Abstract
Cellulase containing nanobiocatalysts have been useful as an extraction tool based on their ability to disrupt plant cell walls. In this work, we investigate the effect of nanoparticle composition and chemical linkage towards immobilized cellulase activity. Cellulase nanoconstructs have been prepared, characterized and compared for their loading efficiencies with standard assays and enzyme kinetics and correlate well with the cognate loading efficiencies. Application of the cellulase-immobilized nanoparticles on onion skins results in release of a distinctive composition of polyphenols. The aglycosidic form of quercetin is the dominant product of onion skin hydrolysis affected by cellulase nanobiocatalysts. Chitosan-coated iron oxide nanoparticles with APTES-conjugated cellulase are found to be most effective for polyphenol release and for transformation of glycosidic to aglycosidic form of quercetin. These results shed light on the activity of immobilized cellulase beyond their role in cell wall disruption and are important for the practical application of cellulase nanobiocatalysts.
Keywords: Biochemistry, Biotechnology, Bioengineering
1. Introduction
Hydrolytic enzymes have been widely used in the food industry for improving quality of processed food products and for enabling efficient extraction of natural products [1, 2]. Factors such as cost, reusability and stability constrain the industrial applications of these and other enzymes. The immobilization of cellulolytic enzymes on solid supports has been researched as a strategy for enhancing enzyme efficacy and lowering the cost of food processing. Cellulase immobilized on magnetic nanoparticles is found to display superior thermal and pH stability as compared to the bare enzymes [3]. Such constructs permit variation of the activity of enzyme based on loading efficiency [4]. Further, the use of magnetic iron oxide nanoparticles is important towards reusability of the cellulase [3, 4, 5]. A variety of substrates have been used for immobilization of cellulase either through covalent attachment or physical adsorption [6, 7]. The surface character of the substrate in conjunction with the attachment methodology contribute significantly towards the efficiency of tethered cellulases [8]. The vast majority of covalent immobilization approaches employ linkers such as glutaraldehyde and/or aminopropyltriethoxysilane (APTES). This is understandable considering the relative ease of linker chemistry and applicability across a range of substrates [9, 10]. Linker-free covalent approaches for immobilization of cellulases are arguably less-accessible albeit providing superior loading characteristics of the enzyme [11, 12]. Nevertheless, the compatibility of substrates with immobilized cellulases is often viewed from the perspective of ease of conjugation. In this regard, there is a dearth of studies that systematically compare the efficacy of immobilized cellulase across different substrates.
Immobilized cellulase has emerged as a valuable extraction tool owing to the green conditions of operation [13, 14]. The enzyme facilitated breakdown of plant cell walls has been found to release a variety of natural products including pigments and polyphenols [5, 15]. In recent years, interest in food phenolics has increased significantly owing to their antioxidant capabilities based on free-radical scavenging and metal-chelating potential [16, 17]. Epidemiological studies have sought to draw correlations between the consumption of polyphenol-rich foods and lower incidence of heart, gastrointestinal, liver and neurological diseases, cancer, atherosclerosis and obesity [18, 19]. Interaction of bioactive compounds such as polyphenols with disease targets is often investigated under conditions that may not faithfully represent the complexity of the natural product. For example, the majority of dietary polyphenols are present in their glycosidic forms that are poorly absorbed as compared to the aglycosidic counterparts [20, 21]. The presence of polyphenols in aglycosidic and glycosidic forms complicates estimation of their bioavailability [22]. The most abundant sources of polyphenols are expensive and do not form a significant component of daily diet.
In this work, we investigate the effect of nanoparticle composition and chemical linkage towards immobilized cellulase activity on onion skins. Quercetin is the major polyphenol in onion skins and is present in a plethora of glycosidic forms [23]. The main objectives of our work include (1) assessing the products of hydrolysis of onion skins with respect to glycosidic and aglycosidic forms of polyphenols, and (2) screen and incorporate suitable components for the most efficient nanobiocatalyst containing cellulases for onion skin hydrolysis.
2. Materials and methods
2.1. Material
APTES (3-Aminopropyltriethoxysilane, 98%), Glutaraldehyde (25 % in H2O), Glutaraldehyde, Formic acid, Quercetin (>98%), Quercetin-3,4′-diglucoside, Quercetin 3-β-D-glucoside, Citric acid monohydrate, Disodium hydrogen phosphate, Trans-trans muconic acid, Tetradecanedioic acid, Dodecanedioic acid, DIC, Oxyma, EDC-HCl, NHS, Tetraethyl orthosilicate (TEOS 98%), Ammonium hydroxide solution 31.5% (NH4OH), Tetraethyl orthosilicate (TEOS 98%), Ammonium hydroxide (NH4OH), Chitosan (C6H11NO4) with MW 60 kDa, tween-80, CTAB, and Cellulase microcrystalline powder from Aspergillus niger were obtained from Sigma Aldrich (Bangalore, India). FeCl2 was obtained from LOBA Chemie, KOH from SRL, Ethanol Methanol, Ultra-pure water, Acetic acid, Butanol (C4H9OH), and Propanol (C3H7OH) were obtained from Merck. Onion (Allium cepa) was obtained from local groceries. All glassware were soaked in piranha solution (concentrated H2SO4 and 35% H2O2, 3:1 v/v) for 1 h and rinsed with deionized water before use.
2.2. Preparation of iron oxide magnetic nanoparticles (IOMNPs)
Iron Oxide (Fe3O4) magnetic nanoparticles (IOMNPs) were prepared by the alkaline hydrolysis of ferrous ions. 1M KOH solution was added to 50 ml of 0.05 M FeCl2 solution until the pH reached 8, with continuous stirring and then kept for precipitation for 2 hours. Black coloured precipitate was separated and washed two times with ultrapure water and two times with ethanol on a vacuum filter, dried and stored in vacuum desiccator for further use [24].
