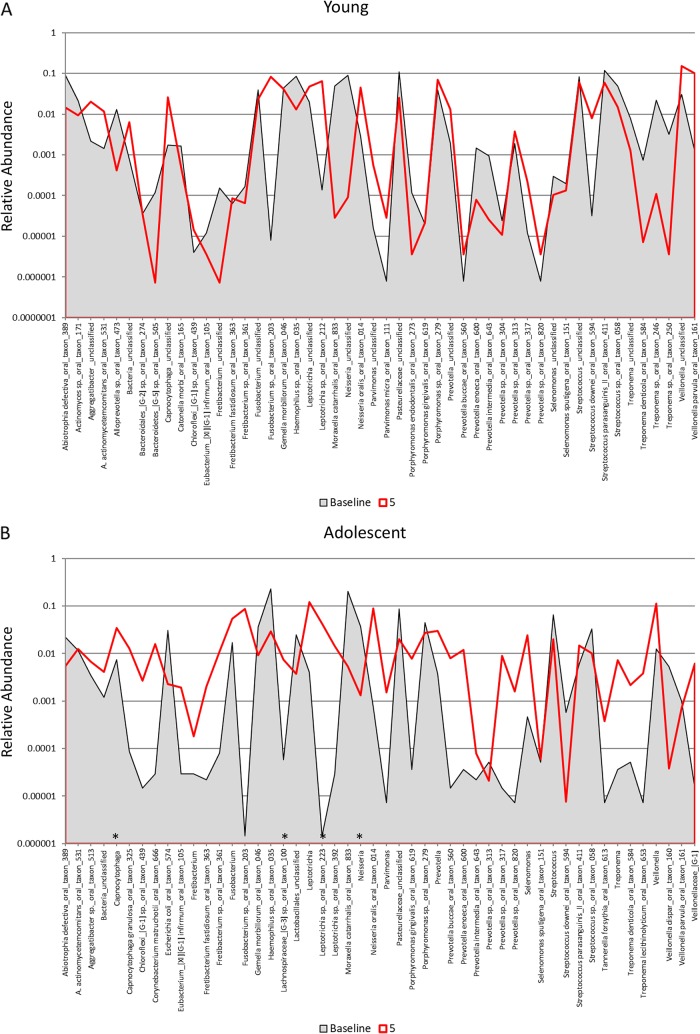
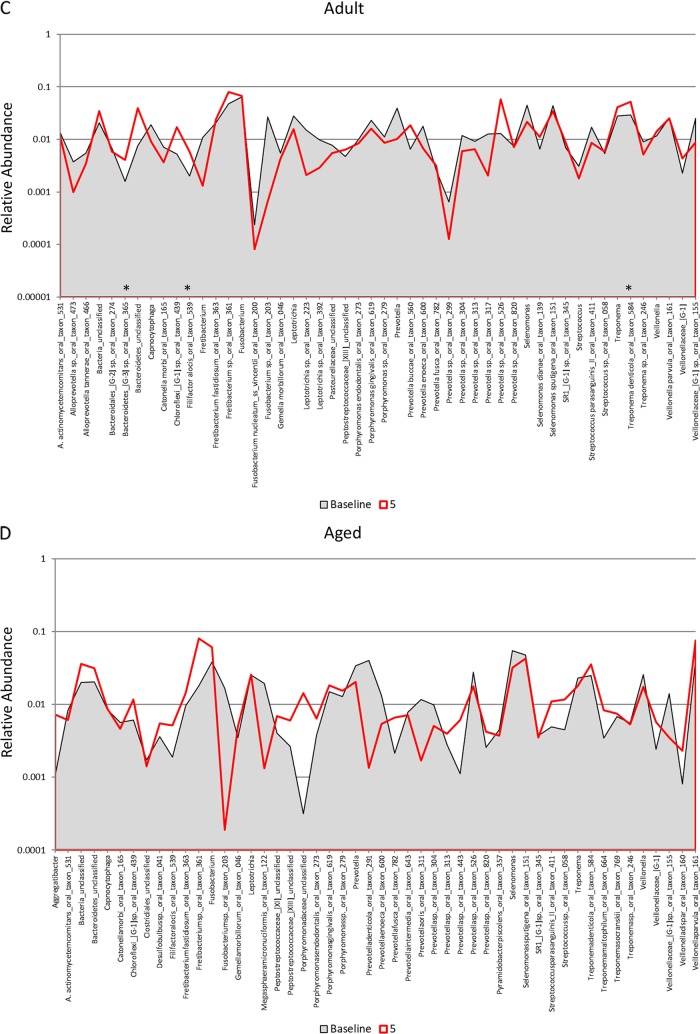
Distribution and relative abundance of signals for the top 50 OTUs in samples from young (A), adolescent (B), adult (C), and aged (D) animals. The plots compare the readouts for each OTU/species for baseline samples compared to samples collected 5 months and 2 months following removal of the ligatures with clinical resolution. The asterisks denote a significant difference from the baseline with resolution (5 months) at a P value of at least <0.05 using Z-score normalization.
FIG 7.
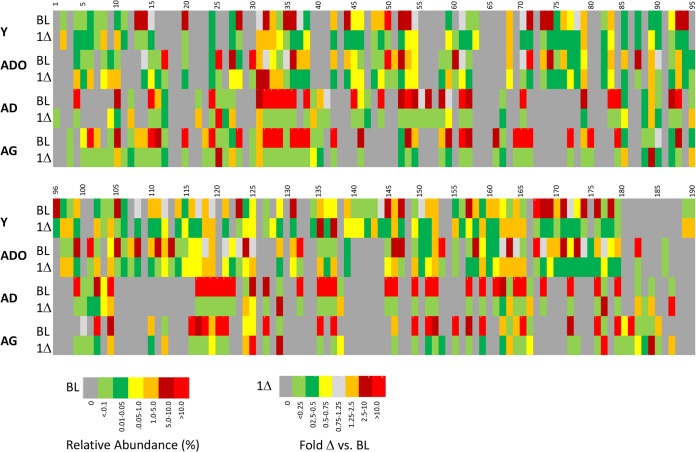
Heat map of relative abundances of OTU reads for all age groups at baseline and the change from baseline to 1 month of periodontitis (1Δ). The identifications of the bacterial OTUs are presented in Table 1.
