Cyclic di-AMP (c-di-AMP) is a recently discovered second messenger in bacteria. The cellular level of c-di-AMP in Streptococcus pyogenes is predicted to be controlled by the synthase DacA and two putative phosphodiesterases, GdpP and Pde2.
KEYWORDS: DacA, GdpP, Pde2, SpeB, Streptococcus pyogenes, c-di-AMP, virulence
ABSTRACT
Cyclic di-AMP (c-di-AMP) is a recently discovered second messenger in bacteria. The cellular level of c-di-AMP in Streptococcus pyogenes is predicted to be controlled by the synthase DacA and two putative phosphodiesterases, GdpP and Pde2. To investigate the role of c-di-AMP in S. pyogenes, we generated null mutants in each of these proteins by gene deletion. Unlike those in other Gram-positive pathogens such as Staphylococcus aureus and Listeria monocytogenes, DacA in S. pyogenes was not essential for growth in rich media. The DacA null mutant presented a growth defect that manifested through an increased lag time, produced no detectable biofilm, and displayed increased susceptibility toward environmental stressors such as high salt, low pH, reactive oxygen radicals, and cell wall-targeting antibiotics, suggesting that c-di-AMP plays significant roles in crucial cellular processes involved in stress management. The Pde2 null mutant exhibited a lower growth rate and increased biofilm formation, and interestingly, these phenotypes were distinct from those of the null mutant of GdpP, suggesting that Pde2 and GdpP play distinctive roles in c-di-AMP signaling. DacA and Pde2 were critical to the production of the virulence factor SpeB and to the overall virulence of S. pyogenes, as both DacA and Pde2 null mutants were highly attenuated in a mouse model of subcutaneous infection. Collectively, these results show that c-di-AMP is an important global regulator and is required for a proper response to stress and for virulence in S. pyogenes, suggesting that its signaling pathway could be an attractive antivirulence drug target against S. pyogenes infections.
INTRODUCTION
Streptococcus pyogenes, also known as group A streptococcus (GAS), causes a variety of human diseases, such as mild superficial infections (e.g., impetigo and pharyngitis), invasive diseases (e.g., necrotizing fasciitis and cellulitis), toxigenic diseases (e.g., toxic shock syndrome and scarlet fever), and postinfectious autoimmune diseases (e.g., rheumatic fever, rheumatic heart disease, and poststreptococcal glomerulonephritis) (1). The primary infection sites of S. pyogenes are the upper respiratory mucosal epithelium and the superficial layers of the epidermis. Occasionally, S. pyogenes penetrates the bloodstream or deep tissues and causes severe invasive diseases. S. pyogenes infections still remain a major public health concern in both developed and developing countries. A recent survey estimated that S. pyogenes causes 1.78 million new cases of severe group A streptococcal diseases each year globally. Over 18 million people suffer from the severe streptococcal diseases, resulting in over half a million annual deaths (2). In the United States, more than 30 million cases of streptococcal pharyngitis (strep throat) occur each year.
To cause diverse diseases successfully, S. pyogenes must be able to sense the unique environmental signals from infection sites and adapt to the host tissues through regulation of various cellular activities, including virulence factor biogenesis. Thus, a detailed understanding of the signaling pathway by which cellular activities, including the biogenesis of cell components and virulence factors, are regulated will provide insights into the initial colonization, successive invasion, and spread of streptococcal infections.
Cyclic nucleotides that act as second-messenger molecules play key roles in signaling pathways that sense environmental changes such as stress, temperature, nutrition, and pH in both prokaryotes and eukaryotes (3–5). As second messengers, these cyclic nucleotides are involved in the transmission of the signals to effector molecules (3, 6). Cyclic di-AMP (c-di-AMP) is a new addition to the growing list of second messenger nucleotides and has been identified in Gram-positive bacteria, including Listeria monocytogenes, Bacillus subtilis, Staphylococcus aureus, and Streptococcus spp., and in a few Gram-negative bacteria, such as Chlamydia trachomatis and Borrelia burgdorferi (3, 7–13). c-di-AMP has been implicated in diverse cellular processes in bacteria. Its main role in bacteria is osmoregulation, but c-di-AMP also plays a distinctive role in each bacterium (for a review, see reference 14). For example, c-di-AMP plays a role in fatty acid synthesis in Mycobacterium smegmatis (15), in the growth of S. aureus under low-potassium-ion conditions (16), in the sensing of DNA integrity in B. subtilis (17–19), and in cell wall homeostasis in S. aureus and B. subtilis (8, 20–22). Although roles of c-di-AMP have been shown to be critical in many pathogenic bacteria, neither its environmental stimuli nor the mechanisms controlling cellular processes and virulence are well understood (11, 16).
c-di-AMP is synthesized by diadenylate cyclases (DACs). DAC enzymes catalyze the synthesis of a single molecule of c-di-AMP from two molecules of ATP or ADP through a condensation reaction (5, 10, 23–25). Four classes of DACs have been identified so far: DisA, DacA (also called CdaA), CdaS, and CdaM. All DAC proteins possess the conserved diadenylate cyclase domain (DAC domain), the only known domain to synthesize c-di-AMP, which commonly contains DGA and RHR motifs (26, 27). Some bacteria produce multiple DAC enzymes. For example, B. subtilis produces three enzymes, DisA, CdaA, and CdaS (28), and Clostridium spp. produce two DACs, CdaA and DisA. However, most other bacteria possess only one c-di-AMP synthase. M. tuberculosis produces only MtDisA, a DisA homolog (29). Mycoplasma pneumoniae produces only CdaM, which is closely related to the DAC domain of CdaS in B. subtilis (30). The Gram-positive pathogens S. pyogenes, S. pneumoniae, S. aureus, and L. monocytogenes produce only DacA, which is the most common c-di-AMP synthase among the four DAC enzymes discovered so far, as it is found in a wide variety of bacteria (10, 12, 31).
The c-di-AMP phosphodiesterases (PDEs) degrade c-di-AMP, converting it into the linear form of phosphoadenyl adenosine (pApA), which can then be further degraded into two molecules of AMP (32, 33). Three classes of PDEs have been discovered thus far: GdpP, Pde2, and PgpH (34, 35). The presence of each class of PDEs varies by bacterial species, but most bacteria produce two PDEs. L. monocytogenes produces GdpP and PgpH, while Streptococcus and Staphylococcus species produce GdpP and Pde2 (34).
Previously, we studied the role of one of the PDEs, GdpP, in S. pyogenes (36). We created a gdpP in-frame deletion strain, ΔGdpP, that is predicted to produce an increased level of c-di-AMP. ΔGdpP exhibited a defect in the production of the virulence factor SpeB, and its virulence was attenuated in a mouse model of subcutaneous infection.
SpeB is a cysteine protease secreted in the stationary phase by S. pyogenes. It accounts for more than 90% of the total secreted proteins in many S. pyogenes strains, including HSC5 (37, 38). SpeB directly cleaves host molecules such as fibronectin (39), vitronectin (39) immunoglobulins (40–42), C3b (43), and plasminogen (44). It also indirectly damages host molecules by activating host matrix metalloproteases (45). SpeB can disturb host immune functions by activating host immune-modulating molecules such as kinins (46) and interleukin-1β (IL-1β) (47). SpeB also liberates streptococcal cell surface virulence factors such as M protein, protein F, and C5a peptidase, possibly for dissemination (48). SpeB is initially translated as a preproprotein (43 kDa) containing a signal sequence and a pro region. During secretion, the signal sequence is cleaved off, and SpeB is secreted as the pro-SpeB zymogen (∼40 kDa), which is enzymatically inactive. The zymogen is folded and processed to the active protease (28 kDa) through cleavages in the pro region, mainly through autocatalysis (36, 49).
In addition to GdpP, S. pyogenes produces two other enzymes that are involved in c-di-AMP biosynthesis and degradation: the c-di-AMP synthase DacA (SPy_1036, based on the SF370 reference gene locus [50]) and the phosphodiesterase Pde2 (SPy_0720). Here, we studied the influence of these enzymes on the growth and virulence of S. pyogenes by analyzing gene deletion strains to decipher the roles of c-di-AMP. The data reveal that dysregulation of the c-di-AMP signaling pathway results in pleiotropic effects on growth, biofilm formation, resistance to stress, virulence gene expression, and overall pathogenicity.
RESULTS
Deletion of S. pyogenes genes involved in the synthesis or degradation of c-di-AMP.
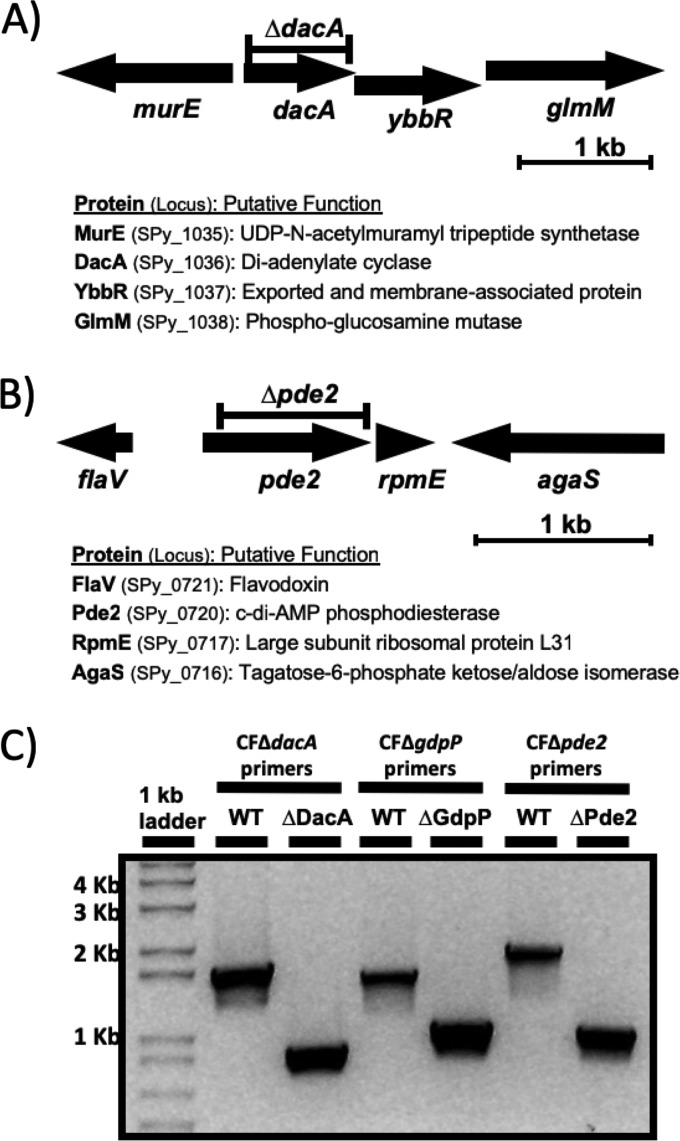
The Gram-positive human pathogen S. pyogenes is predicted to produce only a single c-di-AMP synthase, DacA (10). The gene for DacA is located immediately upstream of the genes encoding YbbR and GlmM, and these three genes form an operon according to our RNA deep-sequencing analysis (51) (Fig. 1A). Since GlmM, a phosphoglucosamine mutase involved in the production of glucosamine 1-phosphate, a building block of cell walls, is essential for growth in bacteria (24, 52), we deleted dacA through an in-frame deletion strategy that does not interfere with the expression of downstream genes within an operon. This was achieved by including the 106-bp region directly upstream of dacA containing the putative promoter within the deletion construct so that expression of the operon is maintained in the merodiploid state. Since DacA in some Gram-positive pathogens was essential for growth in rich media (52–55), we sequenced the chromosome of the in-frame dacA deletion strain, ΔDacA, by high-throughput whole-genome sequencing to examine whether any significant secondary mutations that interfere with the c-di-AMP signaling pathway occurred during the in-frame deletion process. ΔDacA was found to contain two mutations relative to the wild-type HSC5: a 789-bp in-frame deletion in dacA, as expected, and a single nucleotide polymorphism (SNP). The SNP changed an ATG encoding methionine at residue 336 to ATA encoding isoleucine (M336I) in ptsI (spy_1372). The mutation in ptsI appears not to interfere with the function of PtsI because this SNP is also present in GCP754 (PstS−), which was shown to utilize multiple carbon sources that a PtsI− mutant was unable to use (56). This result demonstrates that DacA in S. pyogenes is not essential for growth in rich media. Pde2, a recently discovered c-di-AMP PDE, was also successfully in-frame deleted (Fig. 1B and C). A mutant with a deletion of the remaining PDE gene, ΔGdpP, whose creation process has been previously described (36), was utilized for this study. A double mutant with the deletion of both PDE genes, ΔGdpPΔPde2, was also generated by introducing the gdpP in-frame deletion construct into ΔPde2.