2.3. APTES functionalization of IOMNPs
50 mg of nanoparticles were dispersed in 50 mL of 10% APTES (3-aminopropyltriethoxysilane) solution in methanol, sonicated for 10 min at room temperature then vortexed at 1000 rpm overnight. The modified particles were magnetically collected in another tube, washed 3 times with methanol, dried and stored in vacuum desiccator [5].
2.4. Attachment of chemical linkers on IOMNPs and cellulase immobilization
For glutaraldehyde cross linking, 40 mg of APTES modified IOMNPs were taken in a separate container. 8 mL of 2.5% glutaraldehyde solution was added separately in each container and sonicated for 10 min. These solutions were gently shaken for 1 hr at room temperature. Unbound glutaraldehyde was removed by washing with PBS (pH 7.0). Cellulase solution (0.5% in PBS) was added and the mixture was shaken continuously overnight. Subsequently, the Cellulase attached IOMNPs were washed three times with water and resuspended in PBS buffer. This mixture was then directly used for the hydrolysis activity of Cellulase [5, 24].
For trans, trans-muconic acid, tetradecanedioic acid and docosanedioic acid cross linking, in tube ‘A’ 20 mg of APTES modified IOMNPs were dispersed in 10 ml DMF solvent by sonication for 15 min. In tube ‘B’ 170 mM trans-trans muconic acid, 170 mM DIC and 170 mM Oxyma in 10 ml DMF solvent were combined and kept on shaking for 15 min. The suspension in tube ‘A’ was added drop wise into tube ‘B’ while shaking for 2 hours. After this the reaction mixture was washed three times with DMF and dried in vacuum oven for 14 hours at 50 °C [25]. In tube ‘A’ 10 mg linker attached MNPs were re-dispersed in 8 ml water by sonication. In tube ‘B’ 85 mM EDC-HCl and 85 mM NHS were dissolved in 2 ml water. Solutions of tube ‘B’ were mixed into tube ‘A’ and vortexed for 1 hour. Thereafter, the reaction mixture was washed 3 times with water, followed by magnetic decantation. This was followed by addition of 10 ml 0.5% cellulase solution in PBS and kept on a shaker at 1000 rpm at room temperature for overnight. The cellulase conjugated IOMNPs were washed three times with water and re-suspended in PBS buffer. This mixture was directly used for the hydrolysis activity of cellulase.
2.5. Preparation of silica nanoparticles (SNPs)
Silica nanoparticles (SNPs) were synthesized using Stöber method [26]. Briefly, SNPs were prepared by hydrolysis and condensation of TEOS in absolute ethanol and water as solvent using ammonium hydroxide (NH4OH) as catalyst. 40 ml ethanol was added into 2.5 ml of deionized water and reaction was heated up to 55 °C. Subsequently, 1.8ml NH4OH was added followed by continuous stirring, 1.55 ml TEOS and 4 ml ethanol were added with continuous stirring for 5 hrs. SNPs were centrifuged at 7000 rpm for 10 min and washed four times with ethanol, vacuum dried and stored in vacuum desiccators for further use.
2.6. Preparation of silica coated iron oxide magnetic nanoparticles (S-IOMNPs)
Silica coating was performed on prepared IOMNPs by addition of anhydrous ethanol and deionised water. 50 mg of MNPs were taken initially followed by 80 mL of anhydrous ethanol and 20 mL of deionised water. This mixture was sonicated for approximately 30 min. The pH of the resulting mixture was maintained around 9.0 throughout by addition of ammonium hydroxide (NH4OH). 200 μL of TEOS was added and reaction was continuously stirred for 4 hr in an environment of nitrogen (N2) gas. The prepared S-IOMNPs were washed three times with deionised water and ethanol. S-IOMNPs were dried and kept in vacuum desiccators for further use [27].
2.7. Preparation of chitosan coated iron oxide magnetic nanoparticles (C-IOMNPs)
0.25 g of IOMNPs was dispersed in CTAB (0.5% in deionized water-solution A). Then, 100 ml chitosan solution (0.02%) in acetic acid solution was added drop-wise into solution A. The mixture was continuously stirred at 1000 rpm for 1 h at room temperature. C-IOMNPs were washed three times with ethanol and deionized water. Finally, the obtained C-IOMNPs were dried overnight into vacuum oven at 60 °C and stored in vacuum desiccators for further use. APTES functionalization [5, 24], cross linking of APTES functionalized nanoparticles (SNPs, S-MNPs and C-MNPs) with glutaraldehyde and cellulase immobilization was done by the similar procedure that was used for IOMNPs [5, 24].
2.8. Immobilized-enzyme loading efficiency measurement by bicinchoninic acid (BCA) assay
We used the BCA assay to determine the loading efficiency of cellulase on different nanoparticles. The BCA assay works on the principle of reduction of Cu2+ ions from copper sulphate (CuSO4) to Cu+ by peptide bonds present in the cellulase enzyme [28]. The total amount of Cu2+ ions reduced is proportional to the amount of enzyme thereby allowing determination of the loading of enzyme on nanoparticles. A purple colored product formed with absorbance at 562 nm is indicative of the chelated Cu+ ions. Here, 200 μL of different nanobiocatalysts (10 mg/ml) were incubated with 200 μL BCA reagent in 96 well-plates at 37 °C for 30 min and then absorption was taken at 562 nm. Considering bare enzyme loading efficiency at 100 %, different nanobiocatalysts loading efficiency were measured.