TABLE 1.
Identification of bacterial OTUs from Fig. 7
| Identification | OTU |
|---|---|
| 1 | Abiotrophia defectiva_oral_taxon_389 |
| 2 | Actinomyces |
| 3 | Actinomyces_unclassified |
| 4 | Actinomyces sp._oral_taxon_171 |
| 5 | Actinomyces sp._oral_taxon_180 |
| 6 | Aggregatibacter_unclassified |
| 7 | Aggregatibacter actinomycetemcomitans_oral_taxon_531 |
| 8 | Aggregatibacter sp._oral_taxon_513 |
| 9 | Alloprevotella rava_oral_taxon_302 |
| 10 | Alloprevotella sp._oral_taxon_308 |
| 11 | Alloprevotella sp._oral_taxon_473 |
| 12 | Alloprevotella sp._oral_taxon_912 |
| 13 | Alloprevotella tannerae_oral_taxon_466 |
| 14 | Atopobium |
| 15 | Atopobium parvulum_oral_taxon_723 |
| 16 | Atopobium sp._oral_taxon_199 |
| 17 | Bacteria_unclassified |
| 18 | Bacteroidaceae_[G-1]_ sp._oral_taxon_272 |
| 19 | Bacteroidales_[G-2] sp._oral_taxon_274 |
| 20 | Bacteroidales_unclassified |
| 21 | Bacteroides |
| 22 | Bacteroides heparinolyticus_oral_taxon_630 |
| 23 | Bacteroidetes_[G-3] |
| 24 | Bacteroidetes_[G-3] sp._oral_taxon_280 |
| 25 | Bacteroidetes_[G-3] sp._oral_taxon_365 |
| 26 | Bacteroidetes_[G-5] sp._oral_taxon_505 |
| 27 | Bacteroidetes_[G-5] sp._oral_taxon_511 |
| 28 | Bacteroidetes_unclassified |
| 29 | Bergeyella_unclassified |
| 30 | Bergeyella sp._oral_taxon_907 |
| 31 | Burkholderiaceae_unclassified |
| 32 | Burkholderiales_unclassified |
| 33 | Butyrivibrio sp._oral_taxon_094 |
| 34 | Campylobacter_unclassified |
| 35 | Campylobacter rectus_oral_taxon_748 |
| 36 | Campylobacter sp._oral_taxon_044 |
| 37 | Capnocytophaga_unclassified |
| 38 | Capnocytophaga granulosa_oral_taxon_325 |
| 39 | Capnocytophaga sp._oral_taxon_326 |
| 40 | Cardiobacterium |
| 41 | Cardiobacterium hominis_oral_taxon_633 |
| 42 | Catonella morbi_oral_taxon_165 |
| 43 | Chloroflexi_[G-1] sp._oral_taxon_439 |
| 44 | Clostridiales_unclassified |
| 45 | Clostridiales_[F-1][G-1] sp._oral_taxon_093 |
| 46 | Clostridiales_[F-1][G-2] sp._oral_taxon_402 |
| 47 | Clostridiales_[F-2][G-1] sp._oral_taxon_075 |
| 48 | Clostridiales_[F-2][G-2] sp._oral_taxon_085 |
| 49 | Corynebacterium durum_oral_taxon_595 |
| 50 | Corynebacterium matruchotii_oral_taxon_666 |
| 51 | Desulfobulbus sp._oral_taxon_041 |
| 52 | Desulfomicrobium orale_oral_taxon_703 |
| 53 | Desulfovibrio fairfieldensis_oral_taxon_605 |
| 54 | Dialister invisus_oral_taxon_118 |
| 55 | Eikenella |
| 56 | Eikenella corrodens_oral_taxon_577 |
| 57 | Escherichia coli_oral_taxon_574 |
| 58 | Eubacterium_[XI][G-1] infirmum_oral_taxon_105 |
| 59 | Eubacterium_[XI][G-3] brachy_oral_taxon_557 |
| 60 | Filifactor alocis_oral_taxon_539 |
| 61 | Firmicutes_unclassified |
| 62 | Fretibacterium_unclassified |
| 63 | Fretibacterium fastidiosum_oral_taxon_363 |
| 64 | Fretibacterium sp._oral_taxon_361 |
| 65 | Fusobacterium_unclassified |
| 66 | Fusobacterium nucleatum_ss_vincentii_oral_taxon_200 |
| 67 | Fusobacterium periodonticum_oral_taxon_201 |
| 68 | Fusobacterium sp._oral_taxon_203 |
| 69 | Gemella morbillorum_oral_taxon_046 |
| 70 | Haemophilus_unclassified |
| 71 | Haemophilus sp._oral_taxon_035 |
| 72 | Johnsonella ignava_oral_taxon_635 |
| 73 | Johnsonella sp._oral_taxon_166 |
| 74 | Kingella sp._oral_taxon_459 |
| 75 | Lachnoanaerobaculum |
| 76 | Lachnoanaerobaculum sp._oral_taxon_083 |
| 77 | Lachnoanaerobaculum sp._oral_taxon_496 |
| 78 | Lachnoanaerobaculum umeaense_oral_taxon_107 |
| 79 | Lachnospiraceae_[G-2]_unclassified |
| 80 | Lachnospiraceae_[G-2] sp._oral_taxon_088 |
| 81 | Lachnospiraceae_[G-3] sp._oral_taxon_100 |
| 82 | Lachnospiraceae_[G-8] sp._oral_taxon_500 |
| 83 | Lachnospiraceae_[XIVa]_unclassified_unclassified |
| 84 | Lactobacillales_unclassified_unclassified |
| 85 | Lactobacillus salivarius_oral_taxon_756 |
| 86 | Leptotrichia_unclassified |
| 87 | Leptotrichia goodfellowii_oral_taxon_845 |
| 88 | Leptotrichia hongkongensis_oral_taxon_213 |
| 89 | Leptotrichia sp._oral_taxon_212 |
| 90 | Leptotrichia sp._oral_taxon_223 |
| 91 | Leptotrichia sp._oral_taxon_225 |
| 92 | Leptotrichia sp._oral_taxon_392 |
| 93 | Leptotrichiaceae_unclassified |
| 94 | Leptotrichiasp._oral_taxon_498 |
| 95 | Megasphaera |
| 96 | Megasphaeramicronuciformis_oral_taxon_122 |
| 97 | Mitsuokellasp._oral_taxon_131 |
| 98 | Mogibacterium vescum_oral_taxon_008 |
| 99 | Moraxella catarrhalis_oral_taxon_833 |
| 100 | Moryella sp._oral_taxon_097 |
| 101 | Mycoplasma |
