Legionella pneumophila and other Legionella species replicate intracellularly using the Icm/Dot type IV secretion system. In L. pneumophila this system translocates >300 effectors into host cells and in the Legionella genus thousands of effectors were identified, the function of most of which is unknown.
KEYWORDS: Icm/Dot, Legionella, PI3P-binding domains, effectors
ABSTRACT
Legionella pneumophila and other Legionella species replicate intracellularly using the Icm/Dot type IV secretion system. In L. pneumophila this system translocates >300 effectors into host cells and in the Legionella genus thousands of effectors were identified, the function of most of which is unknown. Fourteen L. pneumophila effectors were previously shown to specifically bind phosphoinositides (PIs) using dedicated domains. We found that PI-binding domains of effectors are usually not homologous to one another; they are relatively small and located at the effectors' C termini. We used the previously identified Legionella effector domains (LEDs) with unknown function and the above characteristics of effector PI-binding domains to discover novel PI-binding LEDs. We identified three predicted PI-binding LEDs that are present in 14 L. pneumophila effectors and in >200 effectors in the Legionella genus. Using an in vitro protein-lipid overlay assay, we found that 11 of these L. pneumophila effectors specifically bind phosphatidylinositol 3-phosphate (PI3P), almost doubling the number of L. pneumophila effectors known to bind PIs. Further, we identified in each of these newly discovered PI3P-binding LEDs conserved, mainly positively charged, amino acids that are essential for PI3P binding. Our results indicate that Legionella effectors harbor unique domains, shared by many effectors, which directly mediate PI3P binding.
INTRODUCTION
Legionella pneumophila is a Gram-negative bacterial species found in aquatic environments (1). In nature, L. pneumophila multiplies inside a variety of protozoan cells that serve as its environmental reservoir (2, 3). When people inhale aerosols containing L. pneumophila, the bacteria infect alveolar macrophages and can cause a life threatening pneumonia called Legionnaires’ disease (4, 5). Instead of being degraded by defense mechanisms of protozoa and human macrophages, L. pneumophila subverts host cell processes and grows within a specialized Legionella-containing vacuole (LCV) (6, 7). To establish the LCV inside eukaryotic cells, L. pneumophila modulates host cell functions using the Icm/Dot type IV secretion system. The Icm/Dot system is essential for L. pneumophila pathogenesis, since it delivers effector proteins into the host cells, and these effectors directly modulate various host cell processes, resulting in the biogenesis of the LCV (reviewed in references 8 to 12). The genome of L. pneumophila encodes more than 300 different effectors and in most cases the deletion of individual effector-encoding genes, or even groups of several effector-encoding genes in a single mutant, has a modest effect on the ability of the bacteria to survive and multiply in both amoeba and mammalian host cells (13–15). The function of most of the L. pneumophila effectors remains unknown, but several effectors were shown to modulate important cellular processes, including vesicular trafficking, apoptosis, ubiquitination, autophagy, signal transduction, lipid metabolism, host gene expression, and others (reviewed in references 12, 16, and 17).
Phosphoinositides (PIs) are low-abundance phospholipids, composed of diacylglycerol (DAG) and phosphatidylinositol. The carbohydrate moiety of PI compounds can be phosphorylated at the positions 3′, 4′, and/or 5′, thus forming seven different PI lipids. Distinct PIs, together with other regulatory factors, define organelle identity, such as phosphatidylinositol 3-phosphate (PI3P), which is present mainly on early endosomes, and phosphatidylinositol 4-phosphate (PI4P), which is enriched at the Golgi complex (18, 19). In addition, PIs play a major role in the trafficking processes between organelles and are crucial regulators of eukaryotic signal transduction (18, 19). PIs mediate their functions in part through the binding of their head groups to cytosolic proteins or cytosolic domains of membrane proteins. Thus, they can regulate and/or recruit proteins to specific organelles. Typically, the binding of proteins to PIs involves electrostatic interactions with the negative charges of the phosphate(s) on the inositol ring. Specific PI-binding domains have been described in eukaryotic proteins (20–22), some of which specifically bind PI3P or PI4P. The FYVE domain (named after the four proteins harboring the domain: Fab1p, YOTB, Vac1p, and EEA1) is between 60 and 80 residues long, and its PI3P-binding site is formed by the positively charged amino acid residues in the conserved WxxD, (R/K)(R/K)HHCR, and RVC sequence (23–25). The Phox homology (PX) domain is generally between 110 and 140 residues long, and its consensus binding motif for PI3P binding is R(Y/F)x23–30Kx13–23R (26, 27). The Pleckstrin homology (PH) domain is a widespread domain family harbored by a variety of proteins which bind different PIs, including PI4P; it is generally between 100 and 120 residues long and is highly homologous in terms of three-dimensional structure and secondary structure, and the interaction with PIs is mediated by lysine and arginine residues (28, 29).
Beside their important role in cellular organelle identity and signal transduction, PIs also play a major role during bacterial infection (30). Several L. pneumophila effectors, most of which contribute to the establishment of the LCV, were shown to bind specific PIs, usually PI3P or PI4P (31). The first effector which was reported to bind PI4P was SidC (32), which was followed by SidM, and other effectors that all bind specifically to PI4P (33). In addition, several effectors, most of which are localized to the LCV, were shown to bind PI3P, and two effectors were shown to bind both PI3P and PI4P (see Table 1 and references therein). Altogether, 14 L. pneumophila effectors were shown to bind specific PIs, but in most cases the domains required for PI binding were found not homologous to one another. Only three L. pneumophila effectors that bind PI4P (SidM, Lem4, and Lem28) have a C-terminal homologous domain (here named PI4P-M). The two homologous effectors SidC and SdcA bind PI4P via the same domain (here named PI4P-C), which is not homologous to the PI4P-M domain. Other effectors that bind PI3P or both PI3P and PI4P are not homologous to one another, not even at their PI-binding domain. It should be noted that PI3P and PI4P are present on the LCV, and their temporal and spatial dynamics are involved in LCV formation. The L. pneumophila wild-type strain was found to acquire PI3P within a minute after uptake by Dictyostelium discoideum and to gradually lose this PI within 2 h. During these 2 h, PI4P steadily accumulated on the LCV of wild-type L. pneumophila and was maintained on it for at least 8 h (34). The temporal localization of these two PIs is likely to dictate the time postinfection in which specific effectors are localized to the LCV.
TABLE 1.
PI4P- and PI3P-binding L. pneumophila effectors
| Locus taga | Name | Size (aa) | Position of PI-BD (aa)b | PI bindingc | Localizationd | Effector function | Reference(s) |
|---|---|---|---|---|---|---|---|
| lpg0160 | RavD | 325 | 190–325 | PI3P | LCV | Helps to prevent encounters of the LCV with lysosomes | 59 |
| lpg0695 | AnkX | 949 | 491–949 | PI3P, PI4P | LCV | Phosphocholinates Rab1 and Rab35 | 60–64 |
| lpg0940 | LidA | 729 | 60–180 | PI3P, PI4P | LCV | Modulates Rab1 to render it persistently active | 33, 65–69 |
| lpg1101 | Lem4 | 322 | 201–322 | PI4P | LCV | Phosphotyrosine phosphatase | 70, 71 |
| lpg1683 | RavZ | 502 | 329–423 | PI3P | AP | Irreversible deconjugation of Atg8/LC3 | 40, 42, 72 |
| lpg1978 | SetA | 644 | 401–644 | PI3P | LCV | Glucosyltransferase, subverts vesicle trafficking | 73, 74 |
| lpg2222 | LpnE | 375 | ND | PI3P | LCV | Binds polyphosphate 5-phosphatase OCRL1 | 75–77 |
| lpg2311 | RidL | 1,167 | 867–1167 | PI3P | LCV | Binds the retromer subunit Vps29 and inhibits retrograde trafficking | 78–81 |
| lpg2464 | SidM/DrrA | 647 | 544–647 | PI4P | LCV | AMPylates Rab1 and Rab35, activates Rab1 | 33, 82–85 |
| lpg2510 | SdcA | 908 | 605–771 | PI4P | LCV | E3 ubiquitin ligase, recruits ER to the LCV | 32 |
| lpg2511 | SidC | 917 | 609–776 | PI4P | LCV | E3 ubiquitin ligase, recruits ER to the LCV | 32, 43, 86–90 |
| lpg2603 | Lem28 | 434 | 326–434 | PI4P | LCV | ND | 70 |
| lpw03701 | LtpD | 679 | 472–626 | PI3P | LCV | Binds a phosphatase involved in PI production | 91 |
| lpp0356 | LtpM | 639 | 469–639 | PI3P | LCV | Glycosyltransferase | 92 |
lpp0356 is found in L. pneumophila strain Paris, and lpw03701 is found in L. pneumophila strain 130b and not in L. pneumophila strain Philadelphia-1.