2.9. Monitoring catalytic activity of cellulase nano-biocatalysts by dinitrosalicylic acid (DNS) assay
The catalytic activity of nanobiocatalysts was determined by DNS assay using the established approach [29] Briefly, the reducing activity of glucose produced by action of cellulase on substrate cellulose was measured by use of DNS reagent. The absorbance at 540 nm was correlated with catalytic activity of the prepared constructs.1 g of cellulose powder was taken in 4 mL of pH 5.5 citrate buffer, 10 mM. Two mL of enzyme (10 mg/mL) or nanobiocatalyst suspension were added to this followed by incubation at 50 °C for 1 h. 3 mL of dinitrosalicylic acid (DNS) reagent was added after the incubation and the samples were heated at 100 °C for 5 min. Subsequently, the samples were allowed to cool and were centrifuged at 5000 rpm at room temperature for 5 min. The UV–visible absorbance of the supernatant was measured at 540 nm. The absorbance at 540 nm was correlated with catalytic activity.
2.10. Monitoring catalytic activity of cellulase nano-biocatalysts by HPLC
100 gm of onion skins was mechanically ground in an electric grinder, until a highly viscous paste was formed. This paste was sonicated for 1 hour at 50 °C in a 250 ml conical flask. 100 ml of anhydrous methanol was added to the suspension and kept in a shaker incubator at 50 °C. The flask was sonicated for 1 hour and kept on shaker for 2 hours. The mixture was centrifuged at 7000 rpm at 4 °C for 10 minutes. The suspension was filtered using 0.45 micron PVDF syringe filter. This was stored in dark bottles at -20 °C. For the hydrolysis of 1 mL of onion extract or 50 mg cellulose (in 1ml citrate buffer, pH 5.5, 10 mM), 1ml of enzyme or nanobiocatalyst (10 mg/ml) was used. The mixture was kept on a thermomixer for different time of periods (1–8 hours) at 50 °C and 500 rpm. The nanobiocatalyst was removed by using a bar magnet and tested for reusability by application on the next batch of cellulose/onion extract. The supernatants were analysed by HPLC followed by the reported method, In short; isocratic mobile phase, Methanol (50): water (50): Formic acid (0.1) was chosen and at the flow rate of 0.5 mL/min. RP-C18 column was used, the injection volume for HPLC analysis was 20 μL and the column was maintained at room temperature. Chromatograms were recorded at 350 nm for each analysis. Above mentioned hydrolysis and HPLC procedure was also used for the reusability and kinetic studies.
2.11. Instruments for characterization
SEM characterizations were performed on a field emission electron scanning microscope (JEOL JSM-7600f, USA). Attachment of linkers and enzyme on nanoparticles was investigated by FT-IR Spectrometer, (PerkinElmer, Spectrum Two, UATRA two, USA). Spectra were recorded in the range of 490–4000 cm−1. UV-visible spectra were recorded using Spectrophotometer (JASCO V-750, Japan). Hydrolysis analysis was done under HPLC (SHIMADZU RID-20A, Japan). Crystallinity studies were performed using XRD (BRUKER D8 Discover, USA). Hydrodynamic sizes and zeta potential of nanoconstructs were measured by DLS (MALVERN NANO ZS, Zetasizer Nano Series, UK).
3. Results and discussion
3.1. Synthesis and characterization of different cellulase-immobilized nanoparticles
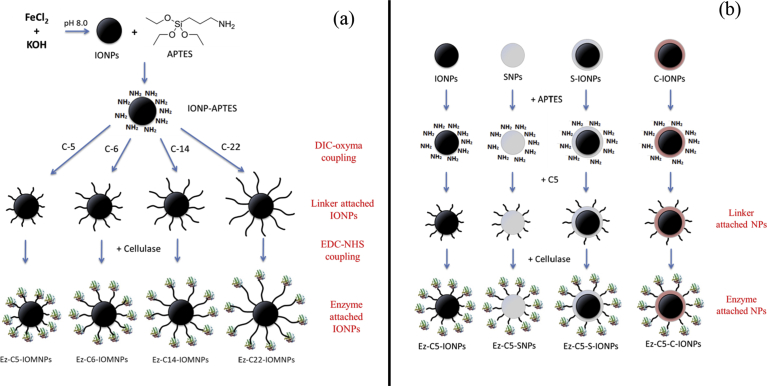
Iron oxide, silica, silica-coated iron oxide and chitosan-coated iron oxide nanoparticles were prepared as described in the Experimental and represented in Fig. 1. The zeta potentials of bare and coated nanoparticles were determined at pH 7 and were as follows: bare iron oxide (-12.6 mV), silica (-39.35 mV), silica-coated iron oxide (-37.5 mV) and chitosan-coated iron oxide (+24.2 mV).
Fig. 1.
Schematic representation for synthesis of cellulase-immobilized nanoparticles of varying (a) linkers, and (b) identities.