| 102 | Neisseria_unclassified |
| 103 | Neisseria oralis_oral_taxon_014 |
| 104 | Neisseriaceae_unclassified_unclassified |
| 105 | Oribacterium_unclassified |
| 106 | Oribacterium sp._oral_taxon_078 |
| 107 | Parvimonas_unclassified |
| 108 | Parvimonas micra_oral_taxon_111 |
| 109 | Pasteurellaceae_unclassified |
| 110 | Peptococcus sp._oral_taxon_167 |
| 111 | Peptostreptococcaceae_[XI][G-1] sp._oral_taxon_383 |
| 112 | Peptostreptococcaceae_[XI][G-4] |
| 113 | Peptostreptococcaceae_[XI][G-5] sp._oral_taxon_493 |
| 114 | Peptostreptococcaceae_[XI]_unclassified |
| 121 | Porphyromonas endodontalis_oral_taxon_273 |
| 122 | Porphyromonas gingivalis_oral_taxon_619 |
| 123 | Porphyromonas sp._oral_taxon_279 |
| 124 | Prevotella_unclassified |
| 125 | Prevotella buccae_oral_taxon_560 |
| 126 | Prevotella dentalis_oral_taxon_583 |
| 127 | Prevotella enoeca_oral_taxon_600 |
| 128 | Prevotella fusca_oral_taxon_782 |
| 129 | Prevotella intermedia_oral_taxon_643 |
| 130 | Prevotella marshii_oral_taxon_665 |
| 131 | Prevotella oris_oral_taxon_311 |
| 132 | Prevotella saccharolytica_oral_taxon_781 |
| 133 | Prevotella sp._oral_taxon_299 |
| 134 | Prevotella sp._oral_taxon_304 |
| 135 | Prevotella sp._oral_taxon_313 |
| 136 | Prevotella sp._oral_taxon_315 |
| 137 | Prevotella sp._oral_taxon_317 |
| 138 | Prevotella sp._oral_taxon_376 |
| 139 | Prevotella sp._oral_taxon_443 |
| 140 | Prevotella sp._oral_taxon_526 |
| 141 | Prevotella sp._oral_taxon_820 |
| 142 | Prevotellabaroniae_oral_taxon_553 |
| 143 | Prevotelladenticola_oral_taxon_291 |
| 144 | Prevotellaoralis_oral_taxon_705 |
| 145 | Prevotellaoulorum_oral_taxon_288 |
| 153 | Solobacterium moorei_oral_taxon_678 |
| 154 | SR1_[G-1] |
| 155 | SR1_[G-1] sp._oral_taxon_345 |
| 156 | SR1_[G-1] sp._oral_taxon_874 |
| 157 | Streptococcus_unclassified |
| 158 | Streptococcus anginosus_oral_taxon_543 |
| 159 | Streptococcus downei_oral_taxon_594 |
| 160 | Streptococcus mutans_oral_taxon_686 |
| 161 | Streptococcus parasanguinis_II_oral_taxon_411 |
| 162 | Streptococcus sp._oral_taxon_058 |
| 163 | Syntrophomonadaceae_[VIII][G-1] sp._oral_taxon_435 |
| 164 | Tannerella |
| 165 | Tannerella forsythia_oral_taxon_613 |
| 167 | Treponema_unclassified |
| 168 | Treponema denticola_oral_taxon_584 |
| 169 | Treponema lecithinolyticum_oral_taxon_653 |
| 170 | Treponema maltophilum_oral_taxon_664 |
| 171 | Treponema socranskii_oral_taxon_769 |
| 172 | Treponema sp._oral_taxon_237 |
| 173 | Treponema sp._oral_taxon_239 |
| 174 | Treponema sp._oral_taxon_246 |
| 175 | Treponema sp._oral_taxon_249 |
| 176 | Treponema sp._oral_taxon_250 |
| 177 | Treponema sp._oral_taxon_255 |
| 178 | Treponema sp._oral_taxon_258 |
| 179 | Treponema sp._oral_taxon_263 |
| 180 | Treponema sp._oral_taxon_270 |
| 181 | Treponema sp._oral_taxon_508 |
| 182 | Veillonella_unclassified |
| 183 | Veillonella dispar_oral_taxon_160 |
| 184 | Veillonella parvula_oral_taxon_161 |
| 185 | Veillonella sp._oral_taxon_780 |
| 186 | Veillonellaceae_unclassified |
| 187 | Veillonellaceae_[G-1]_unclassified |
| 188 | Veillonellaceae_[G-1] sp._oral_taxon_129 |
| 189 | Veillonellaceae_[G-1] sp._oral_taxon_155 |
| 190 | Veillonellaceae_[G-1] sp._oral_taxon_145 |
Figure 3A to D provide an overview of the distribution of the top 50 OTUs, comparing baseline to periodontitis (3 mo.) and to the microbiome of resolved sites (after removal of the ligature [5 months]). Striking profile differences are noted between the changes across the age groups. The proportions of a large number of the species in the top 50 OTUs were lower at baseline and in resolved disease microbiomes in the young (n = 23) samples. In contrast, in the adolescent animals, microbiomes at baseline and 3 months postligation showed many similarities in reads for individual OTUs, while the proportions of 18 of the OTUs were elevated in the 5-month resolution samples. Both the adult and aged samples showed considerably fewer changes in overall OTU reads for the top 50 bacteria. Additionally, profiles for resolution were more similar to baseline patterns in the adults, while they trended toward the characteristics of the disease samples (3 months) in the aged animals. These differences are also reflected in Fig. 5, with significantly more frequent larger increases or decreases in the young and adolescent animals than in the adult and aged animals.