PI-BD, phosphoinositide-binding domain; ND, not determined.
PI3P, phosphatidylinositol 3-phosphate; PI4P, phosphatidylinositol 4-phosphate.
LCV, Legionella-containing vacuole; AP, autophagosomes.
The Legionella genus includes more than 50 species, most of which are pathogenic, and they all contain the conserved Icm/Dot type IV secretion system (35, 36). In a previous study, we predicted thousands of effectors in 41 Legionella species, and this high number of effectors made it possible to identify conserved effector domains in these Legionella species (35). Legionella effector domains (LEDs) are conserved across effector orthologous groups, but they are not present in other known proteins. LEDs represent the building blocks of numerous Legionella effectors, yet their function is unknown. We were interested to determine whether some of these LEDs function as PI-binding domains. To this end, we looked for LEDs which have properties similar to known PI-binding domains in effectors. We found three such LEDs, which are present in a dozen L. pneumophila effectors and in more than 200 predicted effectors in the 41 Legionella species examined. Examination of these L. pneumophila effectors for PI binding revealed that they specifically bind PI3P, thus almost doubling the number of L. pneumophila effectors that bind PIs and establishing the importance of LEDs for discovering effector function.
RESULTS
Identification of common features of L. pneumophila effector PI-binding domains.
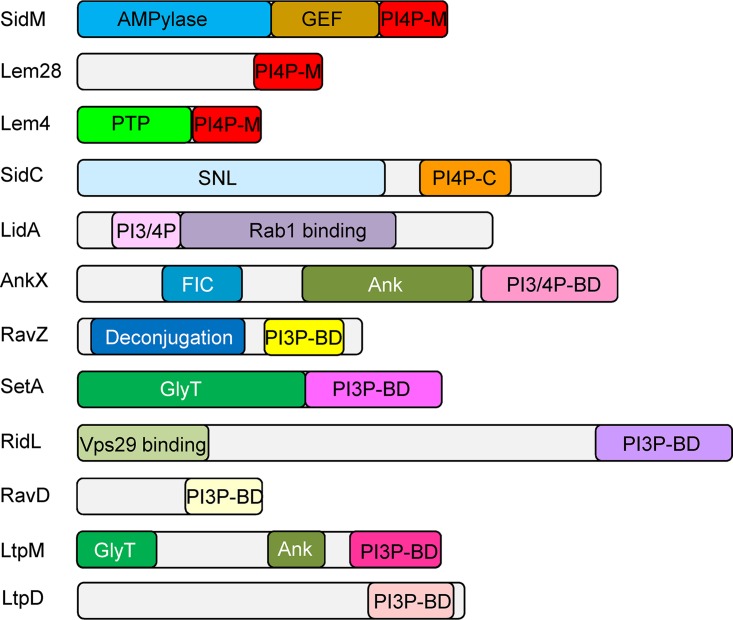
Currently there are 14 L. pneumophila effectors which were shown to specifically bind PI3P and/or PI4P (Table 1 and references therein). In most cases the domains which bind the same PI (or different PIs) are not homologous, suggesting that this function was developed multiple times during evolution. Analysis of 12 L. pneumophila effectors for which the PI-binding domain was characterized to some extent (Table 1 and Fig. 1) revealed several common features of these nonhomologous PI3P or PI4P effector-binding domains: (i) most of these domains are located at the C-terminal ends of the effectors (all except in LidA), (ii) these domains are usually small and comprise about 100 amino acids (similar to the eukaryotic PI-binding domains), and (iii) most of the effectors harboring these PI-binding domains also harbor either catalytic or protein binding domains at their N-terminal regions (Fig. 1). We used these three features to discover novel PI-binding LEDs in Legionella effectors, as described below.
FIG 1.
Domain architecture of L. pneumophila effectors that bind PIs. The known domains of each effector that contains a PI-binding domain are shown. The domains presented are as follows: PI4P-M, PI4P-binding domain of SidM; PI4P-C, PI4P-binding domain of SidC; PI3P-BD, PI3P-binding domain; PI3/4P-BD, PI3P and PI4P-binding domain; deconjugation, a domain related to cysteine proteases, required for Atg8 deconjugation reaction; AMPylase, AMPylation of Rab1; GEF, GTP-GDP exchange factor for Rab1; GlyT, glycosyltransferase; SNL, N-terminal ubiquitin ligase domain; FIC, filamentation induced by cAMP, used by AnkX for phosphocholination of Rab1; Ank, ankyrin repeat; PTP, protein tyrosine phosphatase; Vps29 binding, domain that binds the Vps29 retromer subunit. Domains with the same name and different colors perform the same function but are not homologous.
Identification of LEDs whose features resemble the features of known PI-binding domains.
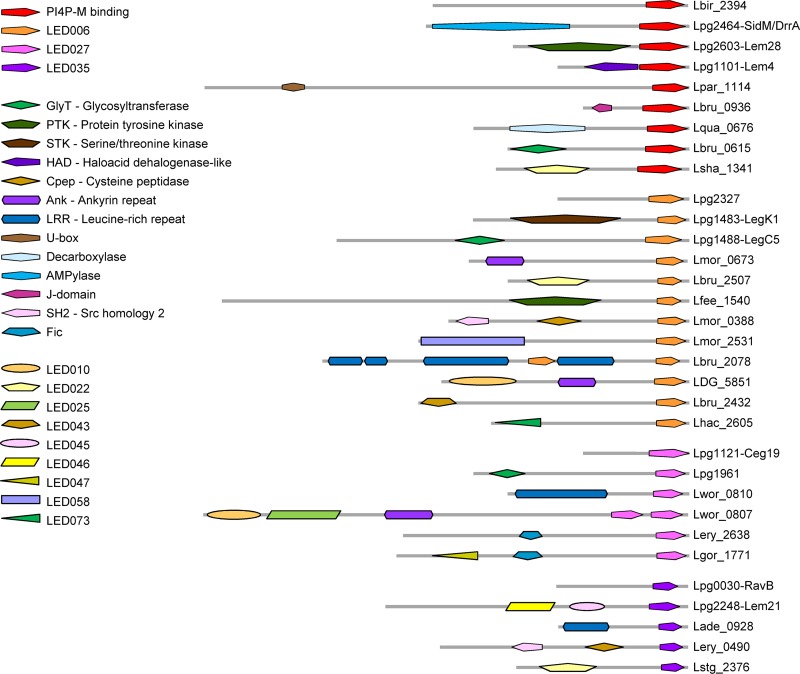
In a previous study, we identified a large number of effectors in the Legionella genus, and this information made it possible to identify and analyze conserved LEDs (35). This analysis revealed 99 LEDs, consisting of 53 well-characterized mostly eukaryotic domains which were previously described (9, 37) and 46 newly discovered conserved domains which share no homology with any known domains (35). We used the three features described above to examine the 46 novel LEDs in order to identify LEDs harboring the same properties. Our analysis revealed three such LEDs: LED006, LED027, and LED035. These three LEDs have the same characteristics: they are always located at the C-terminal ends of the effectors, they are small (about 100 amino acids [aa]), and in some effectors they are found together with catalytic or protein binding domains (Fig. 2). In addition, we compared the protein architectures (different domain combinations) of Legionella effectors which harbor these three novel LEDs to the characterized PI4P-binding domain present in SidM/DrrA and two other effectors (see the introduction). This comparison indicated that in some effectors the PI4P-M-binding domain and these three LEDs are found together with the same catalytic/protein binding domains (Fig. 2 and see Fig. S1 in the supplemental material). Importantly, these three LEDs are highly prevalent in the 41 Legionella species examined (found in 230 effectors; see Table 2), and the architectures presented in Fig. 2 are only of representative effectors that contain additional known domains or LEDs. Most of the effectors containing these three predicted PI-binding LEDs do not contain additional LEDs or any known domains. This architecture is found in several L. pneumophila effectors and in numerous effectors in the Legionella genus (this architecture is presented by the uppermost effector shown for each LED in Fig. 2). The three LEDs identified have average size similar to the PI4P-M-binding domain and have the same distance from the end of the effector, as well as a similar degree of homology among the sequences of individual LEDs (Table 2). The similar characteristics found between the three predicted PI-binding LEDs and the validated PI4P-M-binding domain suggest that the newly identified LEDs might also bind PIs.