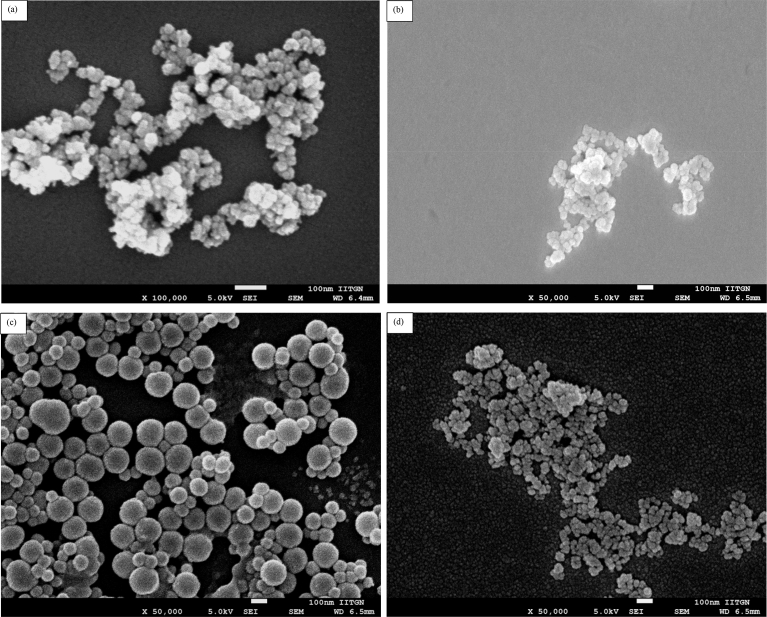
These measurements are compatible with previous literature reports. [30, 31] Size of the nanoparticles was characterized by scanning electron microscopy (SEM) and dynamic light scattering (DLS). The diameter of the IOMNPs, SNPs, S-IOMNPs and C-IOMNPs was found from SEM (Fig. 2) to be in the range of 45–60 nm, 35–55 nm, 80–150 nm and 30–50 nm, respectively.
Fig. 2.
SEM image of bare (a) IOMNPs, (b) SNPs), (c) S-IOMNPs) and (d) C-IOMNPs. Scale bar is 100 nm in all the images.
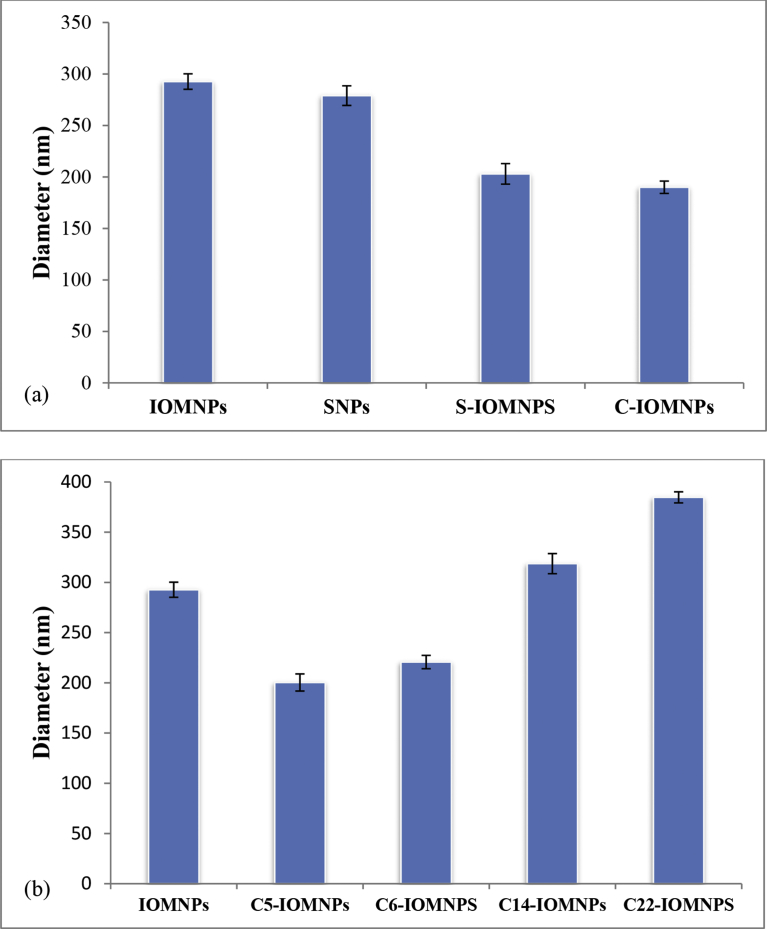
Their hydrodynamic sizes were found from DLS to be, 292 ± 7 nm, 279.5 ± 17, 203 ± 10 and 190 ± 6, respectively (Fig. 3a). Coating of silica on IOMNPs increases the particle size [32] while the coating of chitosan decreases the same owing to the affinity of chitosan for negative surface charge of IOMNPs. This observation is consistent with previous reports regarding the dispersion behavior of biomolecule-conjugated nanoparticles [5, 33]. Magnetic dipole-dipole interactions and van der Waal's forces generated from residual magnetic moments have been suggested as leading to agglomeration of the particles. Stabilization of the magnetic nanoparticles has been sought through electrostatic repulsion that can be achieved by coating the particles with an electrical double layer. Chitosan is one among several non-magnetic substances that has been used to stabilize the magnetic nanoparticles. Chemical modification with APTES and glutaraldehyde (C5 linker) reduced the particle sizes [34]. The average hydrodynamic diameter for C5-IOMNPs is 239 ± 17 nm. Increase in the length of linkers leads to a progressive increase in the size of IOMNPs. The size of C5-IOMNPs, C6-IOMNPs, C14-IOMNPs and C22-IOMNPs are 200 ± 8 nm, 220 ± 6 nm, 318 ± 10 nm and 384 ± 5 nm, respectively as determined by DLS (Fig. 3b).
Fig. 3.
DLS bar graphs of, (a) different nanoparticles, and (b) of IOMNPs with different linkers.