Since samples from the adult and aged groups demonstrated coverage with the top 50 OTUs that ranged from 60% to 86% at the individual-animal level (Fig. S2), we compared the 200 OTUs that predominated across the samples from all 4 groups and accounted for 85% to 98% of the total reads detected (Fig. S2). Important similarities in the profiles of baseline samples from young and adolescent animals, with high proportions of unclassified Streptococcus, Aggregatibacter actinomycetemcomitans, Haemophilus species, unclassified Pasteurellaceae, Streptococcus parasanguinis, Moraxella catarrhalis, and unclassified Neisseria bacteria, were also noted. Clear changes occurred in the diversity of the microbiome within 1 month of disease progression in the young and adolescent animals, including the emergence of Fusobacteria, Leptotrichia, and Veillonella at this time point. Estimation of the diversity of the microbiome also showed a somewhat less diverse microbiome in the disease samples from adult and aged animals. Interestingly, the adolescent samples appeared to move toward the diverse nature of the disease microbiomes exhibited by the adult and aged animals.
DISCUSSION
The human microbiome at various sites in/on the human body is gaining increasing importance with regard to overall health and disease (30–36). Additionally, individual gut bacterial species have been implicated in the development of normal host responses (32), nutritional needs (37), and helping to create microenvironments that are more resistant to pathogens (35, 38). However, these types of interactions between the host and resident bacteria have not been well elucidated related to the acquired microbiomes in the various niches of the oral cavity. There is a large amount of literature documenting the characteristics of the oral microbiome at various sites (e.g., tongue, mucosa, teeth, and supra- and subgingival areas) (17, 39–42) as well as defining characteristics of the subgingival microbial ecology of periodontal health and changes that occur with gingivitis and periodontitis (8), evolving from cultivation to identification of one or more targeted species, to next-generation sequencing linked to human oral microbiome databases (43–45). However, these studies have generally limited their focus to populations of adults.
The emerging importance of a more robust understanding of the variations in commensal bacteria within these consortia, which is crucial for disease resistance, and the role of microbiome dysbiosis driving disease through dysregulated host responses require additional examination (16). To accomplish some of these goals, we have implemented a nonhuman primate model of periodontal disease that has been clearly described to demonstrate clinical, microbiological, and immunological features of this disease in humans (46–48). Moreover, the characteristics of the disease in nonhuman primates, including Macaca fascicularis, Macaca mulatta, Macaca nemestrina, and Papio anubis, display features of naturally occurring human disease, such as the general lack of disease in younger individuals and an increased extent and severity of disease with aging (49–52). Additionally, this model has been extensively used to evaluated the kinetics and progression of disease using a ligature-induced periodontitis model (53) that has been adapted to rodents (54, 55) and rabbits (56). However, to more fully examine the microbiome changes that occur with disease initiation and progression, only nonhuman primates provide a “microbiomic” analogy to humans. Historically, we and others have documented characteristics of a targeted oral microbiota in primates with periodontal health and changes that occur in periodontitis (53, 57, 58). Nevertheless, rather minimal data are available documenting the breadth and depth of the oral microbiome in these animal species, although a recent report by Ocon et al. (25) clearly demonstrates the component similarity and genomic correspondence of the microbiomes between humans and macaque monkeys. As important to this investigation, the vast majority of these data have been derived from adult or aged monkeys, with little information on the oral microbiology of younger animals (59).
This investigation evaluated the oral microbiomes of healthy and diseased sites in nonhuman primates across the life span. We show that younger animals demonstrated substantial inflammation and bleeding following ligature placement and exhibited significantly lower pocketing at baseline and maximum destructive disease (i.e., probing pocket depth). This finding is consistent with our previous data demonstrating increased measures of inflammation and pocketing, albeit still considered clinically periodontally healthy, in older animals (58, 60, 61). However, the rates of change from baseline in the younger and older animals were similar over the initiation and progression of disease. In the current paradigm of periodontitis featuring a dysbiotic microbiome, the clinical differences with aging in nonhuman primates, even those with periodontal health, were reflected in microbiome differences at the phylum and species levels across the age groups. A striking finding was the increased microbiome diversity in the adult and aged animals versus younger animals, even at baseline with a clinical healthy periodontium, with only 20% to 30% of the top 200 OTUs represented in the young/adolescent animals and two-thirds of the OTUs represented in the adult/aged animals. This diversity difference remained in the young animals even after ligation, while the profiles in the adolescent animals began to approach 60% of the OTUs by 1 month postligation, similar to the maintenance of 60% to 70% of the OTUs during disease in the older animals. This type of finding has been reported for human periodontitis with lower diversity in disease samples, albeit findings vary (62–65). It has also been reported that while members of the oral microbiome may differ across individuals, the functional genomics of the bacterial population are more similar in health, with substantial changes in the gene expression profiles of commensals with disease (9, 20, 66).
Also observed was a rapid and extensive difference in the microbiome profiles from baseline to 2 weeks in the young and adolescent animals. This contrasted with changes, but an apparently more stable microbial ecology, in the adult and aged animals during the initiation of disease. This finding likely reflects the dynamics of the establishing oral microbiome in the young animals as well as developmental changes in the adolescent animals that do not occur in the older groups. Consistent with these findings would be a consideration of a more extensive malleability in the microbiome in young individuals that may reflect differences in maternal transmission of the microbiome components at the individual level (57) as well as be amenable to positive manipulation to enhance the establishment of a more disease-resistant microbiome, such as the suggested administration of pre- and probiotic therapeutics (67, 68).