FIG 1.
Construction of the ΔDacA and ΔPde2 mutants. (A) dacA genomic context and in-frame deletion construct. The arrows indicate individual open reading frames and their orientations. dacA is coexpressed with downstream genes ybbR and glmM. The predicted proteins encoded by these open reading frames and their putative functions are shown below the gene organization. The region indicated by the bar above dacA was deleted to make the dacA in-frame deletion mutant ΔDacA, which has a deletion from bp 25 through 813 (Ile9 through Leu271) in dacA. (B) pde2 genomic context and in-frame deletion construct. The region indicated by the bar above pde2 was deleted to make the pde2 in-frame deletion mutant ΔPde2, which has a deletion from bp 82 through 912 (Pro28 through Arg304) in pde2. (C) PCR confirms the deletion of dacA, gdpP, and pde2 in the corresponding mutant strains ΔDacA, ΔGdpP, and ΔPde2, respectively. PCR was performed using the chromosomal DNA extracted from the wild type or each mutant as the template and the primers listed in Table 1. The smaller PCR bands from the mutants relative to the larger bands from the wild type in agarose gel electrophoresis indicate gene deletions. DNA marker sizes are shown at the left side of the gel picture. The following strains were used: wild type (WT), dacA deletion mutant (ΔDacA), gdpP deletion mutant (ΔGdpP), and pde2 deletion mutant (ΔPde2).
Measurement of c-di-AMP in the gene deletion strains confirms that DacA is the sole c-di-AMP synthase and that GdpP and Pde2 are phosphodiesterases.
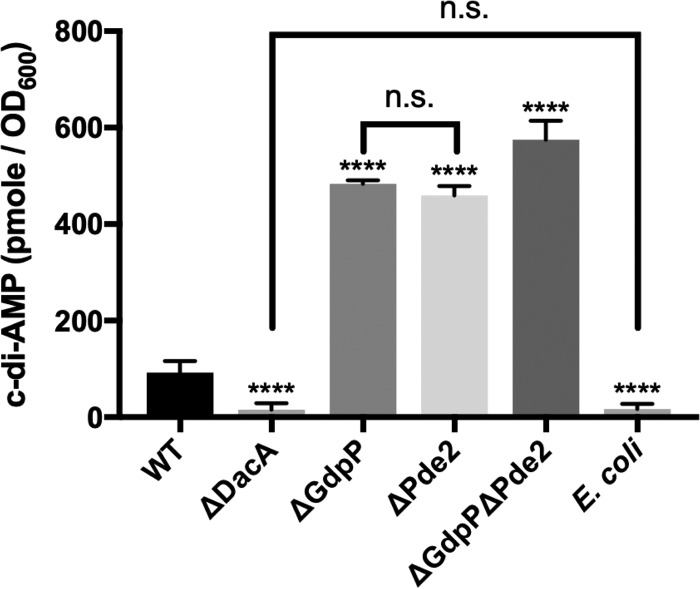
The quantities of c-di-AMP produced by strains were measured by competitive enzyme-linked immunosorbent assay (ELISA) previously described (57, 58). As expected, the deletion of the gene for DacA, which is predicted to be the sole c-di-AMP in S. pyogenes, abolished the production of c-di-AMP. The c-di-AMP amount in ΔDacA was not different from that in Escherichia coli, which does not produce c-di-AMP (Fig. 2). The phosphodiesterase deletion mutants, ΔGdpP and ΔPde2, produced increased amounts of c-di-AMP (∼5 times more) compared to that produced by the wild type, and the increased amounts were similar each other. The amount of c-di-AMP in the double mutant ΔGdpPΔPde2 increased even further, but the degree was not additive.
FIG 2.
ΔDacA does not produce c-di-AMP, while the PDE mutants produce larger amounts of c-di-AMP than the wild type. The concentration of c-di-AMP (pmole/OD600) in each strain was measured with a competitive ELISA. c-di-AMP levels in the ΔDacA mutant were equivalent to those in the negative-control bacterium E. coli, indicating that the deletion of dacA abolished the production of c-di-AMP. In contrast, all of the PDE mutants, ΔGdpP, ΔPde2, and ΔGdpPΔPde2, produced larger amounts of c-di-AMP than the wild type. The significance of difference between values from any two strains was evaluated by one-way analysis of variance (ANOVA) followed by the Tukey test. All the mutants showed a significant difference compared to the wild type (****, P < 0.0001), but the comparisons between ΔDacA and E. coli and between ΔGdpP and ΔPde2 did not show significant differences (n.s., no significance). The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), gdpP deletion mutant (ΔGdpP), pde2 deletion mutant (ΔPde2), gdpP pde2 double deletion mutant (ΔGdpPΔPde2), and E. coli strain TOP10 (E. coli).
Loss of DacA or Pde2 results in growth defects.
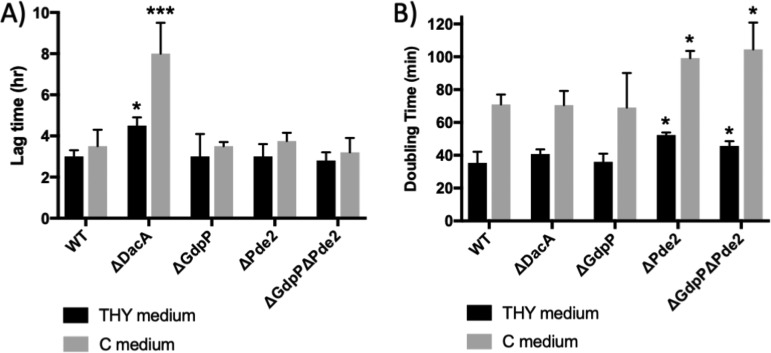
When the lag times and generation times of the mutants were measured in Todd-Hewitt medium with 0.2% yeast extract (THY medium) (protein and carbohydrate rich) and C medium (only protein rich), ΔDacA and ΔPde2 showed growth defects (Fig. 3). In the case of ΔDacA, almost half of the times when cells were inoculated (inoculum size, 1% [vol/vol]), they failed to grow. Even when it did grow, the lag time of ΔDacA was significant longer (∼1.5 times in THY medium and ∼2.1 times in C medium) than that of the wild type (Fig. 3A). Once it grew, its growth rate was not significantly different from that of the wild type in either medium (Fig. 3B). The increased lag time of ΔDacA could be due to a low survival rate at the stationary phase. Serial dilutions of overnight cultures were spread onto THY agar plates, and CFUs were counted to calculate survival rates. The survival rate of ΔDacA was 11.2% ± 8.0% (mean ± standard deviation) of that of the wild type, unlike the other mutants whose survival rates were similar to that of the wild type. These results indicate that even though DacA is not essential in these media, it still plays a critical role in growth. ΔPde2 also showed a growth defect. Although the lag time did not increase, the generation time for ΔPde2 increased to ∼1.5 times those of the wild-type in both media. However, the other phosphodiesterase mutant, ΔGdpP, did not show a significant growth defect (Fig. 3), which is in agreement with our previous result (36).
FIG 3.
ΔDacA and ΔPde2 show growth defects. The lag times (A) and doubling times (B) of the mutants, ΔDacA, ΔGdpP, ΔPde2, and ΔGdpPΔPde2, grown in THY or C medium were measured. The doubling times of ΔDacA were not significantly different from those of the wild type in both media tested, but ΔDacA exhibited an increase in lag time and sometimes failed to grow. In contrast, ΔPde2 showed a longer doubling time than the wild type. The data are the means and standard errors of the means derived from three independent experiments. The asterisks above the bars indicate significance of lag time or growth rate differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between a mutant and the wild type as calculated by one-way ANOVA followed by Dunnett’s multiple-comparison test. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), pde2 deletion mutant (ΔPde2), gdpP deletion mutant (ΔGdpP), and gdpP pde2 double deletion mutant (ΔGdpPΔPde2).
c-di-AMP influences the production of a major virulence factor SpeB in S. pyogenes.
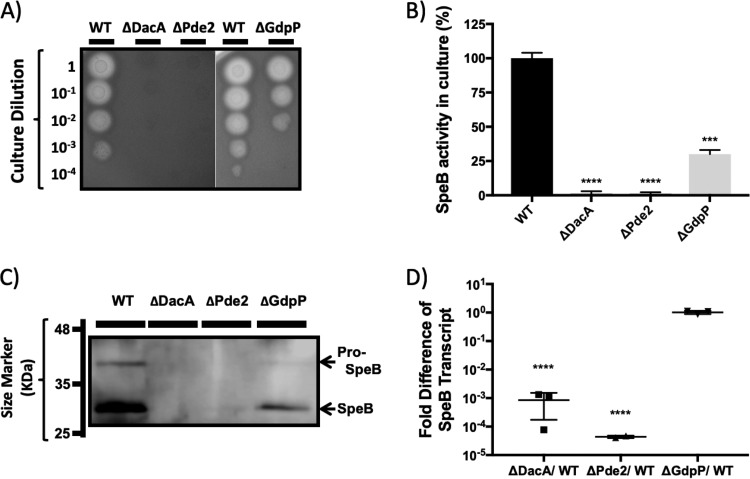
All of the mutants with deletion of a gene involved in c-di-AMP synthesis or degradation presented a defect in the production of the secreted protease SpeB (Fig. 4). As we reported previously, the production of SpeB by ΔGdpP was reduced relative to that by the wild type. ΔDacA and ΔPde2 showed a much more severe defect than ΔGdpP in SpeB production. Both the ΔDacA and ΔPde2 strains showed no protease activity on protease indicator plates (Fig. 4A) and in liquid culture (Fig. 4B) and did not produce detectable zymogen or processed SpeB as examined by Western blotting (Fig. 4C). Interestingly, even though DacA and Pde2 have opposing activities of c-di-AMP synthesis and degradation, respectively, their phenotypes in relation to SpeB production were very similar. As expected, the double mutant ΔGdpPΔPde2 did not produce SpeB either (data not shown).