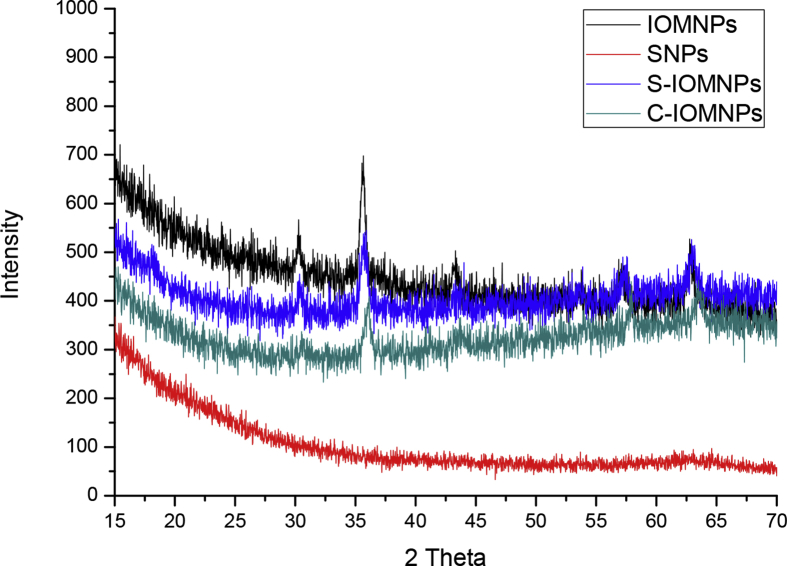
Crystalline structures of IOMNPs, SNPs and S-IOMNPs and C-IOMNPS were determined by XRD patterns. The XRD profile of IOMNPs displayed diffraction peaks at 30.28, 35.657, 43.39, 53.7, 57.31 and 62.91 (2θ) (Fig. 4). These were characteristic of the typical cubic Fe3O4 (JCPDS, # 65-3107) and was confirmed by ICDD database showing 100% purity. The peaks were assigned to the (220), (311), (400), (422), (511) and (440) planes. The characteristic peaks at 2θ, 21.228 and 33.158 were absent in the XRD profile suggesting the absence of other oxides namely goethite (FeOOH) and hematite (Fe2O3) further highlighting the purity of IOMNPs [32]. No distinct change in the crystal structure was observed after the coating of silica on IOMNPs (S-IOMNPs) and for chitosan coated IOMNPs. However, the intensity of diffraction peaks were lower possibly due to surface modification by amorphous material [35, 36]. The XRD pattern of SNPs corresponds to characteristic amorphous nature of silica.
Fig. 4.
XRD pattern of IOMNPs (black), SNPs) (red), S-IOMNPs (blue) and C-IOMNPs (green).
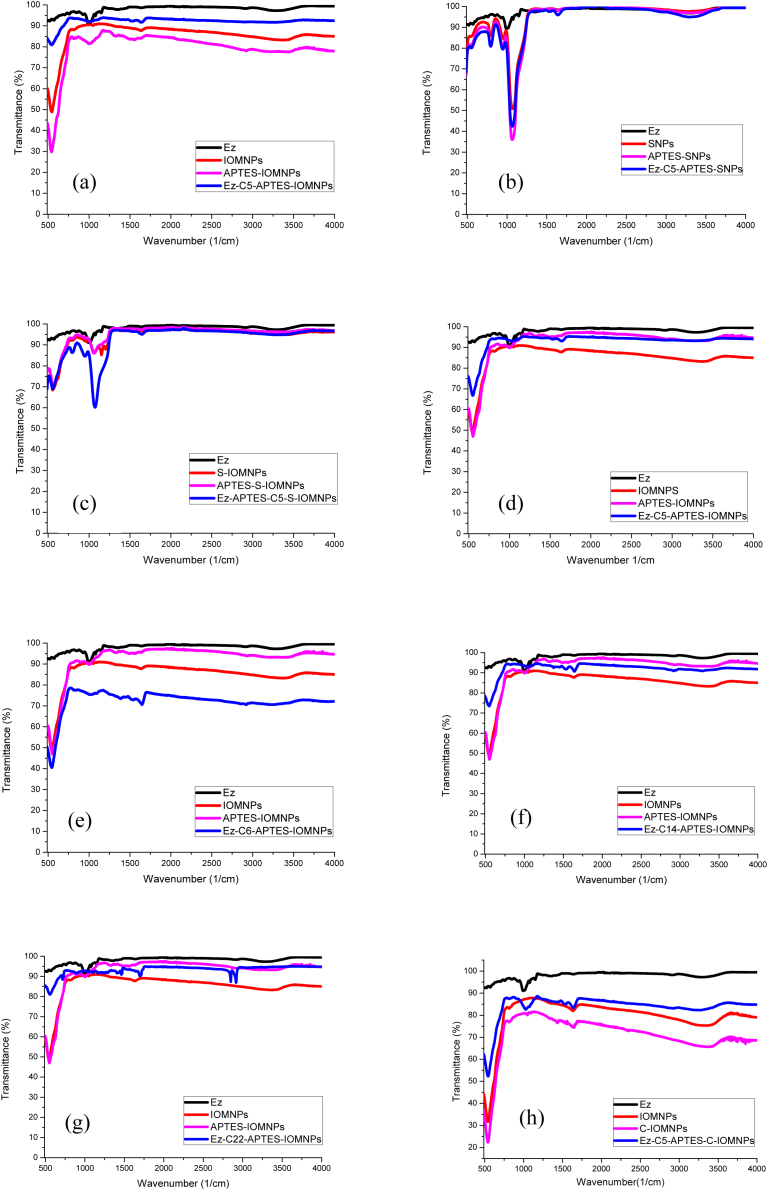
The attachment of linkers and immobilization of cellulase enzyme to the different nanoparticles such as IOMNPs, SNPs, S-MNPs and C-IOMNPs was investigated by FT-IR. The broad adsorption peaks at around 580 cm−1 are characteristic peak of Fe–O bond (Fig. 5) [36]. The presence of amine groups on different nanoparticles was evident by the presence of two broad peaks at 3435 and 1600 cm−1, which are assigned to the N–H stretching vibration and N–H bending respectively [37]. The covalent attachment of aminopropylsilane groups is mainly due to their self-polycondensation leading to a highly cross-linked polysiloxane film entrapping each magnetic nanoparticle. Thus, Fe–O–Si bonds are masked in the FT-IR spectrum by overlapping Fe–O vibrations of magnetite at 580 cm−1 [38].