As the disease initiated and progressed, differences in microbiome features were noted in all age groups. In young, adolescent, and adult animals, the proportions of 25% to 30% of the top 50 OTUs increased, while only 10% changed in the aged animals. Examination of existing literature from human studies suggested a similar difference in healthy and periodontitis samples, with the majority of the data being derived from patients most consistent with the adult group of the monkeys. A recent review by Belibasakis (69) suggested some alterations in the oral microbiome with aging, potentially related to altered immune capabilities, although definitive variations in microbiomes of oral diseases with aging are not dramatic in humans. Thus, two concepts derived from these findings suggest that a more dynamic periodontal microbiome is preferable and that an enhanced disease risk in aging may reflect the intransigence of the aged microbiome. Alternatively, the increased disease in aging individuals may be expressed due to the host responding to a very established inflexible microbiome and developing a less robust protective response to these “familiar” species. Further studies linking the characteristics of the host response with specific oral microbiome changes related to aging will need to be developed to address these options.
Finally, it is clear that standard nonsurgical therapy of scaling and root planing lowers the microbial burden in periodontitis and decreases inflammation as an approach to controlling the disease (44, 62, 64). However, while the majority of the population responds well to this treatment, a portion of the population continues to demonstrate disease, with little predictive understanding of who constitutes this “nonresponding” group (70–73). In humans, it is difficult to implement a prospective periodontitis protocol, and the longitudinal dynamics of the microbiome have generally been defined by comparing disease microbiomes to those posttreatment. While changes from disease to health may be identical to the changes occurring from health to disease, the limitations of the human model are that it cannot elucidate the earliest conversion in the microbiome that initiates a disease process, nor is it able to identify the early transition of the host response to the shifting microbiome that could differentiate susceptibility from resistance to progression to disease. Moreover, comparison of the microbiome characteristics with disease initiation and progression in different age groups of humans has not been reported and would be difficult to accomplish. Finally, in these human studies, an assumption is made that the microbiome of a treated site that is now “clinically improved” is fundamentally the same as the microbiome in a site that has never expressed disease. Regarding the findings for the microbiome in sites that have clinically resolved, overall, the resolved sites differed from baseline healthy sites prior to ligation. The results showed that the patterns of young samples appeared similar to baseline patterns, while the patterns of adolescent samples differed to a greater degree from the baseline patterns and appeared more similar to the profiles with disease. The profiles of the adult and aged microbiomes appeared more resistant to substantial changes reflected during both disease and resolution phases. These findings suggest an increased risk for future disease episodes at these sites that retained some dysbiotic traits, even with clinical improvement. The aging-related differences in the oral microbiome, even with periodontal health, including increases in both the diversity and abundance of bacterial species that have been associated with human periodontal disease (e.g., Porphyromonas, Treponema, and Prevotella, etc.), suggest that there could be a fundamentally increased risk for the onset of disease with aging microbiomes reflecting local environmental changes in the periodontal sulcus.
Using this nonhuman primate model, we provide some of the first data documenting the early and progressing disease microbiome changes, comparison of these differences across the life span, and evaluation of the microbiomes of healthy and disease-resolved sites. Critical questions remain regarding a gap in knowledge delineating the earliest microbiome and host transcriptome response changes in the local sites at the initiation of disease. An improved understanding of the kinetics and dynamics of this interaction might be of strategic value in the development of new therapeutic approaches to improve the toolbox of modalities and enable more evidence-based precision dentistry in periodontics.
MATERIALS AND METHODS
Nonhuman primate model and oral clinical evaluation.
Rhesus monkeys (Macaca mulatta) (n = 36; 19 females and 17 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. The animals were distributed by age into four groups (9/group): ≤3 years (young), 3 to 7 years (adolescent), 12 to 16 years (adult), and 18 to 23 years (aged). The nonhuman primates are typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified; Harlan Teklad). The diet is supplemented with fruits and vegetables, and water is provided ad libitum in an enclosed corral setting.
A protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico enabled anesthetized animals to be examined for clinical measures of periodontal health/disease, including probing pocket depth (PPD) and bleeding on probing (BOP), as we have described previously (74). Animals entered into the study population were determined to be periodontally healthy (BOP and PPD) using full-mouth measures with 4 sites/tooth. Clinical changes with ligature-induced disease were compared to baseline measures of all maxillary and mandibular premolars and 1st and 2nd molars that were subsequently the ligated teeth (see Fig. S3 in the supplemental material). The ligature-induced periodontitis model was implemented as we have described previously, by tying 3-0 silk sutures around the necks of maxillary and mandibular premolar and 1st- and 2nd-molar teeth in each animal (58, 61). As noted previously, removal of ligatures leads to decreases in inflammation and BOP to normal levels and stabilization of any pocket probing depths in the nonhuman primates (61). Thus, the clinical changes at 5 months (2 months after ligature removal) reflect a naturally occurring resolution without any treatment intervention. This is not dissimilar from the results with scaling and root planing in humans, in which the microbial burden and local inciting factors driving the destructive inflammatory response are modulated by the clinical treatment. Removal of the ligatures appears to exert a similar, if not identical, change in the local subgingival challenge. Bacterial samples were obtained by using a curette placed below the gingival margin and removing plaque samples, which were immediately placed in sterile phosphate-buffered saline (PBS) and stored at −80°C for further analysis, prior to clinical evaluation or gingival tissue sampling (58, 75).
Analysis of the oral microbiome.
Bacterial plaque samples in sterile PBS were centrifuged for 10 min at 14,000 × g to pellet the bacterial cells. DNA was extracted from the pelleted cells using MagNA Pure LC DNA isolation kit III (Roche Applied Science) in a MagNA Pure LC automated nucleic acid purification system (Roche Applied Science). DNA was quantified using a Nanodrop instrument (Thermo Fisher).