FIG 2.
Architectures of effectors containing the PI4P-M-binding domain and the three predicted PI-binding LEDs. Each protein architecture containing the PI4P-M-binding domain, LED006, LED027, or LED035 is represented by a single effector. Several known domains, as well as unknown LEDs, were found in the same architecture with two or three of the domains known or predicted to bind PIs (see summary in Fig. S1). The novel LEDs were originally described elsewhere (35). For each of the analyzed C-terminal domains, the upper effector represents a large group of effectors containing the domain with no other known domain or LED. The effectors presented are indicated on the right by their locus tag or gene name.
TABLE 2.
Properties of the PI4P-M binding domain and the predicted PI-binding LEDs
| Domain | Avg size (aa) | Avg distance from C terminus (aa) | No. of LEOGs in the genusa | No. of effectors in L. pneumophila | No. of effectors in the genusb | Avg E value to the LED consensus |
|---|---|---|---|---|---|---|
| PI4P-M | 99 | 10 | 12 | 3 | 36 | 4.8 × 10−8 |
| LED006 | 69 | 20 | 32 | 8 | 138 | 6.9 × 10−8 |
| LED027 | 82 | 48 | 11 | 3 | 51 | 3.7 × 10−8 |
| LED035 | 66 | 44 | 7 | 3 | 41 | 2.5 × 10−21 |
LEOGs, Legionella effector orthologous groups.
The LEDs identified in the L. pneumophila validated effectors and the predicted effectors in the 41 Legionella species examined are described in supplementary Table 9 in reference 35.
The three novel LEDs are found in 14 L. pneumophila effectors.
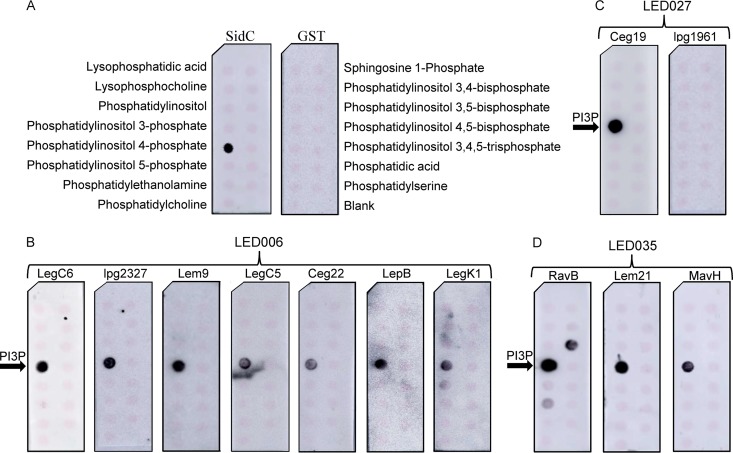
We decided to focus our study on all of the L. pneumophila effectors in which these three novel LEDs were identified (Table 3). (i) LED006 was found in eight L. pneumophila effectors (CegC2, LegK1, Ceg22, LegC5, Lem9, LegC6, LepB, and lpg2327), one of which (LepB) was previously shown to be localized to the LCV (38), and its N-terminal domain specifically converts PI3P into PI3,4P (39), but there was no indication that it binds PIs at its C-terminal end. Two of the other effectors containing LED006 (LegK1 and LegC5) also contain catalytic domains, i.e., a serine/threonine kinase domain and a glycosyltransferase domain, respectively, but there was no information about their ability to bind PIs. (ii) LED027 was found in three L. pneumophila effectors (RavZ, Ceg19, and lpg1961), one of which (RavZ) was previously shown to bind PI3P and to be localized to autophagosomes (40). The PI3P-binding region of RavZ and the amino acids of LED027 overlap. The other two effectors harboring this LED were not studied before. (iii) LED035 was also found in three L. pneumophila effectors (RavB, Lem21, and MavH), none of which was studied prior to our study. During the course of this study, Kubori et al. characterized Lem21 (also named LotA) and found that it binds PI3P at the region which covers the amino acids of LED035 (41). The 13 effectors that were not examined for PI binding prior to our study and contain one of these three novel LEDs were examined as described below.
TABLE 3.
L. pneumophila effectors harboring the predicted PI-binding LEDs
| LED | Locus tag | Name | Effector size (aa) | Position of LED (aa) | Other domaina | Known function(s)b | Reference(s) |
|---|---|---|---|---|---|---|---|
| LED006 | lpg0126 | CegC2 | 1,102 | 1027–1084 | |||
| lpg1483 | LegK1 | 529 | 450–522 | STK | Activates NF-κB | 93, 94 | |
| lpg1484 | Ceg22 | 269 | 189–260 | ||||
| lpg1488 | LegC5 | 865 | 750–838 | GlyT | Inhibits eEF1A | 95, 96 | |
| lpg1491 | Lem9 | 413 | 298–386 | ||||
| lpg1588 | LegC6 | 672 | 556–644 | ||||
| lpg2327 | 297 | 181–269 | |||||
| lpg2490 | LepB | 1,294 | 1210–1283 | GAP for Rab1 on the LCV | 38, 39, 97–99 | ||
| LED027 | lpg1121 | Ceg19 | 256 | 146–250 | Vesicle trafficking | 73 | |
| lpg1683 | RavZ | 502 | 329–423 | Deconjugation of Atg8/LC3 on autophagosomes | 40, 42, 72 | ||
| lpg1961 | 510 | 432–499 | GlyT | ||||
| LED035 | lpg0030 | RavB | 304 | 201–263 | |||
| lpg2248 | Lem21 | 744 | 622–702 | LED45, 46 | Deubiquitinase | 41 | |
| lpg2425 | MavH | 268 | 158–217 |
STK, serine/threonine kinase; GlyT, glycosyltransferase.
GAP, GTase activating protein; LCV, Legionella-containing vacuole.
LEDs from eleven L. pneumophila effectors bind PI3P in vitro.
To determine whether the LEDs of these 13 effectors bind PIs in vitro, we generated N-terminal glutathione S-transferase (GST) fusions from all of these 13 effectors that included only the LED sequence region (Table 3). We were able to express and purify 12 of these fusions (all except CegC2) and assayed their binding to PIs and other lipids immobilized onto nitrocellulose membranes (Fig. 3A). Under these conditions, all seven L. pneumophila effectors harboring LED006 were found to specifically bind PI3P (Fig. 3B). One of the two L. pneumophila effectors harboring LED027 (Ceg19) also bound PI3P, and the other effector (lpg1961) did not bind any lipid (Fig. 3C) (RavZ, which harbors LED027, was previously reported to bind PI3P [42] and was therefore not examined). In addition, all the L. pneumophila effectors harboring LED035 were found to bind PI3P, and one of them (RavB) also bound PI3,5P and, to a much weaker extent, PI5P (Fig. 3D). These results validate that LED006, LED027, and LED035 are indeed PI-binding domains and showed that they all specifically bind PI3P. These LEDs are found in 14 L. pneumophila effectors and in 230 effectors in the 41 Legionella species examined, all of which are thus predicted to bind PIs.
FIG 3.
The three novel LEDs bind specifically to PI3P in vitro. Affinity-purified GST fusion proteins (150 pmol) of SidC and GST controls (A), LED006 from LegC6, lpg2327, Lem9, LegC5, Ceg22, LepB, or LegK1 (B), LED027 from Ceg19 or lpg1961 (C), and LED035 from RavB, Lem21, or MavH (D) were assayed for binding to different synthetic lipids (100 pmol) immobilized to nitrocellulose membranes by a protein-lipid overlay assay using an anti-GST antibody. The different lipids immobilized to the strip are indicated in panel A. PI3P, phosphatidylinositol 3-phosphate.
LED027 and LED035 bind PI3P more strongly than LED006.