Fig. 5.
FT-IR Spectra of enzyme immobilized nanoparticles with the linker C5 (a) IOMNPs, (b) SNPs,(c) S-IOMNPs and enzyme immobilized IOMNPs with the linker (d) C5, (e) C6, (f) C14 and (g) C22; (h) comparative FT-IR spectra of IOMNPs, C-IOMNPs and C5-APTES-C-IOMNPs.
On the other hand, formation of silane layer on the surface of nanoparticles was confirmed by peaks at 1049 and 1018 cm−1 which correspond to the Si–OH and Si–O–Si groups, respectively. The peaks observed at 2918 and 2846 cm−1 are assigned to the alkyl chain (–CH2) stretching vibrations due to the symmetric and the asymmetric CH2 stretching modes,respectively [39]. For SNPs, the peak at 3423 cm−1 is assigned to surface hydroxyl groups (stretching mode) while the band at 1635 cm−1 corresponds to O–H vibration of absorbed water [40]. Peaks at 795 cm−1 and 454 cm−1 are assigned to the stretching vibrations of the mesoporous framework (Si–O–Si) and the peak at 960 cm−1 is attributed to Si–OH bond stretching [41].
The characteristic bands of chitosan on IOMNPs are at 3420 cm−1 (O–H stretching and N–H stretching vibrations), 1645 cm−1 (amide), and 1076 cm−1 (C–O–C stretching vibration) (Fig. 5). These indicated successful generation of Fe3O4–chitosan particles [33]. Peaks at 1635 and 1554 cm−1 correspond to the C O stretching of amide groups and N–H bending of amide groups, respectively, that confirm the covalent attachment of enzyme on nanoparticles [35].
3.2. Immobilized-enzyme loading efficiency
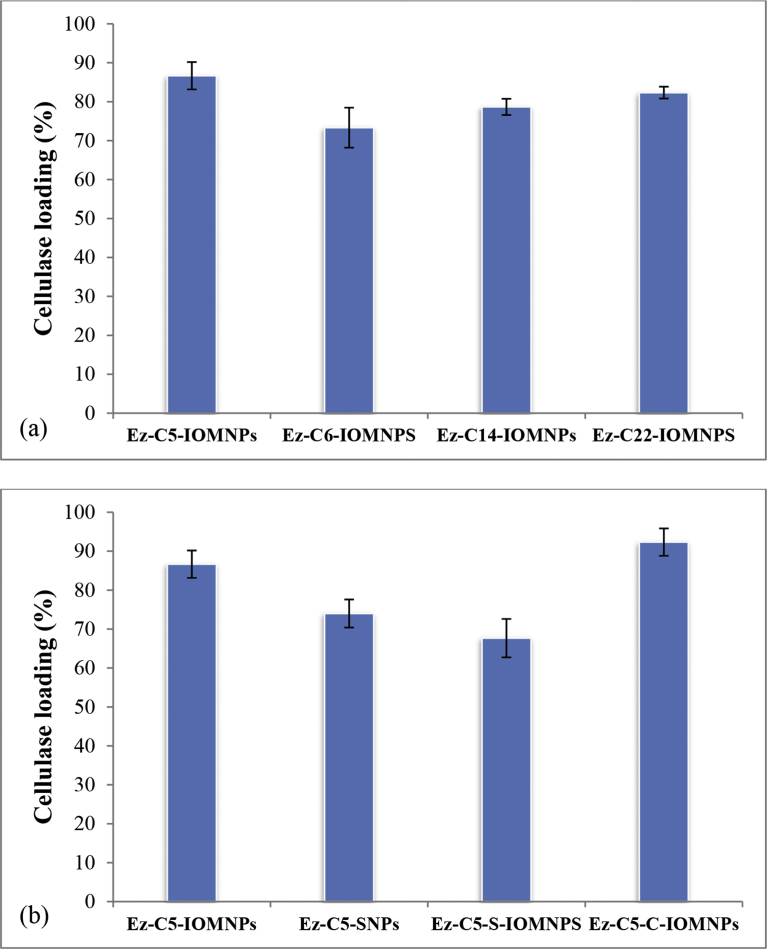
The loading of cellulase on C5-IOMNPs, C6-IOMNPs, C14-IOMNPs and C22-IOMNPs were determined by BCA assay to be 86%, 73%, 79% and 82%, respectively (Fig. 6a). This pattern of enzyme loading is indicative of a specific linker chain length as being more conducive towards packing efficiency on the nanoparticle surface. The interplay of surface charge and linker length in achieving optimal surface coverage has been reported previously [42]. Interestingly, linker lengths were not found to have a significant effect on loading efficiency of small organic molecules on magnetic iron oxide nanoparticles [43]. The larger moiety namely cellulase conjugated in the present work may be better at amplifying differences in the linker length-associated surface coverage on nanoparticles. Activity of xylanase immobilized on magnetite nanoparticles have been previously reported to vary significantly with linker length [44]. Chitosan coated magnetic nanoparticles (C5-C-IOMNPs) exhibit the highest loading efficiency (92%) while C5-SNPs and C5-S-IOMNPs display enzyme loading of 74% and 67%, respectively (Fig. 6b). The improvement in loading efficiency from SNPs to chitosan coated magnetic nanoparticles can be attributed to the favourable surface charge in the latter that enables better packing of the alkyl linkers [45].