A dual-index paired-end sequencing approach for sequencing (76) using a MiSeq sequencer was employed to determine the total composition of the oral microbiota from each sample. First, DNA from each sample was amplified using 16S universal primers, which amplify 254 bp from the V4 region of the 16S rRNA gene. Each primer consisted of the appropriate Illumina adapter, an 8-nucleotide (nt) index sequence to identify each sample, a 10-nt pad sequence, a 2-nt linker, and the gene-specific 16S forward or 16S reverse primer sequence. The resulting 254-bp 16S rRNA gene amplicons generated from each sample were purified and pooled in equimolar concentrations. These amplicon libraries were mixed with Illumina-generated PhiX control libraries and used for shotgun library construction using the Nextera XT genomic library construction protocol. Sequencing of each DNA fragment was done using two sequence reads and two index reads by three sequence primers. The overall process of cluster generation, sequencing, image processing, demultiplexing, and quality score calculation was performed on the MiSeq instrument (Illumina).
Barcoded sequences from the MiSeq run were trimmed and aligned to similar 16S rRNA sequences from the SILVA v4 rRNA database (https://www.arb-silva.de/) using the MOTHUR (v.1.38.0) pipeline (77). Artificial erroneous reads were corrected using the pre.cluster mothur command, and chimeric sequences were removed using the UCHIME (78) command in the MOTHUR pipeline, followed by removal of nonarchaeal/bacterial sequences. Sequences were clustered into phylotypes based on their sequence similarity, and these binned phylotypes were assigned to their respective taxonomic classifications using the Human Oral Microbiome Database (HOMD V13) (http://www.homd.org/index.php?name=seqDownload&file&type=R). As noted, we used the HOMD to classify the OTUs. Since the study classified bacteria in the oral microbiome of rhesus monkeys by comparing them to bacterial species present in humans (i.e., HOMD), there were bacterial sequences identified at the genus level but unclassified by further species comparison since they are not sufficiently similar to existing entries present in the HOMD. The cutoff percentage used in the MOTHUR software (classify.seqs command) for classifying sequences is 80%, which may leave some OTUs unclassified or classified only at the genus level.
The nonparametric species richness estimates ACE and Chao1 and diversity indexes (Shannon and Simpson) were calculated using the MOTHUR pipeline. Goods’ coverage and species richness (observed phylotypes) were also calculated. Rarefaction curves and principal-coordinate analysis (PCoA) plots were also drawn to show α-diversity within the samples and β-diversity between the samples. Statistical differences of bacterial OTUs were determined with the t test at a P value of at least <0.05.
Data availability.
The raw data were deposited at the NIH NCBI under BioProject accession number PRJNA516659.
Supplementary Material
ACKNOWLEDGMENTS
We thank the technical support personnel of the UK Healthcare Genomics Core Laboratory in the College of Medicine at the University of Kentucky for analysis of the samples.
This work was supported by grants P30 GM103538 and P20 GM110788 and the Center for Oral Health Research in the University of Kentucky College of Dentistry.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00067-19.
REFERENCES
- 1.Leite FRM, Peres KG, Do LG, Demarco FF, Peres MAA. 2017. Prediction of periodontitis occurrence: influence of classification and sociodemographic and general health information. J Periodontol 88:731–743. doi: 10.1902/jop.2017.160607. [DOI] [PubMed] [Google Scholar]
- 2.Eke PI, Wei L, Thornton-Evans GO, Borrell LN, Borgnakke WS, Dye B, Genco RJ. 2016. Risk indicators for periodontitis in US adults: NHANES 2009 to 2012. J Periodontol 87:1174–1185. doi: 10.1902/jop.2016.160013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonetti MS, Chapple IL, Jepsen S, Sanz M. 2015. Primary and secondary prevention of periodontal and peri-implant diseases: introduction to, and objectives of the consensus from the 11th European Workshop on Periodontology consensus conference. J Clin Periodontol 42(Suppl 16):S1–S4. doi: 10.1111/jcpe.12382. [DOI] [PubMed] [Google Scholar]
- 4.Loos BG, Papantonopoulos G, Jepsen S, Laine ML. 2015. What is the contribution of genetics to periodontal risk? Dent Clin North Am 59:761–780. doi: 10.1016/j.cden.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Lopez R, Hujoel P, Belibasakis GN. 2015. On putative periodontal pathogens: an epidemiological perspective. Virulence 6:249–257. doi: 10.1080/21505594.2015.1014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol 80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, Tonetti MS, Wade WG, Zaura E. 2016. The oral microbiome—an update for oral healthcare professionals. Br Dent J 221:657–666. doi: 10.1038/sj.bdj.2016.865. [DOI] [PubMed] [Google Scholar]
- 9.Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. 2015. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med 7:27. doi: 10.1186/s13073-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyle J, Chapple I. 2015. Molecular aspects of the pathogenesis of periodontitis. Periodontol 2000 69:7–17. doi: 10.1111/prd.12104. [DOI] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz PI, Hoare A, Hong BY. 2016. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc 44:421–435. [PubMed] [Google Scholar]