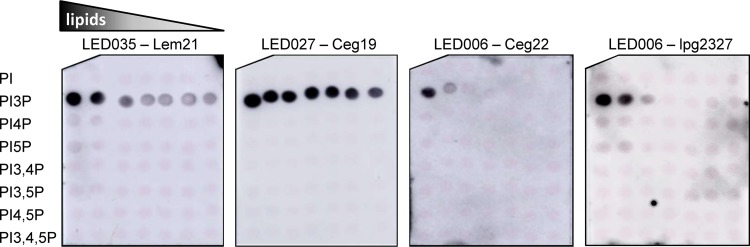
In order to compare whether the binding affinities of the three LEDs to PI3P are similar, we examined the binding of representative effectors containing LED006, LED027, and LED035 to PIs, using PI arrays onto which the lipids were spotted in 2-fold dilution series. Using these PI arrays, we found that LED027 and LED035 (from the effectors Ceg19 and Lem21, respectively) bind exclusively to PI3P with similarly high affinities (Fig. 4). In contrast, under the same conditions, LED006 from the two effectors Ceg22 and lpg2327 (see below for the difference of LED006 between these two effectors) were found to specifically bind PI3P, but with considerably lower affinity than LED027 and LED035 (Fig. 4). These results indicate that even though the examined LEDs have similar properties and bind the same PI, their PI3P-binding affinities are different.
FIG 4.
Three PI3P-binding LEDs bind PI3P in vitro with different affinities. The binding of the affinity-purified GST fusion proteins of several different LEDs—LED027 from Ceg19, LED035 from Lem21, and LED006 from Ceg22 or from lpg2327 (300 pmol)—to 2-fold serial dilutions of PIs (100 to 1.56 pmol) immobilized on nitrocellulose membranes was analyzed by a protein-lipid overlay assay using an anti-GST antibody.
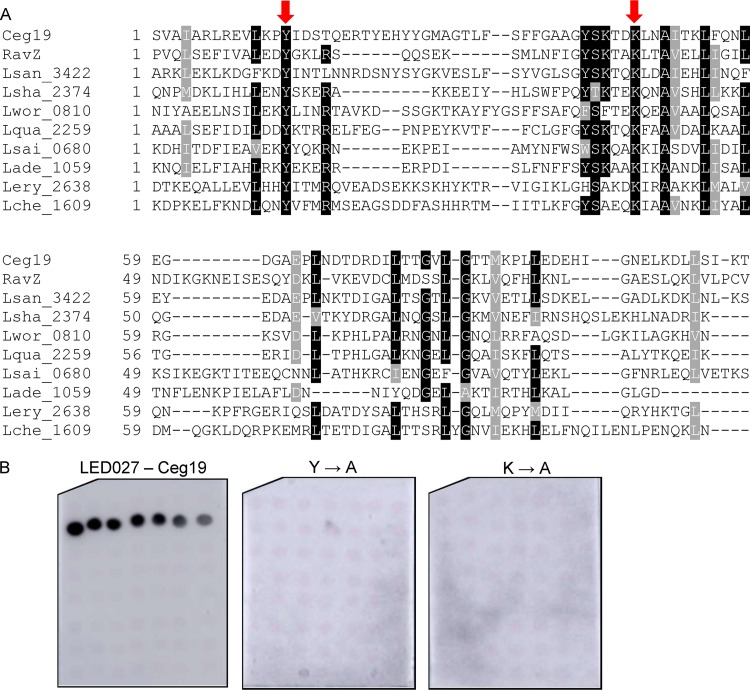
Conserved tyrosine and lysine residues are essential for PI3P binding by LED027.
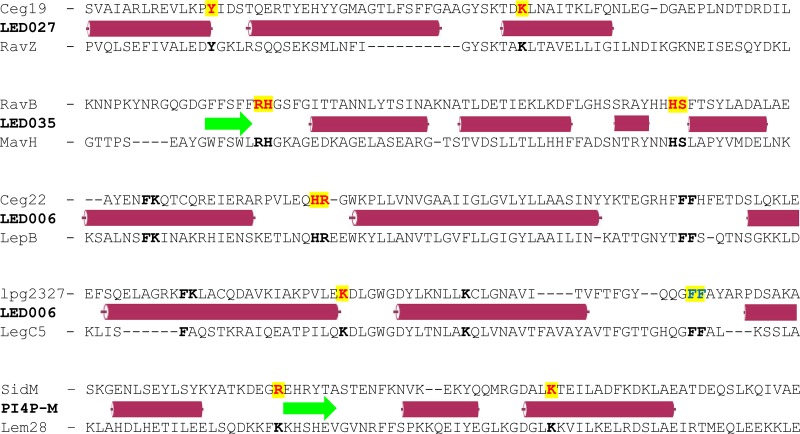
LED027 is one of the two LEDs which were found to bind PI3P with high affinity. In order to identify critical amino acids in LED027 that are required for PI3P binding, we generated a protein multiple sequence alignment of LED027 from effectors and predicted effectors in the Legionella genus. Figure 5A depicts an alignment of LED027 from ten effectors from different orthologous groups (35), including the two L. pneumophila effectors containing LED027 that were found to bind PI3P (Fig. 3 and 4) (42). (lpg1961, which harbors LED027 but did not bind PIs, was excluded from the alignment.) Since PI3P is negatively charged and previous analyses indicated that positively charged amino acids, as well as polar and aromatic amino acids, are involved in PI binding (23, 26), we paid special attention to conserved positively charged amino acids, as well as to conserved polar and aromatic amino acids, found in the LED alignment. This analysis revealed two highly conserved amino acids: tyrosine and lysine (marked by arrows in Fig. 5A). To determine the importance of these amino acids for PI3P binding by Ceg19 (representing LED027), they were individually changed to alanines from Ceg19. GST fusions of these two mutated versions of Ceg19 were purified and examined for PI binding using the PI arrays (Fig. 5B). Both of these mutants completely lost their ability to bind PI3P (with the same amounts of GST-tagged proteins used in both analyses; Fig. 5B and Fig. S2). These results validate the importance of the conserved tyrosine and lysine of Ceg19 for PI3P binding. The same amino acids were also found to be important for PI binding by eukaryotic PI-binding domains (see the introduction).
FIG 5.
Identification of amino acids critical for PI3P binding by Ceg19. (A) Multiple sequence alignment of ten effectors or predicted effectors harboring LED027. The conserved amino acids tyrosine and lysine that were mutated are marked with red arrows. (B) The binding of the affinity-purified GST fusion proteins of the wild-type effector Ceg19 or the two mutants with mutations in the tyrosine and lysine residues (Y160A and K193A) of the same effector (300 pmol) to 2-fold serial dilutions of PIs (100 to 1.56 pmol) immobilized on nitrocellulose membranes was analyzed by a protein-lipid overlay assay using an anti-GST antibody.
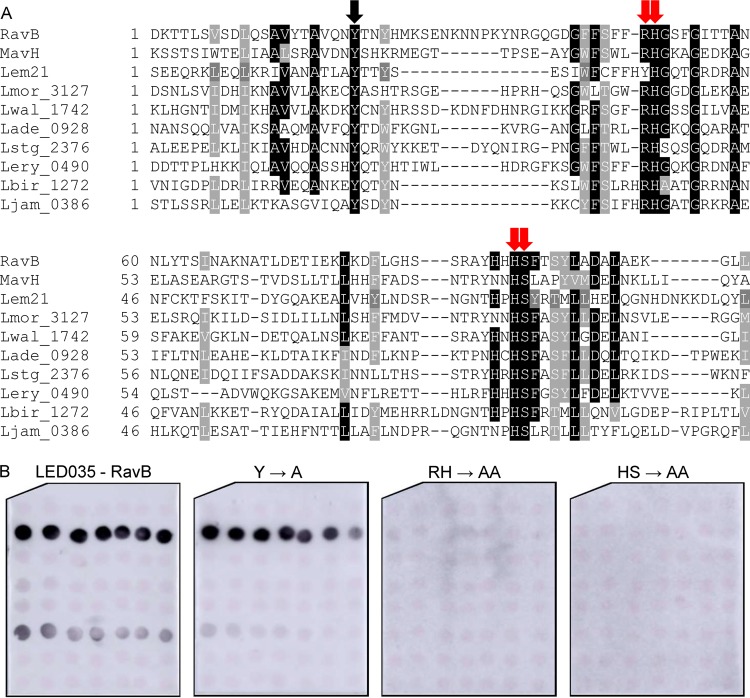
Conserved adjacent arginine-histidine and histidine-serine are required for LED035 PI3P binding.