Fig. 6.
Loading efficiency of (a) nanoconjugates with different chemical linkers and, (b) nanoparticles with different compositions.
3.3. Catalytic activity and kinetics of cellulase nano-biocatalysts
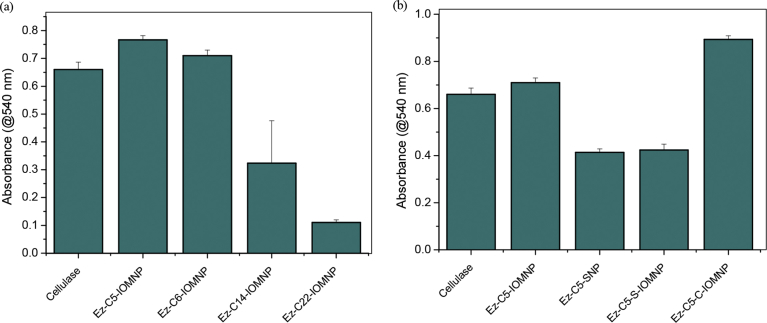
The catalytic activity of nanobiocatalysts was determined by DNS assay. A comparison of catalytic activity of cellulase immobilized on IOMNPs of different linker lengths is shown in Fig. 7a.
Fig. 7.
Catalytic activity of nanobiocatalysts via DNS assay on constructs with (a) different linkers on IOMNPs, and (b) different compositions of nanoparticles.
While C5-IOMNPs display modest improvement in immobilized cellulase activity when compared to the bare enzyme, progressively longer linker lengths are found to result in decreasing enzyme activity. Considering the similar loading efficiencies of cellulase on each of these constructs (Fig. 6), the lower enzyme activity for constructs bearing longer linker lengths is attributable to blockage of enzyme active site by the linker alkyl groups. Heavily loaded enzymes on nanoparticles have been suggested as counterproductive to enzyme activity due to steric blockage of active sites [4]. In this regard, C22-IOMNPs offer significantly greater hydrophobic surface for adhering to immobilized enzymes as compared to the shorter linkers. This possibly manifests in poorer access to the enzyme active site thereby lowering its activity. A comparison of immobilized cellulase activity for different nanomaterials albeit with the same linker length indicates chitosan coated IOMNPs as being most active (Fig. 7b). As shown in Fig. 7b, the variation in activity of cellulase mirrors and in fact amplifies the corresponding loading efficiency of each nanoconstruct. Enzyme kinetics of free cellulase and C5-IOMNPs were evaluated and the kinetic parameters are listed in Table 1. Interestingly, both the Ez-C5-IOMNP and Ez-C5-C-IOMNP exhibit somewhat inferior KM as compared to free cellulase. This may be attributed to hindered access to enzyme active sites. Nevertheless, the enhancement in Vmax sufficiently offsets increased KM and a 30% higher Vmax/KM results for the chitosan-coated nanoconstructs. Similar enhancement in catalytic efficiency of immobilized enzymes albeit with increase in KM has been reported before [46, 47]. Cellulase immobilized on chitosan-coated IOMNPs display superior catalytic efficiency based on the greater Vmax of the nanoconstructs. While magnetic chitosan nanoparticles have been explored previously for cellulase immobilization [48], their relative performance vis-à-vis other nanoconstructs have not been scrutinized. These results clearly demonstrate the importance of linker lengths in conjunction with nanomaterial identity in determining immobilized cellulase activity. We next applied these nanoconstructs on onion skins and monitored the product profile.
Table 1.
Kinetic parameters of free cellulase and nanoconstructs.
| Kinetic Parameter | Free Cellulase | Cellulase@IOMNP | Cellulase@C-IOMNP |
|---|---|---|---|
| KM (mM) | 10.52 | 11.23 | 11.09 |
| Vmax (mM/min) | 19.88 | 25.31 | 26.95 |
| Vmax/KM (min−1) | 1.88 | 2.25 | 2.43 |
3.4. Application of immobilized cellulase on onion skins
We have previously used cellulase-immobilized nanoparticle constructs for the efficient extraction of carotenoidic pigments from peel of oranges [5]. The application of cellulase immobilized nanoparticles on onion skins was studied by HPLC. Overnight digestion of onion skins by nanoconstructs was followed by HPLC analysis of the resulting supernatant.