- 13.Belstrom D, Holmstrup P, Bardow A, Kokaras A, Fiehn NE, Paster BJ. 2016. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J Oral Microbiol 8:30112. doi: 10.3402/jom.v8.30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szafranski SP, Wos-Oxley ML, Vilchez-Vargas R, Jáuregui R, Plumeier I, Klawonn F, Tomasch J, Meisinger C, Kühnisch J, Sztajer H, Pieper DH, Wagner-Döbler I. 2015. High-resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Appl Environ Microbiol 81:1047–1058. doi: 10.1128/AEM.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. mBio 5:e01012-14. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res 69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Feres M, Teles F, Teles R, Figueiredo LC, Faveri M. 2016. The subgingival periodontal microbiota of the aging mouth. Periodontol 2000 72:30–53. doi: 10.1111/prd.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudhakara P, Gupta A, Bhardwaj A, Wilson A. 2018. Oral dysbiotic communities and their implications in systemic diseases. Dent J (Basel) 6:E10. doi: 10.3390/dj6020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, Wang GP. 2015. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol 81:783–793. doi: 10.1128/AEM.02712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. 2014. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J 8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darveau RP. 2009. The oral microbial consortium’s interaction with the periodontal innate defense system. DNA Cell Biol 28:389–395. doi: 10.1089/dna.2009.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon DR, Reife RA, Cebra JJ, Darveau RP. 2004. Commensal bacteria influence innate status within gingival tissues: a pilot study. J Periodontol 75:1486–1492. doi: 10.1902/jop.2004.75.11.1486. [DOI] [PubMed] [Google Scholar]
- 24.Colombo APV, Paster BJ, Grimaldi G, Lourenco TGB, Teva A, Campos-Neto A, McCluskey J, Kleanthous H, Van Dyke TE, Stashenko P. 2017. Clinical and microbiological parameters of naturally occurring periodontitis in the non-human primate Macaca mulatta. J Oral Microbiol 9:1403843. doi: 10.1080/20002297.2017.1403843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ocon S, Murphy C, Dang AT, Sankaran-Walters S, Li CS, Tarara R, Borujerdpur N, Dandekar S, Paster BJ, George MD. 2013. Transcription profiling reveals potential mechanisms of dysbiosis in the oral microbiome of rhesus macaques with chronic untreated SIV infection. PLoS One 8:e80863. doi: 10.1371/journal.pone.0080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bimstein E, Huja PE, Ebersole JL. 2013. The potential lifespan impact of gingivitis and periodontitis in children. J Clin Pediatr Dent 38:95–99. doi: 10.17796/jcpd.38.2.j525742137780336. [DOI] [PubMed] [Google Scholar]
- 27.Bimstein E, Matsson L. 1999. Growth and development considerations in the diagnosis of gingivitis and periodontitis in children. Pediatr Dent 21:186–191. [PubMed] [Google Scholar]
- 28.Bimstein E, Ebersole JL. 1989. The age-dependent reaction of the periodontal tissues to dental plaque. ASDC J Dent Child 56:358–362. [PubMed] [Google Scholar]
- 29.Oliveira RR, Fermiano D, Feres M, Figueiredo LC, Teles FR, Soares GM, Faveri M. 2016. Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res 95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Jazwinski SM. 2018. The gut microbiota and healthy aging: a mini-review. Gerontology 64:513–520. doi: 10.1159/000490615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuong HE, Hsiao EY. 2017. Emerging roles for the gut microbiome in autism spectrum disorder. Biol Psychiatry 81:411–423. doi: 10.1016/j.biopsych.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takiishi T, Fenero CIM, Camara NOS. 2017. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers 5:e1373208. doi: 10.1080/21688370.2017.1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paun A, Yau C, Danska JS. 2017. The influence of the microbiome on type 1 diabetes. J Immunol 198:590–595. doi: 10.4049/jimmunol.1601519. [DOI] [PubMed] [Google Scholar]
- 34.Mulligan CM, Friedman JE. 2017. Maternal modifiers of the infant gut microbiota: metabolic consequences. J Endocrinol 235:R1–R12. doi: 10.1530/JOE-17-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 36.Paun A, Danska JS. 2016. Modulation of type 1 and type 2 diabetes risk by the intestinal microbiome. Pediatr Diabetes 17:469–477. doi: 10.1111/pedi.12424. [DOI] [PubMed] [Google Scholar]
- 37.Buccigrossi V, Nicastro E, Guarino A. 2013. Functions of intestinal microflora in children. Curr Opin Gastroenterol 29:31–38. doi: 10.1097/MOG.0b013e32835a3500. [DOI] [PubMed] [Google Scholar]
- 38.Partida-Rodríguez O, Serrano-Vázquez A, Nieves-Ramírez ME, Moran P, Rojas L, Portillo T, González E, Hernández E, Finlay BB, Ximenez C. 2017. Human intestinal microbiota: interaction between parasites and the host immune response. Arch Med Res 48:690–700. doi: 10.1016/j.arcmed.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Gomez A, Nelson KE. 2017. The oral microbiome of children: development, disease, and implications beyond oral health. Microb Ecol 73:492–503. doi: 10.1007/s00248-016-0854-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakic M, Grusovin MG, Canullo L. 2016. The microbiologic profile associated with peri-implantitis in humans: a systematic review. Int J Oral Maxillofac Implants 31:359–368. doi: 10.11607/jomi.4150. [DOI] [PubMed] [Google Scholar]
- 41.Moon JH, Lee JH. 2016. Probing the diversity of healthy oral microbiome with bioinformatics approaches. BMB Rep 49:662–670. doi: 10.5483/BMBRep.2016.49.12.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaura E, Nicu EA, Krom BP, Keijser BJ. 2014. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol 4:85. doi: 10.3389/fcimb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. 2013. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000 62:95–162. doi: 10.1111/prd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst FE, Paster BJ. 2012. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol 83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Socransky SS, Haffajee AD, Smith C, Dibart S. 1991. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol 18:766–775. doi: 10.1111/j.1600-051X.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 46.Madden TE, Caton JG. 1994. Animal models for periodontal disease. Methods Enzymol 235:106–119. doi: 10.1016/0076-6879(94)35135-X. [DOI] [PubMed] [Google Scholar]
- 47.Oz HS, Puleo DA. 2011. Animal models for periodontal disease. J Biomed Biotechnol 2011:754857. doi: 10.1155/2011/754857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graves DT, Kang J, Andriankaja O, Wada K, Rossa C Jr. 2012. Animal models to study host-bacteria interactions involved in periodontitis. Front Oral Biol 15:117–132. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schou S, Holmstrup P, Kornman KS. 1993. Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J Periodontol 64:497–508. doi: 10.1902/jop.1993.64.6.497. [DOI] [PubMed] [Google Scholar]