Like LED027, LED035 was also found to bind PI3P with high affinity (Fig. 4). Multiple sequence alignment was performed using LED035 from effectors from different ortholog groups, as described above. This sequence analysis revealed five conserved amino acids: a single tyrosine, an adjacent arginine-histidine, and an adjacent histidine-serine (marked by arrows in Fig. 6A). Comparing the sequence alignments of LED027 and LED035 (Fig. 5A and 6A) revealed a conserved tyrosine in both LEDs and, at a similar distance, like the essential lysine in LED027, conserved adjacent arginine-histidine residues were identified in LED035. To determine the importance of these conserved amino acids for PI3P binding of LED035, they were mutated in RavB. Three mutants were constructed in which the single tyrosine or each of the adjacent amino acid pairs were changed to alanines. GST-fusions containing the three mutated RavB were purified and examined for PI binding. This analysis indicated that the mutants with mutations in the adjacent arginine-histidine or the adjacent histidine-serine completely lost the ability to bind PI3P (with the same amounts of GST fusion proteins used in both analyses; Fig. 6B and Fig. S2). However, surprisingly, the mutation in the conserved tyrosine had no effect on PI3P binding (Fig. 6B), even though in Ceg19, which also binds PI3P with high affinity, a conserved tyrosine residue was found to be important for PI3P binding (Fig. 5). These results demonstrate the importance of the two adjacent and conserved amino acids (arginine-histidine and histidine-serine) for the PI3P binding of RavB and show that different LEDs which bind PI3P with high affinity use different amino acids to achieve the same function.
FIG 6.
Identification of amino acids critical for PI3P binding by RavB. (A) Multiple sequence alignment of ten effectors or predicted effectors harboring LED035. The conserved amino acid tyrosine and the adjacent arginine-histidine or histidine-serine that were mutated are marked with red arrows if the mutation affected the PI3P binding or with black arrows if the mutation did not affect PI3P binding (see panel B). (B) Binding of affinity-purified GST fusion proteins of the wild-type effector RavB or the three mutants with mutations in the single tyrosine, the adjacent arginine-histidine, and the adjacent histidine-serine (Y185A, RH213-214AA, and HS257-258AA) of the same effector (300 pmol) to 2-fold serial dilutions of PIs (100 to 1.56 pmol) immobilized on nitrocellulose membranes was analyzed by a protein-lipid overlay assay using an anti-GST antibody.
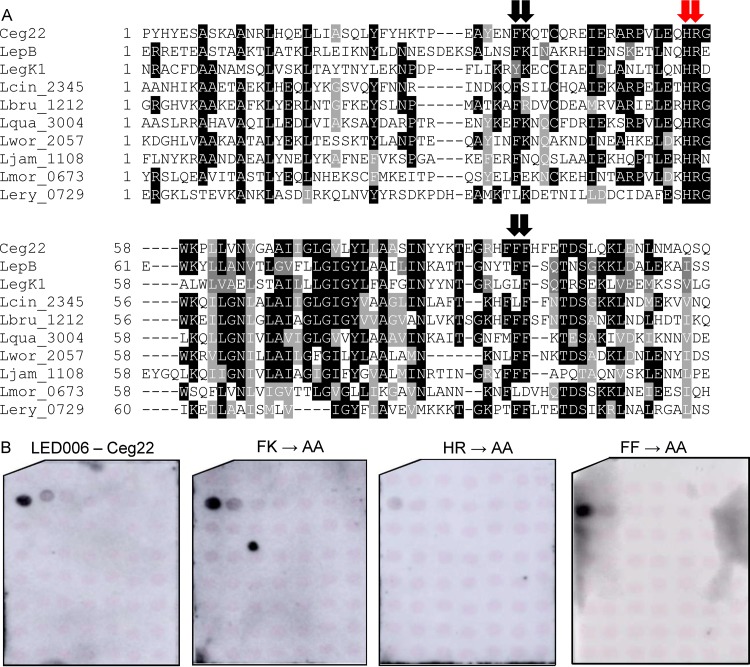
A single lysine or adjacent histidine-arginine residues located at a similar position are required for LED006 PI3P binding.
Of the LEDs we analyzed in this study, LED006 is the most prevalent, both in Legionella predicted effectors and specifically in L. pneumophila validated effectors. However, the binding of LED006 to PI3P is weaker than that of the other PI3P-binding LEDs we identified. To test whether LED006 harbors amino acids essential for PI3P binding similar to LED027 and LED035, a multiple sequence alignment was constructed for the LED region of all the effectors harboring LED006 (Fig. 7A). This analysis revealed several conserved amino acids. Following the importance of positively charged amino acids and aromatic amino acids that we demonstrated for PI3P binding, we decided to examine three pairs of amino acids: phenylalanine-lysine, histidine-arginine, and two adjacent phenylalanine residues (marked by arrows in Fig. 7A). To determine the importance of these amino acids for PI3P binding, these amino acids were changed to alanines in Ceg22. GST fusions of the three mutants were purified, and their examination revealed that only the adjacent histidine-arginine are critical for PI3P binding and that the other mutations did not affect PI3P binding (Fig. 7B and Fig. S2).
FIG 7.
Identification of amino acids critical for PI3P binding by Ceg22. (A) Multiple sequence alignment of ten effectors or predicted effectors harboring LED006. The conserved adjacent phenylalanine-lysine, histidine-arginine, and the two phenylalanine residues that were mutated are marked with red arrows if the mutation affected PI3P binding or with black arrows if the mutation did not affect PI3P binding (see panel B). (B) Binding of affinity-purified GST fusion proteins of the wild-type effector Ceg22 or the three mutants with mutations in the adjacent phenylalanine-lysine, the adjacent histidine-arginine, and the adjacent phenylalanine residues (FK189-190AA, HR207-208AA, and FF245-246AA) of the same effector (300 pmol) to 2-fold serial dilutions of PIs (100 to 1.56 pmol) immobilized on nitrocellulose membranes was analyzed by a protein-lipid overlay assay using an anti-GST antibody.
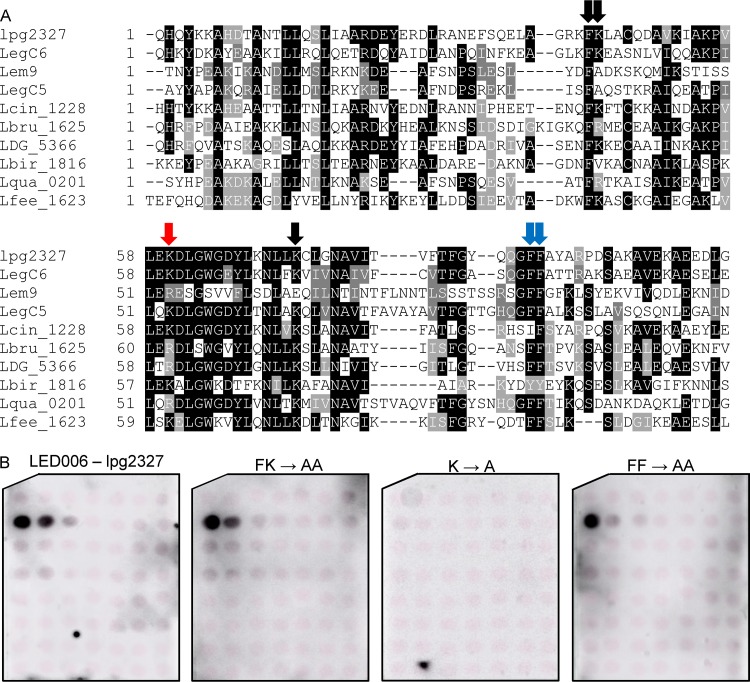
When analyzing the LED006 sequence alignment generated for the seven L. pneumophila effectors that were found to bind PI3P (Fig. S3), we realized that four of them (LegC6, Lem9, LegC5, and lpg2327) lack the histidine-arginine pair that was found to be critical for PI3P binding by Ceg22. These adjacent histidine-arginine residues were absent in 27 of the 138 effectors and predicted effectors harboring LED006 in the 41 Legionella species examined. A multiple sequence alignment of Legionella validated and predicted effectors lacking the histidine-arginine residues revealed that either a lysine or an arginine residue is present in a position similar to the missing histidine-arginine residues (marked by a red arrow in Fig. 8A and boxed in blue in Fig. S3). In addition, 12 amino acids downstream from this position, another conserved lysine residue was identified (marked by single black arrow in Fig. 8A). To determine the importance of these amino acids in the PI3P binding of LED006, these two single lysine residues, the adjacent phenylalanine-lysine residues, and the two adjacent phenylalanine residues (the latter two were also mutated in Ceg22) were changed to alanines in lpg2327. Examination of the PI binding of these mutants revealed that only the lysine residue located at a position similar to the conserved histidine-arginine found in the majority of the effectors harboring LED006 was critical for PI3P binding, and the other mutants constructed had no effect (or a very mild effect in the mutant with a mutation in the two adjacent phenylalanine residues) on PI3P binding (Fig. 8B and Fig. S2). Combined, these results indicate the importance of either adjacent histidine-arginine residues or a lysine residue at a specific position for LED006 PI3P binding (see below).