FIG 4.
ΔDacA and ΔPde2 produce no SpeB due to a transcriptional defect. (A) Protease indicator plates showing an absence of SpeB activity from ΔDacA and ΔPde2. Strains were grown overnight and spotted (2 μl) on 2% skim milk agar plates after serial dilution. Protease activity forms a clear zone around spotted cells. SpeB is the major protease secreted by S. pyogenes HSC5, as a ΔSpeB strain produces no zone of clearance (56). ΔDacA and ΔPde2 show no SpeB activity, and ΔGdpP shows reduced SpeB activity. Each strain name is shown above the picture, and the dilution degree of cultures is indicated at the left side of the picture. (B) Comparison of SpeB activity in the culture supernatants of the mutants. The same culture supernatants for the Western blotting were used to measure relative SpeB activity using fluorescein isothiocyanate (FITC)-casein. (C) Western blot showing no detectable zymogen or processed SpeB protein produced by ΔDacA and ΔPde2. Culture supernatants collected from stationary-phase cultures grown in C medium were used for Western blotting. Anti-SpeB-antibodies are polyclonal rabbit antibodies generated against purified SpeB. Secreted 40-kDa pro-SpeB (the upper band on the Western blot) becomes active 28-kDa SpeB (the lower band) through processing in the culture. Each strain name is shown above the blot, and protein marker sizes are presented at the left side. (D) Disruption of dacA or pde2 results in a severe defect in speB transcription. The relative abundance of the speB transcript during stationary-phase growth in mutant strains was determined using qRT-PCR and compared to that of the wild type. Each column represents the speB transcript abundance in a mutant relative to that in the wild type. The figure shows the means and standard deviations from three independent experiments. Asterisks indicate the significance of differences (***, P < 0.001; ****, P < 0.0001) between a mutant and the wild type as calculated by one-way ANOVA followed by Dunnett’s multiple-comparison test. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), pde2 deletion mutant (ΔPde2), and gdpP deletion mutant (ΔGdpP).
The ΔDacA and ΔPde2 mutants have a transcriptional defect in speB expression.
The speB transcript abundance in the mutants relative to that in the wild type was determined through quantitative reverse transcriptase PCR (qRT-PCR). Both ΔDacA and ΔPde2 expressed 103 to 104 less speB transcript than the wild type (Fig. 4D), indicating that the SpeB expression defect caused by dacA and pde2 deletion occurs at the transcriptional level. The reduction of speB transcript in ΔPde2 was more extreme than that in ΔDacA. In contrast, the speB transcript level in ΔGdpP was equivalent to that in the wild type (Fig. 4D) even though ΔGdpP produced less SpeB in the Western blot assay (Fig. 4C). This indicates that the defect in SpeB production caused by the deletion of gdpP occurs at a posttranscriptional level, as reported previously (36).
Complementation of dacA and pde2 restores SpeB activity.
For a complementation assay, we introduced the dacA and pde2 genes into each corresponding strain, ΔDacA or ΔPde2, using a multicopy streptococcal expression plasmid, pABG5, that can express a gene under control of the rofA promoter (59). When the SpeB activities of the complemented strains, ΔDacA(pDacA) and ΔPde2(pPde2), were measured using protease indicator agar plates, both strains showed restored SpeB activity similar to that of the wild-type control (Fig. 5A and B). However, the growth of ΔPde2 was not restored in ΔPde2(pPde2) (Fig. 5B). The complementation of gdpP also restores SpeB activity, as was shown in our previous report (36).
FIG 5.
Complementation of dacA and pde2 restores the SpeB activity. The dacA and pde2 genes were expressed ectopically in ΔDacA or ΔPde2 using the expression vector pABG5 to complement the deleted genes. The plasmid pABG5, which is a derivative of pLZ12Km, is a multicopy plasmid in S. pyogenes. The resulting plasmids, pDacA and pPde2, were transformed into ΔDacA or ΔPde2 to generate ΔDacA(pDacA) and ΔPde2(pPde2), and the SpeB activities of the complemented strains were determined using protease indicator agar plates. ΔDacA(pDacA) showed growth and SpeB activity similar to those of the wild-type controls (A). On the other hand, ΔPde2(pPde2) showed fully restored SpeB activity but not growth rate (B). The following strains were tested: wild-type, WT; dacA-complemented strain, ΔDacA(pDacA); and pde2-complemented strain, ΔPde2(pPde2).
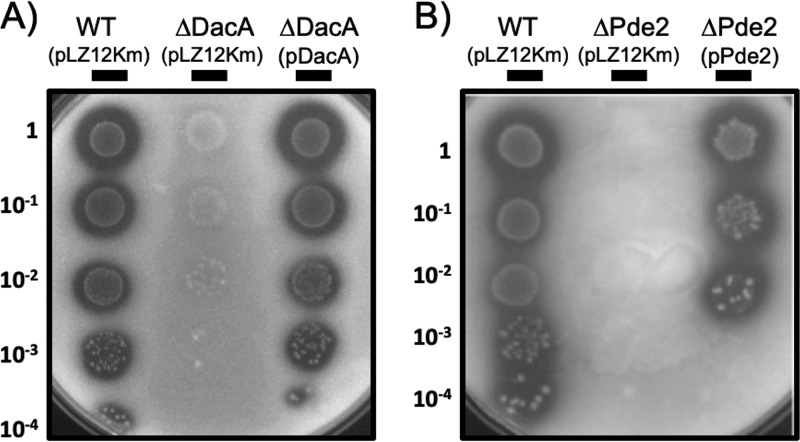
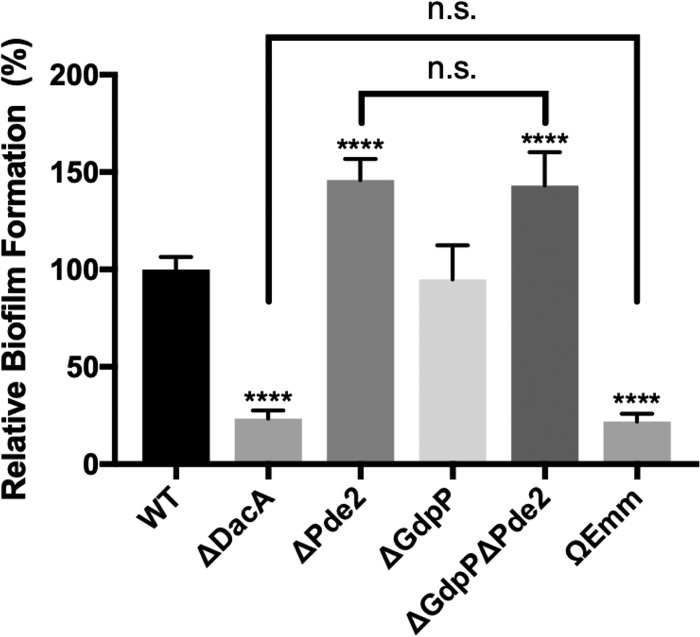
ΔDacA forms no biofilm, and ΔPde2 forms more biofilm than the wild type.
The biofilm formation ability of the mutants was investigated (Fig. 6). The M protein null mutant ΩEmm was used as a negative-control strain, as M protein was previously shown to be required for biofilm formation under the assay conditions (60). ΔGdpP formed the same amount of biofilm as the wild type. ΔDacA formed much less biofilm (∼0.2 times) than the wild type and was as defective as the negative control, ΩEmm. In contrast, ΔPde2 and ΔGdpPΔPde2 formed more biofilm (∼1.4 times) than the wild type. These data highlight the complex relationship that exists between the c-di-AMP level and biofilm formation.
FIG 6.
Deletion of dacA results in decreased biofilm formation, but deletion of pde2 results in increased biofilm formation. The biofilm formation ability of the mutants was examined and compared to that of the wild type. An M protein null mutant, ΩEmm, was used as a negative-control strain. The data are the means and standard deviations of eight data points from two independent experiments. The significance of differences of values between any two strains was calculated by one-way ANOVA followed by the Tukey test. All the mutants except ΔGdpP showed a significant difference compared to the wild type (****, P < 0.0001). The comparisons between ΔDacA and ΩEmm and between ΔPde2 and ΔGdpPΔPde2 did not show a significant difference (n.s., no significance). The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), gdpP deletion mutant (ΔGdpP), pde2 deletion mutant (ΔPde2), gdpP and pde2 double deletion mutant (ΔGdpPΔPde2), and M protein gene (emm)-inactivated mutant (ΩEmm).
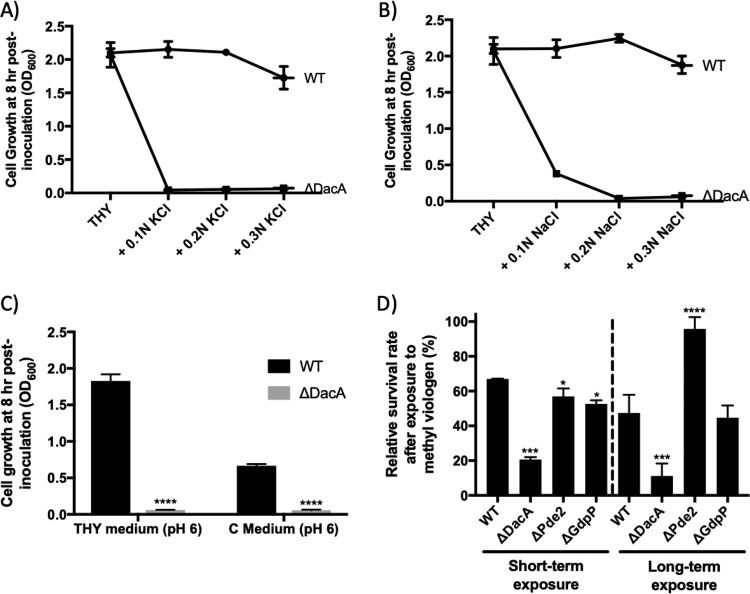
ΔDacA is sensitive to salt, pH, and oxidative stresses.
Cell growth of the mutants in THY medium with various salt concentrations (0.1 N, 0.2 N, and 0.3 N KCl or NaCl) was examined. ΔGdpP and ΔPde2 grew similarly to the wild type under all of the conditions (data not shown). ΔGdpPΔPde2 also grew similarly to the wild type with all of the KCl concentrations but grew less with all of the NaCl concentrations (data not shown). In contrast, ΔDacA displayed a severe growth defect, as it failed to grow under almost all conditions (Fig. 7A and B).
FIG 7.