- 50.Miller DR, Aufdemorte TB, Fox WC, Waldrop TC, Mealey BL, Brunsvold MA. 1995. Periodontitis in the baboon: a potential model for human disease. J Periodontal Res 30:404–409. doi: 10.1111/j.1600-0765.1995.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 51.Persson GR, Engel LD, Moncla BJ, Page RC. 1993. Macaca nemestrina: a non-human primate model for studies of periodontal disease. J Periodontal Res 28:294–300. doi: 10.1111/j.1600-0765.1993.tb02096.x. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez OA, Novak MJ, Kirakodu S, Stromberg AJ, Shen S, Orraca L, Gonzalez-Martinez J, Ebersole JL. 2013. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis 18:249–259. doi: 10.1007/s10495-013-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornman KS, Holt SC, Robertson PB. 1981. The microbiology of ligature-induced periodontitis in the cynomolgus monkey. J Periodontal Res 16:363–371. doi: 10.1111/j.1600-0765.1981.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 54.Liang S, Hosur KB, Domon H, Hajishengallis G. 2010. Periodontal inflammation and bone loss in aged mice. J Periodontal Res 45:574–578. doi: 10.1111/j.1600-0765.2009.01245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grauballe MB, Belstrom D, Ostergaard JA, Paster BJ, Schou S, Flyvbjerg A, Holmstrup P. 2017. Ligature-associated bacterial profiles are linked to type 2 diabetes mellitus in a rat model and influenced by antibody treatment against TNF-alpha or RAGE. Clin Exp Dent Res 3:25–31. doi: 10.1002/cre2.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasturk H, Kantarci A, Ebrahimi N, Andry C, Holick M, Jones VL, Van Dyke TE. 2006. Topical H2 antagonist prevents periodontitis in a rabbit model. Infect Immun 74:2402–2414. doi: 10.1128/IAI.74.4.2402-2414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebersole JL, Holt SC, Delaney JE. 2014. Acquisition of oral microbes and associated systemic responses of newborn nonhuman primates. Clin Vaccine Immunol 21:21–28. doi: 10.1128/CVI.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds MA, Dawson DR, Novak KF, Ebersole JL, Gunsolley JC, Branch-Mays GL, Holt SC, Mattison JA, Ingram DK, Novak MJ. 2009. Effects of caloric restriction on inflammatory periodontal disease. Nutrition 25:88–97. doi: 10.1016/j.nut.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez OA, Nagarajan R, Novak MJ, Orraca L, Gonzalez-Martinez JA, Kirakodu SS, Ebersole JL. 2016. Immune system transcriptome in gingival tissues of young nonhuman primates. J Periodontal Res 51:152–163. doi: 10.1111/jre.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebersole JL, Steffen MJ, Reynolds MA, Branch-Mays GL, Dawson DR, Novak KF, Gunsolley JC, Mattison JA, Ingram DK, Novak MJ. 2008. Differential gender effects of a reduced-calorie diet on systemic inflammatory and immune parameters in nonhuman primates. J Periodontal Res 43:500–507. doi: 10.1111/j.1600-0765.2008.01051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Branch-Mays GL, Dawson DR, Gunsolley JC, Reynolds MA, Ebersole JL, Novak KF, Mattison JA, Ingram DK, Novak MJ. 2008. The effects of a calorie-reduced diet on periodontal inflammation and disease in a non-human primate model. J Periodontol 79:1184–1191. doi: 10.1902/jop.2008.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mombelli A. 2018. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol 2000 76:85–96. doi: 10.1111/prd.12147. [DOI] [PubMed] [Google Scholar]
- 63.Sabharwal A, Ganley K, Miecznikowski JC, Haase EM, Barnes V, Scannapieco FA. 2018. The salivary microbiome of diabetic and non-diabetic adults with periodontal disease. J Periodontol 90:26–34. doi: 10.1002/JPER.18-0167. [DOI] [PubMed] [Google Scholar]
- 64.Liu G, Luan Q, Chen F, Chen Z, Zhang Q, Yu X. 2018. Shift in the subgingival microbiome following scaling and root planing in generalized aggressive periodontitis. J Clin Periodontol 45:440–452. doi: 10.1111/jcpe.12862. [DOI] [PubMed] [Google Scholar]
- 65.Chen C, Hemme C, Beleno J, Shi ZJ, Ning D, Qin Y, Tu Q, Jorgensen M, He Z, Wu L, Zhou J. 2018. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J 12:1210–1224. doi: 10.1038/s41396-017-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frias-Lopez J, Duran-Pinedo A. 2012. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol 194:2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pradeep K, Kuttappa MA, Prasana KR. 2014. Probiotics and oral health: an update. SADJ 69:20–24. [PubMed] [Google Scholar]
- 68.Zarco MF, Vess TJ, Ginsburg GS. 2012. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis 18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 69.Belibasakis GN. 2018. Microbiological changes of the ageing oral cavity. Arch Oral Biol 96:230–232. doi: 10.1016/j.archoralbio.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Slots J. 2017. Periodontitis: facts, fallacies and the future. Periodontol 2000 75:7–23. doi: 10.1111/prd.12221. [DOI] [PubMed] [Google Scholar]
- 71.Cobb CM. 2008. Microbes, inflammation, scaling and root planing, and the periodontal condition. J Dent Hyg 82:4–9. [PubMed] [Google Scholar]
- 72.Darby IB, Hodge PJ, Riggio MP, Kinane DF. 2005. Clinical and microbiological effect of scaling and root planing in smoker and non-smoker chronic and aggressive periodontitis patients. J Clin Periodontol 32:200–206. doi: 10.1111/j.1600-051X.2005.00644.x. [DOI] [PubMed] [Google Scholar]
- 73.Nagarajan R, Miller CS, Dawson D III, Ebersole JL. 2019. Biologic modelling of periodontal disease progression. J Clin Periodontol 46:160–169. doi: 10.1111/jcpe.13064. [DOI] [PubMed] [Google Scholar]
- 74.Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. 2008. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol 15:1067–1075. doi: 10.1128/CVI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Novak MJ, Novak KF, Hodges JS, Kirakodu S, Govindaswami M, Diangelis A, Buchanan W, Papapanou PN, Michalowicz BS. 2008. Periodontal bacterial profiles in pregnant women: response to treatment and associations with birth outcomes in the obstetrics and periodontal therapy (OPT) study. J Periodontol 79:1870–1879. doi: 10.1902/jop.2008.070554. [DOI] [PubMed] [Google Scholar]
- 76.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data were deposited at the NIH NCBI under BioProject accession number PRJNA516659.