FIG 8.
Identification of an amino acid critical for PI3P binding present in a subset of effectors containing LED006. (A) Multiple sequence alignment of ten effectors or predicted effectors harboring LED006. The conserved adjacent phenylalanine-lysine and the two adjacent phenylalanine residues, as well as the single lysine or lysine/arginine that were mutated, are marked with red arrows if the mutation affected PI3P binding, with a blue arrow if the mutation had a weak effect on PI3P binding, or with black arrows if the mutation did not affect PI3P binding (see panel B). (B) Binding of affinity-purified GST fusion proteins of the wild-type effector lpg2327 or the three mutants with mutations in the adjacent phenylalanine-lysine, the single lysine, or the two adjacent phenylalanine residues (FK220-221AA, K237A, and FF268-269AA) of the same effector (300 pmol) to 2-fold serial dilutions of PIs (100 to 1.56 pmol) immobilized on nitrocellulose membranes was analyzed by a protein-lipid overlay assay using an anti-GST antibody.
Comparison of the Legionella PI3P-binding domains identified.
The analysis of the three newly discovered PI3P-binding LEDs presented here makes it possible to compare the domains and amino acids required for PI binding. A comparison that included the three novel LEDs, as well as the PI4P-M-binding domain (Fig. 9), led us to two main findings. The first is that the amino acids essential for PI binding by Legionella effectors are usually positive or aromatic amino acids. This observation also fits the known information about the eukaryotic PX domain, which binds PI3P and harbors the consensus R(Y/F)x23–30Kx13–23R sequence (26, 27); the eukaryotic FYVE domain, which binds PI3P and harbors the consensus WxxD, (R/K)(R/K)HHCR, and RVC sequence (23–25); and the PI4P-C domain, present in the Legionella homologous effectors SidC and SdcA, in which arginine, tryptophan, and phenylalanine residues are required for PI4P binding (43). Our second finding is that the residues essential for PI binding in the newly identified PI3P-binding LEDs and the PI4P-M-binding domain are located at or close to the end of an alpha-helix or a beta-strand structure (Fig. 9). The same was found in the PI4P-C domain (43) and in eukaryotic PI-binding domains (22). There was not even a single case of an essential PI-binding residue located in the middle of an alpha-helix or a beta-strand. There were three cases where we mutated amino acids located in the middle of a secondary structure (phenylalanine and lysine in Ceg22 and lpg2327 and a lysine residue in lpg2327, marked in black in Fig. 9), but their mutagenesis did not affect the PI3P binding of LED006.
FIG 9.
Comparison of the regions of the LEDs that were found to contain amino acids critical for PI binding. The domains of effectors that were found to bind PI3P or PI4P are presented. For each of the three newly identified PI3P-binding LEDs, as well as for the PI4P-M-binding domain, the amino acid sequence of two effectors is presented. The sequence of the upper effector representing each LED mutagenized and the amino acids required for PI3P binding identified in it are presented. The predicted secondary structure (red cylinder, alpha-helix; green arrow, beta-strand [according to JPred]) for each LED is presented between the pair of sequences. In the upper sequences, amino acids in red and yellow were found critical for PI binding, amino acids marked in boldface were mutagenized and found not to be required for PI binding, and amino acids marked in blue and yellow had a small effect on PI3P binding. In the lower sequences, amino acids located at the same position as the ones mutated in the upper sequences are marked in boldface.
The analysis performed with the three newly discovered PI3P-binding LEDs uncovered similarities and differences in their PI-binding properties and demonstrated that the study of LEDs can reveal new information regarding the function mediated by groups of effectors containing the same LEDs.
DISCUSSION
Previous studies have shown that the Legionella genus contains thousands of effectors (35, 36). These effectors are expected to function inside host cells during infection and to modulate host cell processes to the benefit of the bacteria. One of the biggest challenges in the Legionella pathogenesis field is to uncover the functions of these effectors in order to discover all the pathways and processes modulated by these human pathogens during infection. Since the number of effectors is so large, novel approaches are required to uncover effector functions; one such approach was presented here and it involves the study of LEDs (35). About half of the LEDs are uncharacterized protein domains found in various orthologous groups of Legionella effectors. Deciphering the function of these basic effector building blocks will allow us to better understand the effector domain architecture and the function mediated by multiple effectors. Using several properties of known Legionella effector PI-binding domains, we identified here three LEDs that specifically bind PI3P. Our results are a proof of concept that effectors can be studied by characterizing their LEDs. The finding that 12 of the 13 L. pneumophila effectors harboring LED006, LED027, or LED035 that were examined here were found to bind PI3P suggests that the majority of the 230 Legionella effectors (Table 2) harboring these LEDs likely bind PI3P.
It was previously demonstrated, using several L. pneumophila effectors, that PI3P- and PI4P-binding domains are involved in the localization of effectors in the host cell, in most cases to the LCV (Table 1 and references therein). The finding that 11 L. pneumophila effectors containing LED006, LED027, or LED035 bind PI3P considerably expands the repertoire of catalytic domains (such as serine/threonine kinase and cysteine peptidase) and protein binding domains (such as leucine-rich repeat) that might be recruited to the LCV, since they are found together with these three LEDs as part of the same effector (Fig. 2). In addition, our findings can also direct the study of the function of additional LEDs with unknown function. According to our analysis, there is only a single PI-binding LED in each effector. Therefore, LEDs of unknown function that are found on the same effector, together with a PI-binding LED, are most likely catalytic or protein-protein binding LEDs (see the list in Fig. 2). For example, LED022 (present in 58 effectors in the 41 Legionella species examined) is found on the same effectors together with LED006, LED035, or the PI4P-M-binding domain and always at the N terminus (Fig. 2 and Fig. S1). Analysis of LED022, which is on average 150 amino acids long, revealed several highly conserved amino acids, including four adjacent residues (data not shown). These amino acids might serve as part of a catalytic core of this LED, and additional study of this LED might lead to the identification of its function in connection to the PI-binding LEDs associated with it in several effectors.
Several eukaryotic domains were reported to specifically bind PIs in numerous eukaryotic proteins (20, 21). Comparison of these domains to the Legionella PI-binding domains revealed one clear difference. The Legionella PI-binding domains are almost exclusively located at the effector's C-terminal end (in all the effectors examined beside LidA). However, the eukaryotic PX domain which binds PI3P and the PH domain which binds PI4P were shown to be located at either the N-terminal or the C-terminal end of the different proteins in which they were found (26, 27). This difference might be related to the fact that the translocation signal of the Icm/Dot effectors is located at the ∼25 C-terminal amino acids of the effectors (44–47). The C-terminal part of the effectors probably enters the host cells before its N-terminal part; thus, the location of the effectors’ PI-binding domains at the C-terminal end (right next to the translocation signal) might facilitate the localization of these effectors to the LCV even before the entire effector has entered the host cell.
It is well established that Legionella effectors harbor domains typically found in eukaryotic proteins, such as ankyrin, F-box, and U-box domains and more (36, 37, 48, 49). The analysis performed here indicates that the three novel PI3P-binding domains identified, even though not homologous to eukaryotic PI-binding domains, share the same amino acids critical for PI binding (22). This observation suggests either convergent evolution, i.e., bacteria and eukaryotes independently evolved similar mechanisms for PI binding, or that these domains represent an ancient acquisition from eukaryotes that was followed by many changes, leading to lack of homology, except in residues critical for PI binding.
The LEDs, three of which were studied here, are present in numerous effectors and share relatively low degree of homology to each other. However, as we showed here, LEDs present in different effectors can perform the same function. Thus, the study of LEDs, which are the building blocks of effectors, has the potential to uncover functions mediated by numerous effectors, as well as to uncover the way by which a combination of LEDs present in the same effector contribute to the overall functions mediated by effectors during infection.
MATERIALS AND METHODS
Bacterial strains and media.