ΔDacA is more susceptible to environmental stress. (A and B) ΔDacA is sensitive to salt stress. The growth (OD600) of S. pyogenes in THY medium with various concentrations (0.1 N, 0.2 N, and 0.3 N) of KCl (A) or NaCl (B) was measured at 8 h postinoculation, when S. pyogenes typically reaches the stationary phase. (C) ΔDacA is sensitive to low pH. The growth of S. pyogenes in THY or C medium whose pH was lowered to pH 6 was measured at 8 h postinoculation. (D) ΔDacA has increased sensitivity to oxidative stress. The sensitivity of S. pyogenes strains to oxidative stress was tested by challenging them with methyl viologen (MV). Cells were exposed to 50 mM MV for 3 h in short-term exposure and to 1.7 mM MV for 18 h in long-term exposure. The relative survival rate was determined by comparing viable cell counts with and without MV treatment. The data in the figure are the means and standard errors of the means derived from two independent experiments. The significance of the difference between each mutant and the wild type was calculated by two-way ANOVA (A and B) or one-way ANOVA (C and D) followed by Dunnett’s multiple-comparison test. (A and B) The values of all data points except those from THY are different (P < 0.001) from those of the wild type. (C and D) Asterisks above the bars indicate the significance of the difference (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) between the mutant and the wild type. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), pde2 deletion mutant (ΔPde2), and gdpP deletion mutant (ΔGdpP).
Cell growth of the mutants in medium with various pHs was also determined. The wild-type HSC5 grew to a similar optical density at 600 nm (OD600) in medium adjusted to pH 6 as in the original medium (pH ∼7.5), but it did not grow at all at pH 5 and below. ΔGdpP, ΔPde2, and ΔGdpPΔPde2 grew similarly to the wild type at pH 6 (data not shown). However, ΔDacA growth was undetectable at pH 6 (Fig. 7C).
The sensitivity of strains to oxidative stress was examined by challenge with methyl viologen (MV) (paraquat) (Fig. 7D). Two methods were employed for this test: short-term exposure to MV (50 mM for 3 h) and long-term exposure (1.7 mM for 18 h). In both assays, the susceptibility of ΔDacA to oxidative stress was significantly greater than that of the wild type (3.3 times more sensitive at the short-term exposure and 4.3 times at the long-term exposure). In contrast, ΔPde2 was 1.5 times more resistant than the wild type only in the long-term exposure. ΔGdpPΔPde2 behaved like ΔPde2 (data not shown). ΔGdpP behaved like the wild type. These data demonstrate that c-di-AMP contributes to multiple mechanisms of stress resistance, including salt, pH, and oxidative stresses.
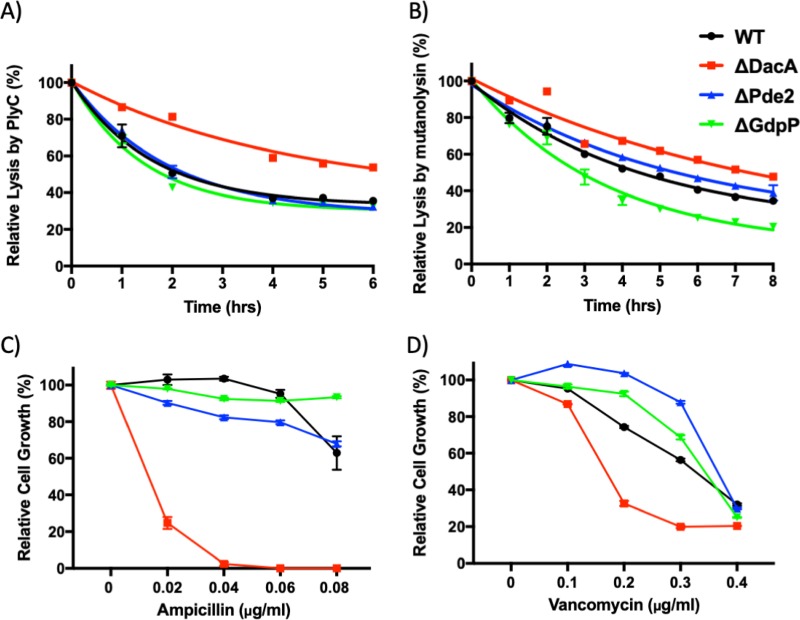
ΔDacA is more resistant to cell wall-degrading enzymes and more susceptible to cell wall-targeting antibiotics than the wild type.
Since c-di-AMP has been demonstrated to influence cell wall properties (8), the rates of lysis of the mutants upon exposure to cell wall-degrading enzymes were examined. S. pyogenes cells were treated with PlyC or mutanolysin, and their relative lysis was determined by measuring cell density (OD600). PlyC, an N-acetylmuramoyl-l-alanine amidase produced by streptococcal phage C1, cleaves the cell wall of S. pyogenes by hydrolyzing the amide bond connecting N-acetylmuramic acid and the first l-alanine of the murein pentapeptide (61, 62). Mutanolysin is an N-acetylmuramidase that hydrolyzes the β-N-acetylmuramyl-(1→4)-N-acetylglucosamine linkage of the bacterial peptidoglycan (64). In this assay, ΔPde2 was comparable to the wild type under both treatments (Fig. 8A and B). ΔGdpP was more susceptible only to mutanolysin treatment. In contrast, ΔDacA was less susceptible to both enzymes than the wild type (Fig. 8A and B).
FIG 8.
Susceptibility of S. pyogenes mutants to cell wall-degrading enzymes and antibiotics inhibiting cell wall synthesis. (A and B) The lysis of mutant strains by the cell wall-hydrolyzing enzymes PlyC (A) and mutanolysin (B) was examined. A stationary-phase culture of each strain was incubated with PlyC (3,000 units/ml) or mutanolysin (100 units/ml) at 37°C. The turbidity (OD600) was measured to determine the degree of cell lysis over time. (C and D) The growth of the mutants treated with sublethal levels of the cell wall-targeting antibiotics ampicillin (C) and vancomycin (D) was monitored. The turbidity (OD600) of overnight growth in C medium containing an antibiotic was measured to determine cell density. The figure present the means and standard deviations of data from experiments performed in triplicate. The significance of the difference between the data points for each mutant and the wild type was calculated by two-way ANOVA followed by Dunnett’s multiple-comparison test. (A) The values of all ΔDacA data points are different (P < 0.001) from those of the wild type. (B) The values of all data points except those for ΔPde2 at 1 h and ΔGdpP at 1 h are different (at least P < 0.05) from those for the wild type. (C) The values of all data points except those for ΔGdpP with 0.02 μg/ml ampicillin, ΔGdpP with 0.06 μg/ml ampicillin, and ΔPde2 with 0.08 μg/ml ampicillin are different (P < 0.001) from those for the wild type. (D) The values of all data points except those for ΔGdpP with 0.1 μg/ml vancomycin are different (at least P < 0.01) from those for the wild type. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), pde2 deletion mutant (ΔPde2), and gdpP deletion mutant (ΔGdpP).
To further investigate the differences in cell walls, the susceptibility of S. pyogenes mutants to the cell wall-targeting antibiotics ampicillin and vancomycin was examined. Cells were grown overnight in C medium containing sublethal concentrations of ampicillin or vancomycin, and their growth was determined by measuring the OD600. Ampicillin acts as an irreversible inhibitor of transpeptidase, which is required to synthesize the bacterial cell wall. Vancomycin inhibits Gram-positive cell wall synthesis by binding to terminal d-alanyl-d-alanine moieties of the N-acetylglucosamine/N-acetylmuramic acid-peptide, which inhibits the action of both transglycosylase and transpeptidase (65). Among the mutants, ΔDacA showed the most severe growth defect in the antibiotic treatment (Fig. 8C and D). Together, these data demonstrate that c-di-AMP plays a significant role in cell wall remodeling.
The virulence of ΔDacA and ΔPde2 is highly attenuated in a murine model of soft tissue infection.
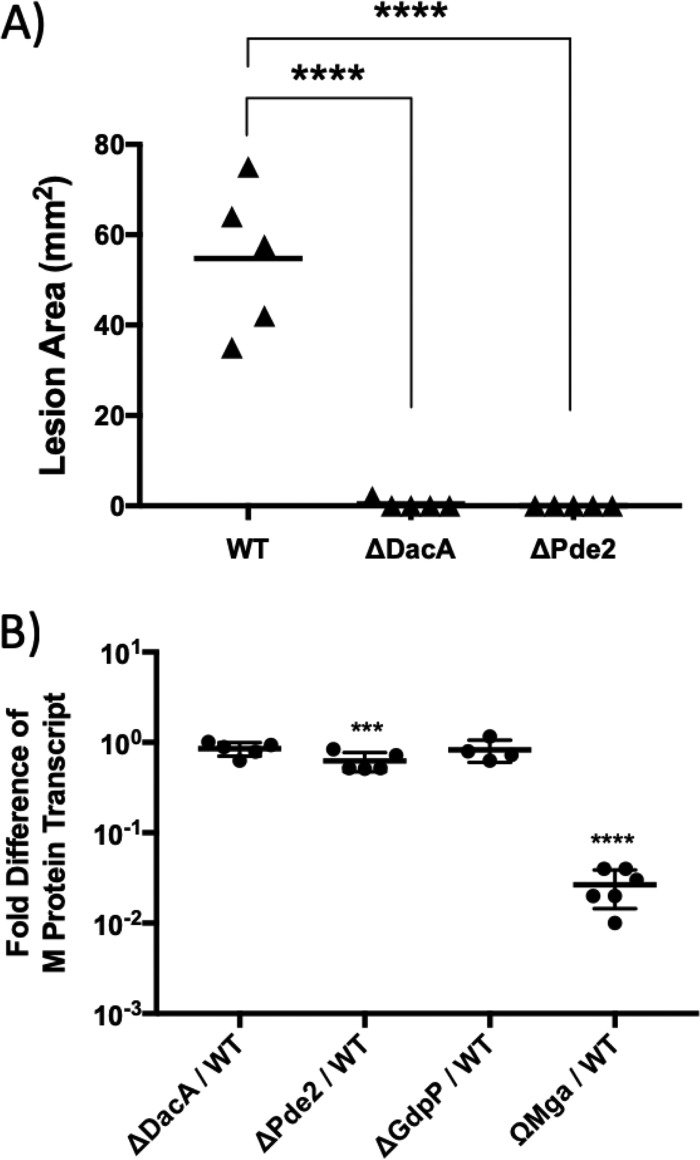
The ability of the ΔDacA and ΔPde2 mutants to cause disease in soft tissue was evaluated using a murine subcutaneous infection model. The sizes of lesions caused by strains were measured at 3 days postinfection, when ulcer formation was maximal. In this infection model, ΔDacA and ΔPde2 did not produce any detectable lesions (Fig. 9A), while the wild type produced significant lesions, demonstrating that the virulence of ΔDacA and ΔPde2 is severely attenuated. We have previously shown that gdpP deletion attenuates S. pyogenes virulence to 50% in the same infection model (36).
FIG 9.
The deletion of dacA or pde2 attenuates the virulence of S. pyogenes. (A) The ability of the ΔDacA and ΔPde2 mutants to cause lesions in a murine subcutaneous infection model is shown. Virulence was evaluated on the basis of the area of the lesion produced at the time when lesion formation was maximal (3 days postinfection). The triangles represent lesion sizes in mice caused by the injection of the wild type, ΔDacA, or ΔPde2. The solid bars indicate the mean values of the ulcer sizes. The asterisks above each bracket indicate the significance of differences (****, P < 0.0001) between each mutant and the wild type as calculated by the Mann-Whitney U test statistic. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), and pde2 deletion mutant (ΔPde2). (B) Relative expression of the M protein transcript by strains. The transcript amount of the M protein gene (emm) in mutant strains relative to that in the wild type was determined with real-time RT-PCR. As a negative control, ΩMga, a mutant with a mga gene disruption, was used. Mga is the major positive transcriptional regulator for emm expression. Asterisks indicate the significance of differences (***, P < 0.001; ****, P < 0.0001) between the mutant and the wild type as calculated by one-way ANOVA followed by Dunnett’s multiple-comparison test. The following strains were tested: wild type (WT), dacA deletion mutant (ΔDacA), pde2 deletion mutant (ΔPde2), gdpP deletion mutant (ΔGdpP), and mga disruption mutant (ΩMga).
M protein, a cell surface-anchored adhesin and antiphagocytic factor that influences virulence in animal models, can occasionally become lost during the process of in-frame deletion (66). When the amounts of M protein transcript in mutants were quantitated through qRT-PCR, they were comparable to that in the wild type, indicating that the M protein expression ability of the strains used in this study was not impaired (Fig. 9B).
DISCUSSION
The sole c-di-AMP synthase DacA in S. pyogenes is not essential for growth in rich media.
Due to the crucial role of c-di-AMP in multiple cellular processes, c-di-AMP is often essential for growth in standard laboratory media in some c-di-AMP-producing bacteria. In B. subtilis, c-di-AMP is essential for growth in rich media due to its involvement in the regulation of potassium ion transport, which is required for growth, but is not essential in a medium containing a low K+ concentration (67). The sole c-di-AMP synthase DacA of L. monocytogenes is essential for growth in a rich medium because the loss of dacA increases production of the alarmone (p)ppGpp, which inhibits growth through inactivation of the transcriptional activator CodY (54). However, DacA is not essential in a minimal medium that favors CodY inactivation.
DacA appears to be the sole c-di-AMP synthase in streptococci, because the deletion of dacA abolishes the production of c-di-AMP in S. pyogenes (this study) and S. mutans (68). However, unlike the case for L. monocytogenes, S. pyogenes dacA was deleted in a rich medium without suppressor mutations in our study, indicating that DacA in S. pyogenes is not essential under standard lab growth conditions. Previous studies have shown that DacA is not essential for growth in some streptococci, such as S. sanguinis and S. mutans (68, 69). Even though DacA is not essential for growth in S. pyogenes, dacA deletion exhibited phenotypes with pleotropic defects, indicating that c-di-AMP plays crucial roles in S. pyogenes growth and virulence.
c-di-AMP appears to influence the expression or activity of a SpeB transcriptional regulator.
This study demonstrates that all the enzymes identified in S. pyogenes that are involved in c-di-AMP synthesis and degradation influence SpeB production. The deletion of dacA and pde2 almost completely abolished SpeB production, while gdpP deletion reduced it. The deletion of gdpP causes SpeB reduction at a posttranscriptional level, which is in agreement with our previous study (36). In contrast, the defect of SpeB production caused by dacA or pde2 deletion is caused at the transcriptional level. Interestingly, although DacA and Pde2 are involved in opposing reactions (synthesis and degradation of c-di-AMP, respectively), their null mutants presented similar phenotypes with respect to SpeB production. This result may indicate that an optimum cellular concentration of c-di-AMP is required for proper SpeB production. Since c-di-AMP regulates the expression of speB at the transcriptional level, it is highly plausible that a SpeB transcription regulator mediates in this control.
The phenotypes of the c-di-AMP PDE mutants ΔGdpP and ΔPde2 are not the same, implying that their cellular roles are different.
Most microorganisms possess two c-di-AMP-hydrolyzing PDEs that may fulfill different cellular roles, such as sensing different signals or displaying different substrate preference. In L. monocytogenes, PgpH is preferentially expressed in broth culture, while GdpP is preferentially expressed during intracellular infection in eukaryotic host cells (34), suggesting that the expression or activity of these enzymes is regulated by different stimuli. S. pyogenes produces two PDEs, GdpP and Pde2. No homolog of PgpH has been identified in Streptococcus spp. GdpP is attached to the membrane through two membrane-spanning helices at its N terminus (34, 35). A PAS (Per-Arnt-Sim) sensory domain follows the membrane-spanning helices. This domain can bind to heme, which inhibits phosphodiesterase activity (70). GdpP contains a catalytic DHH/DHHA1 domain (DHH, Asp-His-His) that cleaves c-di-AMP into pApA. The operon containing gdpP appears to be conserved among all GdpP-producing bacteria. In the operon, gdpP is cotranscribed with the genes of the ribosomal protein L9 (RpL9) and the DNA helicase DnaB (71). Pde2 is a recently discovered c-di-AMP-degrading enzyme containing a DHH/DHHA1 domain. It was first identified in S. pneumoniae (32, 72), and its enzymatic activity and structure have been studied in S. aureus (57), Mycobacterium spp. (73–75), and Borrelia burgdorferi (76). S. pyogenes produces an ortholog with 60.8% amino acid identity to S. pneumoniae Pde2. Pde2 is a cytoplasmic protein that can degrade c-di-AMP but preferentially hydrolyzes linear pApA to AMP, as demonstrated in S. pneumoniae and S. aureus (32, 57). Deletion of pde2 leads to an increase of intracellular pApA levels, which in turn increases the c-di-AMP concentration as pApA inhibits the c-di-AMP-hydrolyzing activity of GdpP (57). Thus, Pde2 also plays a key role in controlling intracellular c-di-AMP levels.
In this study, the PDE gene deletion strains ΔGdpP and ΔPde2 presented phenotypic differences even though their c-di-AMP production ability is the same (Fig. 2). Generally, ΔPde2 exhibited a more severe defect than ΔGdpP. The growth rate of ΔPde2 was significantly lower than that of ΔGdpP (Fig. 3), the defect in SpeB production by ΔPde2 was more severe than that by ΔGdpP (Fig. 4), and virulence of ΔPde2 was more attenuated than that of ΔGdpP (Fig. 9). These data suggest that the cellular roles of the two PDEs are different in S. pyogenes. They may be involved in different cellular processes, their expression or activities may be stimulated by different environmental signals, or their distinct enzymatic activities may cause different phenotypic outcomes. Pde2 is an oligonucleotidase family protein. This family includes nano-RNase proteins that degrade small RNA oligonucleotides such as pApA. pApA, which is accumulated by the deletion of pde2, might play unique cellular roles separate from those of c-di-AMP in S. pyogenes. The pde2 deletion also likely results in elevation of many other nano-RNA molecules in the cell, some of which might have cellular functions. Similar to Pde2, a nano-RNase in Gram-negative bacteria has been implicated in pGpG degradation. Deletion of this protein gene results in elevated levels of pGpG, which in turn inhibits c-di-GMP phosphodiesterases (77).
c-di-AMP appears to regulate the production of a factor other than M protein to control biofilm formation in S. pyogenes.
Just as c-di-GMP has long been known to affect biofilm formation (78), c-di-AMP has recently been recognized as playing a role in biofilm formation in some bacteria. In a B. subtilis mutant lacking both c-di-AMP PDEs, GdpP and PgpH, the expression of genes involved in biofilm formation is reduced significantly (79). In Streptococcus suis, a high c-di-AMP level due to the deletion of gdpP increases biofilm formation (80). In S. mutans, both high and low c-di-AMP levels promote biofilm formation. A DacA null mutation promotes biofilm formation in S. mutans (68), and a null mutation of a c-di-AMP PDE, PdeA, also enhances biofilm formation by upregulating the expression of gtfB, encoding a major glucan-producing enzyme (81). Furthermore, a GdpP null S. aureus mutant produces 3 times more biofilm than the wild type (8). It therefore appears that c-di-AMP’s influence on biofilm formation by bacteria is quite complex. This lack of consistency may be due to the fact that bacterial biofilms are highly complex and nonuniform structures, and there are many different processes that feed into biofilm formation. Our study also demonstrates that c-di-AMP plays a role in biofilm formation in S. pyogenes. The ΔDacA mutant produced almost no biofilm, while ΔPde2 formed 1.4 times more biofilm than the wild type (Fig. 6). It has been shown that M protein plays a crucial role in the initial attachment of S. pyogenes to abiotic surfaces during biofilm formation (60). The wild type and the mutants tested in this study expressed almost the same amount of M protein transcript (Fig. 9B), indicating that c-di-AMP controls the production of a factor or factors other than M protein that influence biofilm formation in S. pyogenes. Further investigation is therefore necessary to elucidate the mechanism(s) by which c-di-AMP influences biofilm formation.
c-di-AMP is required for stress response and influences cell wall properties of S. pyogenes.
c-di-AMP has been reported to be associated with various stress responses in bacteria. For example, S. aureus mutants lacking GdpP are more than 13% smaller than their wild-type counterparts and display an 8-fold increase in resistance against cell wall- and membrane-targeting antibiotics (8, 52). The cellular level of c-di-AMP in S. aureus increases at the poststationary phase, which presumably supports bacterial growth and survival in the later stages of growth (8). PDE mutant strains of L. monocytogenes displaying high c-di-AMP levels produce very weak cell walls, which are unable to resist acid stress and readily lyse (53). PDE mutant strains also gain resistance to antibiotics due to the loss of cell wall homeostasis (53). In S. pneumoniae, alterations in c-di-AMP levels via mutation of either Pde1 (GdpP ortholog) or Pde2 result in poor growth and short-chain formation, as well as altered uptake of potassium ions (31). Deletion of ossG (dacA ortholog) in S. thermophilus increases the susceptibility to oxidative stress created by methyl viologen (paraquat) (82).
In our study, ΔDacA showed increased susceptibility to salts, pH, and oxidative stress. Unlike the wild type, ΔDacA did not grow in almost all media containing more than 0.1 N KCl or NaCl (Fig. 7A and B). Since alteration of c-di-AMP levels changes potassium ion channel activity in other bacteria (52, 83), the growth impairment seen for KCl and NaCl could be due to the role of c-di-AMP in potassium ion homeostasis. Potassium ions are the most abundant cations inside cells and play a critical role in responding to osmotic stress; therefore, dysfunction of the potassium ion transport system could make cells vulnerable to osmotic stress in general. Also, ΔDacA was more sensitive to low pH and oxidative stress than the wild type (Fig. 7C and D). Taken together, these results indicate that c-di-AMP in S. pyogenes plays an important role in stress response and is necessary to adapt to adverse stress conditions.
When S. pyogenes strains were challenged by cell wall-targeting enzymes and antibiotics, once again, ΔDacA showed the most dramatic phenotypes. ΔDacA exhibited increased resistance to both of the cell wall-targeting enzymes tested, PlyC and mutanolysin (Fig. 8A and B). PlyC cuts peptides off the cell wall carbohydrate chain, and mutanolysin cleaves between carbohydrates in the carbohydrate chain. Since these two enzymes target different sites on peptidoglycan cell walls, the c-di-AMP null condition might alter the accessibility of these enzymes to cell walls. Unlike the smaller size of ΔGdpP mutants of S. aureus (8), there were no observable differences in cell sizes or chain lengths in all the mutants we tested (data not shown). In contrast to the case for the cell wall-hydrolyzing enzymes, ΔDacA was more susceptible to sublethal-level cell wall-targeting antibiotics such as ampicillin and vancomycin (Fig. 8C and D). Thus, similar to the case for other Gram-positive bacteria, c-di-AMP likely plays a role in the regulation of cell wall synthesis in S. pyogenes.