The L. pneumophila wild-type strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (50). The Escherichia coli strains used in this work were MC1022 (51) and BL21(DE3) (52). Bacterial media, plates, and antibiotics were previously described (53).
Plasmid construction.
N-terminal GST fusions were constructed for LED006 from eight effectors (CegC2, LegK1, Ceg22, LegC5, Lem9, LegC6, LepB, and lpg2327), for LED027 from two effectors (Ceg19 and lpg1961), and for LED035 from three effectors (RavB, Lem21, and MavH) by amplification of their coding regions and cloning into the pGEX-5x3 vector (Amersham). The resulting plasmids are listed in Data Set S1 in the supplemental material, and the primers used for the cloning are listed in Data Set S2 in the supplemental material.
To construct GST fusions containing site-directed mutations in amino acids predicted to be involved in PI3P binding of the effectors examined, site-directed mutagenesis was performed by the PCR overlap extension approach (54), in a way similar to that described previously (55). The following amino acids were mutated: Ceg22 (FK-189-190, HR-207-208, and FF-245-246), lpg2327 (FK-220-221, K-237, K-250, and FF-268-269), Ceg19 (Y-160 and K-193), and RavB (Y-185, RH-213-214, and HS-257-258). All of the single and double mutants constructed contained changes to a single or double alanine residues, respectively. The primers used for the mutagenesis are listed in Data Set S2 in the supplemental material, and the plasmids resulting from the site-directed mutagenesis are listed in Data Set S1.
Protein purification.
BL21(DE3) strains containing the GST fusion protein encoding plasmids were lysed using B-PER (Thermo Fisher) or sonication. For sonication, the pellet was resuspended in 5 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 0.1% Triton X-100, and 1 mg/ml lysozyme, complete mini-protease inhibitor [Roche]; pH 8), and the cells were then disrupted by sonication in 10-s on/off intervals. For lysis using the B-PER reagent, pellets were resuspended in 5 ml of B-PER reagent supplemented with lysozyme, DNase I, and complete mini-EDTA-free protease inhibitor (Roche) in the concentrations suggested by the manufacturer, and the lysate was then gently mixed for 15 min. For both lysis methods, the soluble fraction was collected by centrifugation at 10,000 × g for 10 min, 0.5 ml of reduced glutathione resin (GE Healthcare) was added, and the mixture was gently shaken at room temperature for 60 min. After incubation, the resin was washed three times with 10 ml of phosphate-buffered saline (140 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 2 mM KH2HPO4 [pH 7.3]). The proteins were eluted using 0.5 mM Tris buffer (pH 8.0) containing 10 mM reduced glutathione.
Lipid overlay assay.
A lipid overlay assay was performed as previously described (56), with few modifications. In brief, the lipid strips or PI arrays (Echelon Biosciences) were blocked for 1 h in TBST (50 mM Tris, 3 mM KCl, 137 mM NaCl [pH 7.5], 0.1% Tween 20) containing 5% skim milk at room temperature and then incubated overnight with the GST fusion protein of choice (150 pmol for the lipid strips and 300 pmol for the PI arrays) at 4°C. On the following day, the membrane was incubated for 2 h at room temperature in TBST containing 5% skim milk and a goat anti-GST antibody (Amersham) diluted 1:2,000. The membrane was then incubated with for 1 h at room temperature in TBST containing 5% skim milk and a horseradish peroxidase-conjugated donkey anti-goat antibody (Jackson Immunoresearch Laboratories, Inc.) diluted 1:10,000. Signal was detected using an Imager 600 (GE Healthcare). Western blot analysis was performed in a similar way.
Bioinformatic analyses.
Protein multiple sequence alignments were performed using the MAFFT program (57) and visualized using BoxShade. Protein secondary structure predictions were performed using JPred (58).
Supplementary Material
ACKNOWLEDGMENTS
We thank Marika Linsky and David Burstein for reading the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00153-19.
REFERENCES
- 1.Burillo A, Pedro-Botet ML, Bouza E. 2017. Microbiology and epidemiology of Legionnaire’s disease. Infect Dis Clin North Am 31:7–27. doi: 10.1016/j.idc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Boamah DK, Zhou G, Ensminger AW, O’Connor TJ. 2017. From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front Cell Infect Microbiol 7:477. doi: 10.3389/fcimb.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev 34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 4.Cunha BA, Burillo A, Bouza E. 2016. Legionnaires’ disease. Lancet 387:376–385. doi: 10.1016/S0140-6736(15)60078-2. [DOI] [PubMed] [Google Scholar]
- 5.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowbotham TJ. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Luo ZQ. 2013. Cell biology of infection by Legionella pneumophila. Microbes Infect 15:157–167. doi: 10.1016/j.micinf.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. 2011. Comparative and functional genomics of Legionella identified eukaryotic-like proteins as key players in host-pathogen interactions. Front Microbiol 2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 11.Isberg RR, O’Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cozy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu J, Luo ZQ. 2017. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15:591–605. doi: 10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo ZQ, Vance RE. 2011. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog 7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. doi: 10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asrat S, de Jesus DA, Hempstead AD, Ramabhadran V, Isberg RR. 2014. Bacterial pathogen manipulation of host membrane trafficking. Annu Rev Cell Dev Biol 30:79–109. doi: 10.1146/annurev-cellbio-100913-013439. [DOI] [PubMed] [Google Scholar]
- 17.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 18.Behnia R, Munro S. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 19.Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 20.Balla T. 2005. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci 118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 21.Hammond GR, Balla T. 2015. Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim Biophys Acta 1851:746–758. doi: 10.1016/j.bbalip.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 23.Kutateladze TG. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta 1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayakawa A, Hayes S, Leonard D, Lambright D, Corvera S. 2007. Evolutionarily conserved structural and functional roles of the FYVE domain. Biochem Soc Symp 74:95–105. doi: 10.1042/BSS2007c09. [DOI] [PubMed] [Google Scholar]
- 25.Stenmark H, Aasland R, Driscoll PC. 2002. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett 513:77–84. doi: 10.1016/S0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 26.Jia Z, Ghai R, Collins BM, Mark AE. 2014. The recognition of membrane-bound PtdIns3P by PX domains. Proteins 82:2332–2342. doi: 10.1002/prot.24593. [DOI] [PubMed] [Google Scholar]
- 27.Teasdale RD, Collins BM. 2012. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J 441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- 28.Lemmon MA. 2007. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp 74:81–93. doi: 10.1042/BSS2007c08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemmon MA, Ferguson KM. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 30.Pizarro-Cerdá J, Kühbacher A, Cossart P. 2015. Phosphoinositides and host-pathogen interactions. Biochim Biophys Acta 1851:911–918. doi: 10.1016/j.bbalip.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Steiner B, Weber S, Hilbi H. 2017. Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int J Med Microbiol 308:49–57. doi: 10.1016/j.ijmm.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. 2009. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber S, Wagner M, Hilbi H. 2014. Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. mBio 5:e00839-13. doi: 10.1128/mBio.00839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert AJ, Pupko T, Shuman HA, Segal G. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Valero L, Rusniok C, Carson D, Mondino S, Perez-Cobas AE, Rolando M, Pasricha S, Reuter S, Demirtas J, Crumbach J, Descorps-Declere S, Hartland EL, Jarraud S, Dougan G, Schroeder GN, Frankel G, Buchrieser C. 2019. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci U S A 116:2265–2273. doi: 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol 187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra AK, Del Campo CM, Collins RE, Roy CR, Lambright DG. 2013. The Legionella pneumophila GTPase activating protein LepB accelerates Rab1 deactivation by a non-canonical hydrolytic mechanism. J Biol Chem 288:24000–24011. doi: 10.1074/jbc.M113.470625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong N, Niu M, Hu L, Yao Q, Zhou R, Shao F. 2016. Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat Microbiol 2:16236. doi: 10.1038/nmicrobiol.2016.236. [DOI] [PubMed] [Google Scholar]
- 40.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubori T, Kitao T, Ando H, Nagai H. 2018. LotA, a Legionella deubiquitinase, has dual catalytic activity and contributes to intracellular growth. Cell Microbiol 20:e12840. doi: 10.1111/cmi.12840. [DOI] [PubMed] [Google Scholar]