The c-di-AMP signaling pathway may serve as an excellent target to develop antivirulence drugs.
Even though c-di-AMP is not essential for growth in rich medium, perturbation of cellular c-di-AMP levels exhibited pleotropic phenotypes, indicating that c-di-AMP plays crucial roles in S. pyogenes physiology. Additionally, the data presented here demonstrate that c-di-AMP regulates the expression of the virulence factor SpeB, indicating that c-di-AMP likely plays a crucial role in the virulence of S. pyogenes (Fig. 4). Indeed, the virulence of ΔDacA and ΔPde2 was almost completely attenuated in a mouse model of subcutaneous infection (Fig. 9). Since c-di-AMP is not essential for growth but abolishes virulence, its signaling pathway may serve as an excellent target to develop antivirulence drugs (84). To develop the antivirulence drugs, it is imperative to understand in detail the mechanisms of how c-di-AMP controls bacterial physiology and virulence, and this study paves the way to further unveil the roles of c-di-AMP signaling networks in the pathogenesis of S. pyogenes.
MATERIALS AND METHODS
Bacterial strains and media.
S. pyogenes HSC5 (emm genotype 14) (37, 38) was employed for all experiments including strain construction. SF370 locus numbers (SPy_####) are used as references for genes in HSC5 (50). Molecular cloning experiments utilized Escherichia coli DH5α or TOP10 (Invitrogen), which was cultured in Luria-Bertani broth. Routine culture of S. pyogenes employed Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (Difco) (THY medium) with incubation at 37°C in sealed tubes without agitation. Unless otherwise indicated, C medium (85) was used to grow S. pyogenes for SpeB activity assay, Western blotting, and RNA preparation for real-time PCR. To produce solid media, Bacto agar (Difco) was added to a final concentration of 1.4% (wt/vol). Since S. pyogenes is an aerotolerant anaerobe that grows better under anaerobic conditions, cultures on solid media were incubated under the anaerobic condition produced by a commercial product (GasPak; catalog no. 260678 [BBL]). When appropriate, antibiotics were added to the media at the following concentrations if they are not specified: kanamycin, 50 μg/ml for E. coli and 500 μg/ml for S. pyogenes; and erythromycin, 500 μg/ml for E. coli and 1 μg/ml for S. pyogenes.
Manipulation of DNA.
Plasmid DNA was isolated via standard techniques and used to transform S. pyogenes or E. coli as described previously (86). Restriction endonucleases, ligases, and polymerases were used according to the recommendations of the manufacturers. Chromosomal DNA was purified from S. pyogenes by using a standard kit (Wizard genomic DNA kit [Promega] or GenElute bacterial genomic DNA kit [Sigma]). When required, DNA fragments were purified using the Mini Elute gel extraction kit (Qiagen) following agarose gel electrophoresis.
Strain construction.
In-frame deletion mutations in chromosomal loci were generated by employing the temperature-sensitive shuttle vector pGCP213 (87). Deletion alleles of dacA and pde2 were generated by PCR through a process of overlap extension PCR (88), and the modified alleles were inserted directly into pGCP213 at the M13F and M13R universal primer binding sites utilizing the overlap extension PCR cloning method described in detail elsewhere (89). These plasmids with the deletion alleles were named pΔDacA and pΔPde2, respectively. The generated plasmids were used to replace the wild-type dacA or pde2 by a method that employs the temperature sensitivity of the plasmid replication origin as was described previously (90). To confirm that ΔDacA does not have significant secondary mutations, whole-genome sequencing of the ΔDacA chromosome was performed (see below for the procedure).
Complementation strains were generated using the streptococcal multicopy plasmid pABG5 (59). For the complementation, the dacA or pde2 gene was amplified by PCR and inserted into pABG5, which can express a gene under control of the rofA promoter. The insertion was performed by the fast cloning method or Gibson assembly (92, 93). The plasmids pDacA and pPde2 were transferred into the corresponding strains to make the complemented strains ΔDacA(pDacA) and ΔPde2(pPde2).
The fidelity of all molecular constructs and mutated chromosomal loci was confirmed by PCR and DNA sequencing (Genewiz or Macrogen). Primers for PCR used in this study are listed in Table 1.
TABLE 1.
Primers used
| Category and namea | Sequenceb | Plasmid created or size (kb) of amplified DNA |
|---|---|---|
| Mutagenic primersc | ||
| ΔDacA-M13F-F1 | tgtaaaacgacggccagtCCATTAGTACCAGTGACGACC | pΔDacA |
| ΔDacA-R2 | CTCCTAGAATTTTCTTATACCATGG_ATCGATACTAGATAAATTATTCATACAACTC | |
| ΔDacA-F3 | GAGTTGTATGAATAATTTATCTAGTATCGAT_CCATGGTATAAGAAAATTCTAGGAG | |
| ΔDacA-M13R-R4 | cacacaggaaacagctatgacTCACTTCAGAATCTAATTTGATACG | |
| ΔPde2-M13F-F1 | tgtaaaacgacggccagtCATTTGCAAGACTAGCTTTGATAG | pΔPde2 |
| ΔPde2-R2 | GCAAACAGCGATGAGTTC_GTCAGGATTTTGATGGCG | |
| ΔPde2-F3 | CGCCATCAAAATCCTGAC_GAACTCATCGCTGTTTGC | |
| ΔPde2-M13R-R4 | cacacaggaaacagctatgacGAACTTTTGACGTCCTGTGTAG | |
| 5pABG5 FC | cgaccaagagagccataaacacc | pDacA |
| 3pABG5 FC | cattttctctcctctcgaattcagttcc | |
| 5DacA-pABG5 FC | gaattcgagaggagagaaaATGAATAATTTATCTAGTATCGATATTAAATTTTTATTAAG | |
| 3DacA-pABG5 FC | ttatggctctcttggtcgCGTTTCATTTAGATTTCCCTCCTAGAATTTTC | |
| 5pABG5Gibson | taacaaagtgcaggggccca | pPde2 |
| 3pABG5Gibson | acacttagaaagccaaataagtatttgataagtgtattctcc | |
| 5Pde2-pABG5Gibson | tacttatttggctttctaagtgtAAAAAGAAAGATTAAAGCATGATAACAACTTTTG | |
| 3Pde2-pABG5Gibson | cccctgcactttgttaCTAAATCTCTTGGCAAACAGCGATG | |
| Analysis primersd | ||
| CFΔdacA-F | AGCACCATCTACAATCATCTGATAAGTAGTG | 1.6 in wild type, 0.8 in ΔDacA |
| CFΔdacA-R | TGGAGGTTAGATAAACTTCCGCACC | |
| CFΔgdpP-F | CCAACGCTGTAGAATGGTATAATCC | 1.6 in wild type, 1.1 in ΔGdpP |
| CFΔgdpP-R | CTAATACAAAGCTAGCCTCAACGTG | |
| CFΔpde2-F | ACATCATGGCCCAATTCTTG | 2.0 in wild type, 1.0 in ΔPde2 |
| CFΔpde2-R | TGCTTATTTTGCTGCGTTTG | |
| RTspeB-F | TGTCGGTAAAGTAGGCGGAC | |
| RTspeB-R | GAGCTGAAGGGTTTAGTGCG | |
| RTemm-F | TTCAGACGCAAGCCGTAAG | |
| RTemm-R | TCTAAATCACGGCGAAGACC | |
| RTgyrA-F | AACAACTCAAACAGGTCGGG | |
| RTgyrA-R | CTCCTTCACGGCTAGATTCG | |
| Primers for PlyC expression | ||
| PlyCA-F | GTACCCGGGAAAGGGAGAATTTATTTAATG | 1.4 |
| PlyCA-R | CCCAAGCTTTCAATGATGATGATGATGATGGTCGACGGCGCTATTTTTAAATGTTATCAAACCAGTTAG | |
| PlyCB-F | GTACCCGGGGAAGTAATTTCCATTCTTGAA | 0.3 |
| PlyCB-R | CCCAAGCTTTTACTTTTTCATAGCCTTTCT | |
Primers are categorized as forward (F) or reverse (R) relative to the direction of a transcript. Forward primers anneal to the noncoding template strand, while reverse primers anneal to the coding strand.
Sequences are shown 5′ to 3′. Uppercase sequences anneal to the HSC5 chromosome, and lowercase sequences anneal to plasmid sequences. Underscores indicate junctions between contiguous DNA regions. Restriction enzyme sites are underlined. The 6× His tag sequence is in bold.
Mutagenesis primers were used for PCRs to amplify DNA segments used to construct plasmids for gene deletion.
Analysis primers were used in regular PCR to confirm (CF) gene deletion or in qRT-PCR (RT) to measure the level of gene transcription.
Generation of the mutants ΔGdpP, ΩEmm, and ΩMga has been described elsewhere (36, 60). The emm gene encodes M protein, and mga encodes Mga, a positive transcriptional regulator for M protein expression. ΩEmm and ΩMga have a gene disruption of emm or mga, respectively, due to a plasmid insertion into the genes through homologous recombination, so they do not express M protein.
Whole-genome sequencing.
The chromosome of the dacA deletion strain ΔDacA was purified using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA) and qualified utilizing Agilent Bioanalyzer DNA 12000 assays, and its concentration was measured via Qubit. Genomic DNA was sonicated to an average size of 175 bp. The fragments were blunt ended, an “A” base was added to 3′ ends, and the fragments were ligated with Illumina’s sequencing adapters to the ends. The ligated fragments underwent amplification incorporating a unique indexing sequence tag. The resulting libraries were sequenced by the Washington University Genome Technology Access Center (https://gtac.wustl.edu/) using the Illumina HiSeq2500 (Illumina, San Diego, CA, USA) as single-end reads extending 50 bases of the library fragments, yielding >4 million reads (approximately 100× coverage). The sequences were compared to that of the wild-type HSC5 using DNAStar SeqMan NGen v 4.0.0 (DNAStar) with the default stringency parameters.
qRT-PCR.
RNA isolation from S. pyogenes cultures and quantitative reverse transcriptase PCR (qRT-PCR) were conducted as described elsewhere (36). The primers for qRT-PCR are listed in Table 1. The gyrase A subunit gene (gyrA) was used as the internal reference gene to normalize the expression level of a specific transcript between samples (94). The reported data represent the means and standard errors from three independent assays, each performed on a different day with a new RNA sample.
Determination of doubling time.
Strains were grown overnight in THY medium and diluted 1:100 into fresh THY or C medium. The optical density at 600 nm (OD600) of the cultures was measured every 30 min. The OD600 values in exponential-growth-phase cells, which show a linear increase over time (x) versus log10 OD600 (y), were used to calculate the time (minutes) needed for the cell density (OD600) to double [doubling time (minutes) = log10 2/(Δlog10 OD600/minutes)]. The reported data represent the means and standard errors derived from four independent assays. Lag times were determined by measuring the time to reach to the exponential phase from inoculation.