- 42.Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM. 2015. The Legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev Cell 34:569–576. doi: 10.1016/j.devcel.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y. 2015. Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog 11:e1004965. doi: 10.1371/journal.ppat.1004965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A 102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubori T, Hyakutake A, Nagai H. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 46.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type IVB secretion signal. Proc Natl Acad Sci U S A 110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 49.Degtyar E, Zusman T, Ehrlich M, Segal G. 2009. A Legionella effector acquired from protozoa is involved in sphingolipid metabolism and is targeted to the host cell mitochondria. Cell Microbiol 11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 50.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol 138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 52.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 53.Segal G, Shuman HA. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun 65:5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 55.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 56.Weber S, Dolinsky S, Hilbi H. 2013. Interactions of Legionella effector proteins with host phosphoinositide lipids. Methods Mol Biol 954:367–380. doi: 10.1007/978-1-62703-161-5_23. [DOI] [PubMed] [Google Scholar]
- 57.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pike CM, Boyer-Andersen R, Kinch LN, Caplan JL, Neunuebel MR. 7 February 2019. Legionella effector RavD binds phosphatidylinositol-3-phosphate and helps suppress endolysosomal maturation of the Legionella-containing vacuole. J Biol Chem doi: 10.1074/jbc.RA118.007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allgood SC, Romero Duenas BP, Noll RR, Pike C, Lein S, Neunuebel MR. 2017. Legionella effector AnkX disrupts host cell endocytic recycling in a phosphocholination-dependent manner. Front Cell Infect Microbiol 7:397. doi: 10.3389/fcimb.2017.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 108:21212–21217. doi: 10.1073/pnas.1114023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goody PR, Heller K, Oesterlin LK, Muller MP, Itzen A, Goody RS. 2012. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J 31:1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campanacci V, Mukherjee S, Roy CR, Cherfils J. 2013. Structure of the Legionella effector AnkX reveals the mechanism of phosphocholine transfer by the FIC domain. EMBO J 32:1469–1477. doi: 10.1038/emboj.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neunuebel MR, Mohammadi S, Jarnik M, Machner MP. 2012. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J Bacteriol 194:1389–1400. doi: 10.1128/JB.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conover GM, Derre I, Vogel JP, Isberg RR. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol 48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 67.Schoebel S, Cichy AL, Goody RS, Itzen A. 2011. Protein LidA from Legionella is a Rab GTPase supereffector. Proc Natl Acad Sci U S A 108:17945–17950. doi: 10.1073/pnas.1113133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng W, Yin K, Lu D, Li B, Zhu D, Chen Y, Zhang H, Xu S, Chai J, Gu L. 2012. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog 8:e1002528. doi: 10.1371/journal.ppat.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng G, An X, Ye S, Liu Y, Zhu W, Zhang R, Zheng X. 2013. The crystal structure of LidA, a translocated substrate of the Legionella pneumophila type IV secretion system. Protein Cell 4:897–900. doi: 10.1007/s13238-013-2100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hubber A, Arasaki K, Nakatsu F, Hardiman C, Lambright D, De Camilli P, Nagai H, Roy CR. 2014. The machinery at endoplasmic reticulum-plasma membrane contact sites contributes to spatial regulation of multiple Legionella effector proteins. PLoS Pathog 10:e1004222. doi: 10.1371/journal.ppat.1004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beyrakhova K, Li L, Xu C, Gagarinova A, Cygler M. 2018. Legionella pneumophila effector Lem4 is a membrane-associated protein tyrosine phosphatase. J Biol Chem 293:13044–13058. doi: 10.1074/jbc.RA118.003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang A, Pantoom S, Wu YW. 2017. Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. Elife 6:e23905. doi: 10.7554/eLife.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heidtman M, Chen EJ, Moy MY, Isberg RR. 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11:230–248. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jank T, Bohmer KE, Tzivelekidis T, Schwan C, Belyi Y, Aktories K. 2012. Domain organization of Legionella effector SetA. Cell Microbiol 14:852–868. doi: 10.1111/j.1462-5822.2012.01761.x. [DOI] [PubMed] [Google Scholar]
- 75.Newton HJ, Sansom FM, Dao J, McAlister AD, Sloan J, Cianciotto NP, Hartland EL. 2007. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun 75:5575–5585. doi: 10.1128/IAI.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voth KA, Chung IYW, van Straaten K, Li L, Boniecki MT, Cygler M. 2018. The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein. FEBS J 286:710–725. doi: 10.1111/febs.14710. [DOI] [PubMed] [Google Scholar]
- 77.Weber SS, Ragaz C, Hilbi H. 2009. The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol 11:442–460. doi: 10.1111/j.1462-5822.2008.01266.x. [DOI] [PubMed] [Google Scholar]
- 78.Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. 2013. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 79.Yao J, Yang F, Sun X, Wang S, Gan N, Liu Q, Liu D, Zhang X, Niu D, Wei Y, Ma C, Luo ZQ, Sun Q, Jia D. 2018. Mechanism of inhibition of retromer transport by the bacterial effector RidL. Proc Natl Acad Sci U S A 115:E1446–E1454. doi: 10.1073/pnas.1717383115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romano-Moreno M, Rojas AL, Williamson CD, Gershlick DC, Lucas M, Isupov MN, Bonifacino JS, Machner MP, Hierro A. 2017. Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL. Proc Natl Acad Sci U S A 114:E11151–E11160. doi: 10.1073/pnas.1715361115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barlocher K, Hutter CAJ, Swart AL, Steiner B, Welin A, Hohl M, Letourneur F, Seeger MA, Hilbi H. 2017. Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5. Nat Commun 8:1543. doi: 10.1038/s41467-017-01512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Del Campo CM, Mishra AK, Wang YH, Roy CR, Janmey PA, Lambright DG. 2014. Structural basis for PI(4)P-specific membrane recruitment of the Legionella pneumophila effector DrrA/SidM. Structure 22:397–408. doi: 10.1016/j.str.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 84.Schoebel S, Blankenfeldt W, Goody RS, Itzen A. 2010. High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep 11:598–604. doi: 10.1038/embor.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 86.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. 2008. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 87.Horenkamp FA, Mukherjee S, Alix E, Schauder CM, Hubber AM, Roy CR, Reinisch KM. 2014. Legionella pneumophila subversion of host vesicular transport by SidC effector proteins. Traffic 15:488–499. doi: 10.1111/tra.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu F, Luo X, Qiu J, Teng YB, Jin J, Smolka MB, Luo ZQ, Mao Y. 2014. The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc Natl Acad Sci U S A 111:10538–10543. doi: 10.1073/pnas.1402605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dolinsky S, Haneburger I, Cichy A, Hannemann M, Itzen A, Hilbi H. 2014. The Legionella longbeachae Icm/Dot substrate SidC selectively binds phosphatidylinositol 4-phosphate with nanomolar affinity and promotes pathogen vacuole-endoplasmic reticulum interactions. Infect Immun 82:4021–4033. doi: 10.1128/IAI.01685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wasilko DJ, Huang Q, Mao Y. 2018. Insights into the ubiquitin transfer cascade catalyzed by the Legionella effector SidC. Elife 7:e36154. doi: 10.7554/eLife.36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harding CR, Mattheis C, Mousnier A, Oates CV, Hartland EL, Frankel G, Schroeder GN. 2013. LtpD is a novel Legionella pneumophila effector that binds phosphatidylinositol 3-phosphate and inositol monophosphatase IMPA1. Infect Immun 81:4261–4270. doi: 10.1128/IAI.01054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levanova N, Mattheis C, Carson D, To KN, Jank T, Frankel G, Aktories K, Schroeder GN. 2018. The Legionella effector LtpM is a new type of phosphoinositide-activated glucosyltransferase. J Biol Chem 294:2862–2879. doi: 10.1074/jbc.RA1118.005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, Liu L, Shao F. 2009. A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc Natl Acad Sci U S A 106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Losick VP, Haenssler E, Moy MY, Isberg RR. 2010. LnaB: a Legionella pneumophila activator of NF-κB. Cell Microbiol 12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shen X, Banga S, Liu Y, Xu L, Gao P, Shamovsky I, Nudler E, Luo ZQ. 2009. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol 11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Belyi Y, Tabakova I, Stahl M, Aktories K. 2008. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 98.Mihai Gazdag E, Streller A, Haneburger I, Hilbi H, Vetter IR, Goody RS, Itzen A. 2013. Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep 14:199–205. doi: 10.1038/embor.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu Q, Hu L, Yao Q, Zhu Y, Dong N, Wang DC, Shao F. 2013. Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP. Cell Res 23:775–787. doi: 10.1038/cr.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.