Analysis of SpeB expression.
The measurement of SpeB expression was performed using a protease indicator medium, Western blot analysis, and SpeB activity assay as described previously (56, 95).
Measurement of biofilm formation.
The degree of biofilm formation by strains was measured as described previously with some modifications (60). Crystal violet (3%, wt/vol) was used instead of safranin to stain S. pyogenes biofilm, which was quantified by measuring the OD600.
Effect of high salt concentration on cell growth.
The ability of S. pyogenes strains to tolerate salt stress was assessed through the addition of various concentrations of KCl and NaCl to media. S. pyogenes cells grown overnight in THY medium were inoculated into fresh THY medium containing various concentrations (0.1 N, 0.2 N, and 0.3N) of KCl or NaCl and incubated at 37°C. Cell growth (OD600) was measured at 8 h postinoculation, when cells reached stationary phase.
Effect of pH on cell growth.
S. pyogenes cells grown in THY medium overnight were inoculated into fresh THY and C medium whose pH was adjusted to 4, 5, or 6 with 2 N HCl. Cell growth (OD600) was measured at 8 h postinoculation, when cells reached stationary phase.
Effect of oxidative stress on cell growth.
Susceptibility to oxidative stress was measured by exposing cells to methyl viologen (MV) (82). Briefly, S. pyogenes strains were grown in THY medium until the OD600 reached 0.6. For short-term exposure, each culture was divided into two 7.5-ml subcultures in 15-ml screw-cap tubes (at a liquid/air space ratio of 1:1), and MV (paraquat; Sigma-Aldrich) was added to one of the subcultures at a final concentration of 50 mM. Cultures were further incubated at 37°C for 3 h, and serial dilutions were spread onto THY agar plates for counting CFU. For long-term exposure, 3-ml subcultures (OD600 = 0.6) were placed in 15-ml screw-cap tubes (at a liquid/air space ratio of 5:1), supplemented or not with 1.7 mM MV, and incubated at 37°C for 18 h. Serial dilutions of cultures were spread onto THY agar plates for CFU counting. MV sensitivity was measured by comparing recoverable CFU values from MV-treated cultures to those from nontreated cultures.
Effect of mutanolysin or PlyC treatment on cell lysis.
S. pyogenes cells grown in THY medium overnight were harvested, washed twice with 10 ml phosphate-buffered saline (PBS), and resuspended in PBS to an OD600 of 1.0. One milliliter of each cell suspension was transferred to a 1.5-ml cuvette, and mutanolysin (100 units; Sigma) or PlyC (3,000 units) (61, 96) was added. The cysteine protease inhibitor E-64 (final concentration, 20 μM) that inactivates secreted SpeB was also added to these solutions. The solutions were incubated at 37°C, and cell lysis was monitored by measuring the OD600 every hour up to 8 h. These experiments were performed in triplicate.
PlyC is a multimeric enzyme composed of one PlyCA subunit and eight PlyCB subunits (61). For the purification of PlyC, the PlyC genes, plyCA and plyCB, were amplified separately from purified phage stocks through PCR with primers listed in Table 1. The 6× histidine tag sequence was embedded in the reverse primer to tag PlyCA at the C terminus. Each amplified gene was inserted behind the arabinose-inducible promoter in pBAD33 (97) and transferred to E. coli TOP10 cells. After each subunit of PlyC was expressed separately in E. coli, cells were lysed with a French pressure cell. The two cell extracts containing PlyCA or PlyCB were mixed and incubated with gentle shaking at 4°C overnight. PlyC was then purified from the mixed cell extract through affinity chromatography using the Talon polyhistidine tag purification resin (Thermo Scientific).
Susceptibility to sublethal concentrations of ampicillin or vancomycin.
The susceptibility of mutant strains to sublethal concentrations of cell wall-targeting antibiotics was monitored. S. pyogenes cells grown in THY medium overnight were harvested and inoculated in fresh C medium containing ampicillin (0.00, 0.02, 0.04, 0.06, or 0.08 μg/ml) or vancomycin (0.0, 0.1, 0.2, 0.3, or 0.4 μg/ml). Cells were then grown overnight at 37°C, and the OD600 of the cultures was measured to determine the final cell density. This experiment was performed in triplicate.
Quantification of c-di-AMP in cell extracts by ELISA.
The quantity of c-di-AMP in cell extracts was measured by competitive enzyme-linked immunosorbent assay (ELISA) as described previously (57, 58). S. pyogenes strains were grown to the exponential phase (OD600 = ∼0.4) in 10 ml THY medium, washed three times with PBS, resuspended in 1 ml PBS, and lysed by PlyC treatment. The clear supernatant of the culture was collected in a fresh tube after centrifugation at 7,000 relative centrifugal force (rcf) at 4°C for 10 min. The cell lysates were then boiled for 10 min. Clear supernatants were collected after centrifugation and stored at −80°C until used to measure intracellular c-di-AMP concentration. Since E. coli does not produce c-di-AMP, E. coli TOP10 cells (Invitrogen) were used as a negative control for the assay. TOP10 cells were cultured to the exponential phase (OD600 = ∼0.4) in 10 ml LB broth with continuous shaking at 37°C, washed three times with PBS, resuspended in 1 ml 50 mM Tris buffer (pH 8), lysed with beads using Fast Prep 24 5G (MP Biomedical) at 6.0 m/s for 40 s, and stored at −80°C until used to measure intracellular c-di-AMP concentration.
CabP protein was purified as previously described (57). Briefly, E. coli BL21(DE3) with the recombinant plasmid pET28b-His-CabPspT from the Grundling lab was grown to an OD600 of ∼0.6 in 600 ml LB medium containing 30 μg/ml kanamycin at 37°C in a shaker. The culture was then incubated at room temperature for an additional 4 hours with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce CabP production. Next, cells were harvested, washed, and resuspended in 20 ml lysis buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM imidazole, and 10% glycerol). The cells were then lysed by bead beating using the Fast Prep 24 5G (6.0 m/s for 40 s twice). The beads and cell debris were removed by centrifugation at 13,000 rcf at 4°C for 20 min, and then 8-ml cell lysates containing His-tagged CabP proteins were loaded onto the column with 1 ml HisPur cobalt resin (Thermo Scientific) and incubated for 1 h. The resin was washed with 10 ml of lysis buffer containing 10 mM imidazole, 20 mM imidazole, and finally 30 mM imidazole, successively. Subsequently, the proteins were eluted in 4 ml lysis buffer containing 250 mM imidazole. The eluted proteins were then dialyzed twice against 1 liter of PBS at 4°C for 1 h and then overnight against 1 liter of PBS containing 10% glycerol. The proteins were concentrated using a Centriprep centrifugal filter unit (Sigma-Aldrich) at 2,000 × g at 4°C until they reached ∼0.2 mg/ml. The protein concentration was measured with the Bradford assay kit (Sigma-Aldrich). Finally, the purity of the protein was determined by SDS-PAGE, and the protein samples were stored in small aliquots at −80°C until used.
The purified CabP was diluted to 50 μg/ml in coating buffer (50 mM Na2CO3, 50 mM NaHCO3, pH 9.6), and 100 μl of the solution was added to each well to coat the wells of a 96-well flat-bottom plate (Dot Scientific Inc.). The coated plates were sealed with plastic wrap and incubated overnight (14 to 18 h) at 4°C. The coated wells were then washed three times with PBS containing 0.05% Tween 20 (PBST) and blocked with 5% bovine serum albumin (BSA). The cell extract samples were diluted (5 times) with 50 mM Tris buffer (pH 8). For preparing a standard curve, c-di-AMP (Biolog) solutions containing 0.5 nmol to 250 nmol were prepared in 50 mM Tris buffer (pH 8). Biotinylated c-di-AMP (Biolog) was added to all the controls and samples to give the final amount of 25 nmol. 100 μl of all the controls and samples were added to the coated wells (in triplicate). The plates were incubated for 2 h at room temperature. Each well of the plates was washed three times with 200 μl PBST. Next, 100 μl of 0.1-μg/ml high-performance streptavidin (Thermo Fisher Scientific) in PBS was added and incubated for 1 h. Wells were washed three times with PBST before adding substrate. One hundred microliters of the substrate (0.5 mg of o-phenylenediamine dihydrochloride [Sigma-Aldrich] in citrate buffer [pH 5] containing 20 μl H2O2) was added to each well and incubated for 30 min at room temperature. Finally, the reactions were stopped with 100 μl of 2 M H2SO4. The OD492 was measured for each well using a plate reader (EL-808 Ultra microplate reader; Bio-Tek Instruments, Inc.). A standard curve was generated to measure the level of c-di-AMP. The concentrations of the samples were plotted along the x axis against OD492 on the y axis, and a polynomial standard curve was generated to find the concentration of c-di-AMP in the samples.
Murine subcutaneous infection.
The ability of S. pyogenes strains to cause disease in soft tissue was evaluated using 6- to 8-week-old SKH1 hairless mice (Charles River Labs) as described previously (36). Each mouse was subcutaneously injected with approximately 1 × 107 CFU in a 100-μl volume into the right flank. The area of the lesion that formed was documented every 24 h by digital photography, and the lesion area was calculated from the digital record using ImageJ (NIH). Any differences in the areas of lesions between experimental groups were tested for significance by the Mann-Whitney U test. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. This animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Indiana State University (ISU). All mice were anesthetized with isoflurane when the lesion sizes were measured and were euthanized by carbon dioxide asphyxiation at the end of the experiment.
Statistical testing.
All statistical tests were performed using GraphPad Prism (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
ACKNOWLEDGMENTS
We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. We also thank Michael Caparon (Washington University) for sharing S. pyogenes mutant strains and Angelica Grundling (Imperial College London) for sharing the plasmid pET28b-His-CabPspT.
This study was supported in part by National Institutes of Health grant 7R15 GM101603-02 and Indiana Academy of Science grant 549237 to Kyu Hong Cho. The Genome Technology Access Center is partially supported by NCI Cancer Center Support grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA grant UL1TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research.
This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
REFERENCES
- 1.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 3.Hengge R, Grundling A, Jenal U, Ryan R, Yildiz F. 2016. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J Bacteriol 198:15–26. doi: 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo YL, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev 42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 5.Pesavento C, Hengge R. 2009. Bacterial nucleotide-based second messengers. Curr Opin Microbiol 12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Huynh TN, Choi PH, Sureka K, Ledvina HE, Campillo J, Tong L, Woodward JJ. 2016. Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol Microbiol 102:233–243. doi: 10.1111/mmi.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrade WA, Firon A, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA, Trieu-Cuot P, Golenbock DT, Kaminski PA. 2016. Group B streptococcus degrades cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell Host Microbe 20:49–59. doi: 10.1016/j.chom.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandara C, Alonso JC. 2015. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair (Amst) 27:1–8. doi: 10.1016/j.dnarep.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Kamegaya T, Kuroda K, Hayakawa Y. 2011. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J Med Sci 73:49–57. [PMC free article] [PubMed] [Google Scholar]
- 11.Pham TH, Liang ZX, Marcellin E, Turner MS. 2016. Replenishing the cyclic-di-AMP pool: regulation of diadenylate cyclase activity in bacteria. Curr Genet 62:731–738. doi: 10.1007/s00294-016-0600-8. [DOI] [PubMed] [Google Scholar]
- 12.Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. 2013. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4:e00018-13. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahmi T, Port GC, Cho KH. 2017. c-di-AMP: an essential molecule in the signaling pathways that regulate the viability and virulence of Gram-positive bacteria. Genes (Basel) 8:197. doi: 10.3390/genes8080197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Li W, He ZG. 2013. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem 288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep 12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. 2006. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. 2013. Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem 288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths JM, O'Neill AJ. 2012. Loss of function of the GdpP protein leads to joint beta-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 56:579–581. doi: 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level beta-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh TN, Woodward JJ. 2016. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol 30:22–29. doi: 10.1016/j.mib.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corrigan RM, Grundling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 25.Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stulke J. 2015. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol 97:189–204. doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 26.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, Stulke J. 2015. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol 197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg J, Dickmanns A, Neumann P, Gunka K, Arens J, Kaever V, Stulke J, Ficner R, Commichau FM. 2015. Structural and biochemical analysis of the essential diadenylate cyclase CdaA from Listeria monocytogenes. J Biol Chem 290:6596–6606. doi: 10.1074/jbc.M114.630418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y, Helmann J. 2012. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. 2012. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One 7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blötz C, Treffon K, Kaever V, Schwede F, Hammer E, Stülke J. 2017. Identification of the components involved in cyclic di-AMP signaling in Mycoplasma pneumoniae. Front Microbiol 8:1328. doi: 10.3389/fmicb.2017.01328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai YL, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai GC. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. 2013. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol 195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manikandan K, Sabareesh V, Singh N, Saigal K, Mechold U, Sinha KM. 2014. Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS One 9:e86096. doi: 10.1371/journal.pone.0086096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh TN, Luo SK, Pensinger D, Sauer JD, Tong L, Woodward JJ. 2015. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A 112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao F, See RY, Zhang DW, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 286:29441. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho KH, Kang SO. 2013. Streptococcus pyogenes c-di-AMP phosphodiesterase, GdpP, influences SpeB processing and virulence. PLoS One 8:e69425. doi: 10.1371/journal.pone.0069425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Port GC, Paluscio E, Caparon MG. 2013. Complete genome sequence of emm type 14 Streptococcus pyogenes strain HSC5. Genome Announc 1:e00612-13. doi: 10.1128/genomeA.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanski E, Horwitz PA, Caparon MG. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect Immun 60:5119–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapur V, Topouzis S, Majesky MW, Li LL, Hamrick MR, Hamill RJ, Patti JM, Musser JM. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog 15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson A, Norgren M. 2003. Cleavage of antigen-bound immunoglobulin G by SpeB contributes to streptococcal persistence in opsonizing blood. Infect Immun 71:211–217. doi: 10.1128/IAI.71.1.211-217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collin M, Olsen A. 2001. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect Immun 69:7187–7189. doi: 10.1128/IAI.69.11.7187-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collin M, Svensson MD, Sjoholm AG, Jensenius JC, Sjobring U, Olsen A. 2002. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect Immun 70:6646–6651. doi: 10.1128/IAI.70.12.6646-6651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terao Y, Mori Y, Yamaguchi M, Shimizu Y, Ooe K, Hamada S, Kawabata S. 2008. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J Biol Chem 283:6253–6260. doi: 10.1074/jbc.M704821200. [DOI] [PubMed] [Google Scholar]
- 44.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. 2006. Trigger for group A streptococcal M1T1 invasive disease. FASEB J 20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 45.Burns EH Jr, Marciel AM, Musser JM. 1996. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun 64:4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herwald H, Collin M, Müller-Esterl W, Björck L. 1996. Streptococcal cysteine proteinase releases kinins: a virulence mechanism. J Exp Med 184:665–673. doi: 10.1084/jem.184.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapur V, Majesky MW, Li LL, Black RA, Musser JM. 1993. Cleavage of interleukin 1 beta (IL-1 beta) precursor to produce active IL-1 beta by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci U S A 90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen M, Bjorck L. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol Microbiol 43:537–544. doi: 10.1046/j.1365-2958.2002.02766.x. [DOI] [PubMed] [Google Scholar]
- 49.Carroll RK, Musser JM. 2011. From transcription to activation: how group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol 81:588–601. doi: 10.1111/j.1365-2958.2011.07709.x. [DOI] [PubMed] [Google Scholar]
- 50.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A 98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tesorero RA, Yu N, Wright JO, Svencionis JP, Cheng Q, Kim JH, Cho KH. 2013. Novel regulatory small RNAs in Streptococcus pyogenes. PLoS One 8:e64021. doi: 10.1371/journal.pone.0064021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corrigan RM, Bowman L, Willis AR, Kaever V, Grundling A. 2015. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J Biol Chem 290:5826–5839. doi: 10.1074/jbc.M114.598300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. 2013. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4:e00282-13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteley AT, Pollock AJ, Portnoy DA. 2015. The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeden MS, Schuster CF, Bowman L, Zhong Q, Williams HD, Grundling A. 2018. Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J Biol Chem 293:3180–3200. doi: 10.1074/jbc.M117.818716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Port GC, Vega LA, Nylander AB, Caparon MG. 2014. Streptococcus pyogenes polymyxin B-resistant mutants display enhanced ExPortal integrity. J Bacteriol 196:2563–2577. doi: 10.1128/JB.01596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowman L, Zeden MS, Schuster CF, Kaever V, Grundling A. 2016. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J Biol Chem 291:26970–26986. doi: 10.1074/jbc.M116.747709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Underwood AJ, Zhang Y, Metzger DW, Bai G. 2014. Detection of cyclic di-AMP using a competitive ELISA with a unique pneumococcal cyclic di-AMP binding protein. J Microbiol Methods 107:58–62. doi: 10.1016/j.mimet.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Granok AB, Parsonage D, Ross RP, Caparon MG. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J Bacteriol 182:1529–1540. doi: 10.1128/JB.182.6.1529-1540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho KH, Caparon MG. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol 57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 61.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. 2006. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci U S A 103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krause RM. 1957. Studies on bacteriophages of hemolytic streptococci. I. Factors influencing the interaction of phage and susceptible host cell. J Exp Med 106:365–384. doi: 10.1084/jem.106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Calandra GB, Cole RM. 1980. Lysis and protoplast formation of group B streptococci by mutanolysin. Infect Immun 28:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiao Y, Srisuknimit V, Rubino F, Schaefer K, Ruiz N, Walker S, Kahne D. 2017. Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat Chem Biol 13:793–798. doi: 10.1038/nchembio.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho KH, Wright J, Svencionis J, Kim JH. 2013. The prince and the pauper: which one is real? The problem of secondary mutation during mutagenesis in Streptococcus pyogenes. Virulence 4:664–665. doi: 10.4161/viru.26767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gundlach J, Herzberg C, Kaever V, Gunka K, Hoffmann T, Weiß M, Gibhardt J, Thürmer A, Hertel D, Daniel R, Bremer E, Commichau FM, Stülke J. 2017. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal 10:eaal3011. doi: 10.1126/scisignal.aal3011. [DOI] [PubMed] [Google Scholar]
- 68.Cheng X, Zheng X, Zhou X, Zeng J, Ren Z, Xu X, Cheng L, Li M, Li J, Li Y. 2016. Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans. Environ Microbiol 18:904–922. doi: 10.1111/1462-2920.13123. [DOI] [PubMed] [Google Scholar]
- 69.Xu P, Ge X, Chen L, Wang X, Dou Y, Xu JZ, Patel JR, Stone V, Trinh M, Evans K, Kitten T, Bonchev D, Buck GA. 2011. Genome-wide essential gene identification in Streptococcus sanguinis. Sci Rep 1:125. doi: 10.1038/srep00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan E, Rao F, Pasunooti S, Pham TH, Soehano I, Turner MS, Liew CW, Lescar J, Pervushin K, Liang ZX. 2013. Solution structure of the PAS domain of a thermophilic YybT protein homolog reveals a potential ligand-binding site. J Biol Chem 288:11949–11959. doi: 10.1074/jbc.M112.437764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lou Y, Helmann J. 2012. A σD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology 158:2732–2741. doi: 10.1099/mic.0.062174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cron LE, Stol K, Burghout P, van Selm S, Simonetti ER, Bootsma HJ, Hermans PWM. 2012. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect Immun 80:2974. doi: 10.1128/IAI.00519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He Q, Wang F, Liu S, Zhu D, Cong H, Gao F, Li B, Wang H, Lin Z, Liao J, Gu L. 2016. Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase. J Biol Chem 291:14386–14387. doi: 10.1074/jbc.A115.699801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang Q, Luo Y, Zheng C, Yin K, Ali MK, Li X, He J. 2015. Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int J Biol Sci 11:813–824. doi: 10.7150/ijbs.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. 2014. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol 93:65–79. doi: 10.1111/mmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye M, Zhang JJ, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang XF. 2014. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun 82:1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gundlach J, Rath H, Herzberg C, Mader U, Stulke J. 2016. Second messenger signaling in Bacillus subtilis: accumulation of cyclic di-AMP inhibits biofilm formation. Front Microbiol 7:804. doi: 10.3389/fmicb.2016.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du B, Ji W, An H, Shi Y, Huang Q, Cheng Y, Fu Q, Wang H, Yan Y, Sun J. 2014. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol Res 169:749–758. doi: 10.1016/j.micres.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 81.Peng X, Zhang Y, Bai GC, Zhou XD, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol 99:945–959. doi: 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thibessard A, Borges F, Fernandez A, Gintz B, Decaris B, Leblond-Bourget N. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl Environ Microbiol 70:2220–2229. doi: 10.1128/AEM.70.4.2220-2229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y, Pham TH, Nhiep THN, Vu NMT, Marcellin E, Chakrabortti A, Wang YL, Waanders J, Lo R, Huston WM, Bansal N, Nielsen LK, Liang ZX, Turner MS. 2016. Cyclic-di-AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis. Mol Microbiol 99:1015–1027. doi: 10.1111/mmi.13281. [DOI] [PubMed] [Google Scholar]
- 84.Maura D, Ballok AE, Rahme LG. 2016. Considerations and caveats in anti-virulence drug development. Curr Opin Microbiol 33:41–46. doi: 10.1016/j.mib.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loughman JA, Caparon M. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J Bacteriol 188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Caparon MG, Stephens DS, Olsen A, Scott JR. 1991. Role of M protein in adherence of group A streptococci. Infect Immun 59:1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nielsen HV, Guiton PS, Kline KA, Port GC, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. 2012. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. mBio 3:e00177-12. doi: 10.1128/mBio.00177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 89.Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.King KY, Horenstein JA, Caparon MG. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J Bacteriol 182:5290–5299. doi: 10.1128/JB.182.19.5290-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reference deleted.
- 92.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. 2011. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol 11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kang SO, Caparon MG, Cho KH. 2010. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun 78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho KH, Caparon MG. 2008. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun 76:3176–3186. doi: 10.1128/IAI.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci U S A 98:4107–4112. doi: 10.1073/pnas.061038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]