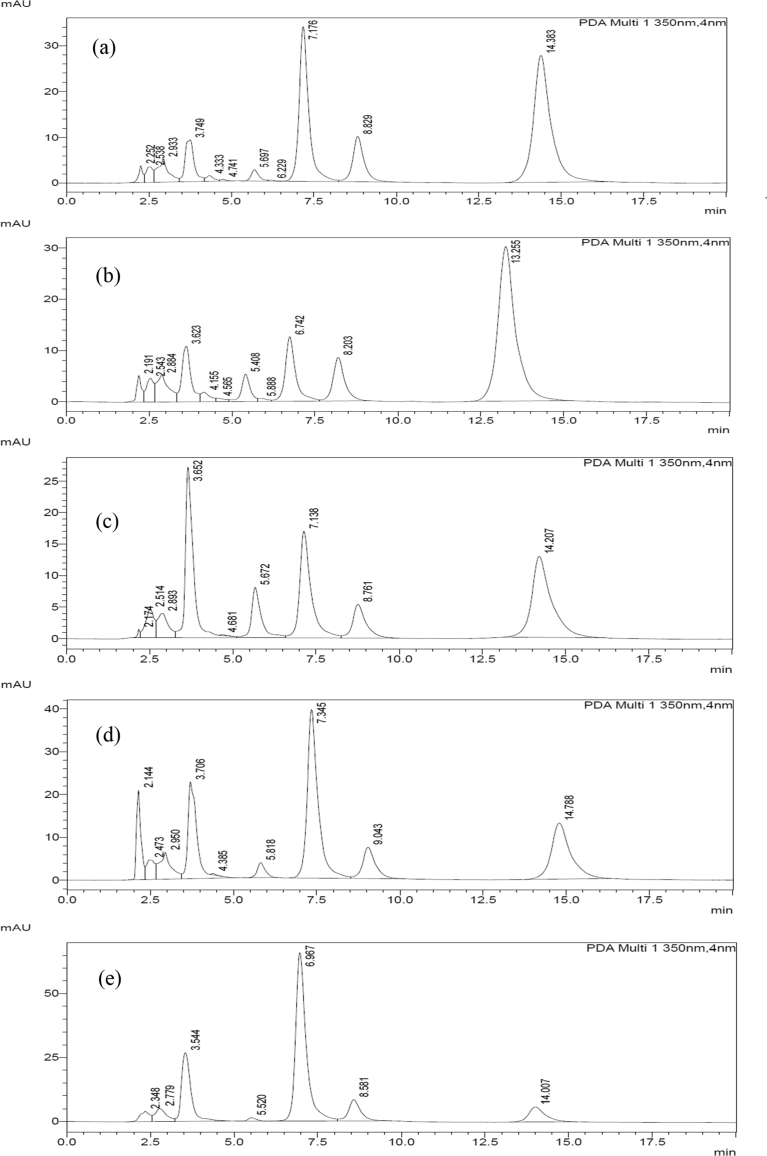
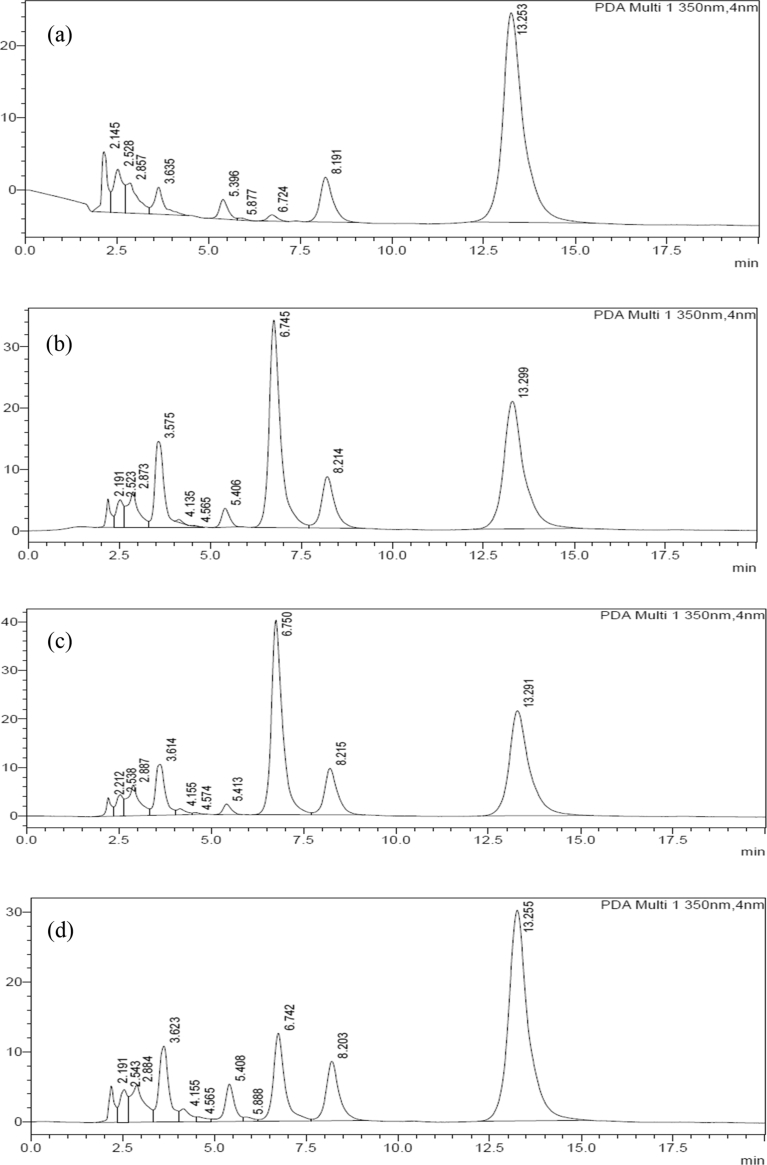
Chromatographic comparison with standards (Fig. 8 and Fig. 9) and mass spectrometric characterization of the extracted compounds led to their identification as quercetin and quercetin di- and monoglucosides (see Table 2). While the extraction of quercetin from onion skins has been explored using a variety of techniques [49] including enzyme-assisted methods [50], the fate of quercetin-glucosides has received less attention. The application of various cellulase immobilized MNPs on onion skins results in distinctive distribution of quercetin in free and glycosidic forms as compared to the use of bare enzyme. A similar inference has been made in a recent report on use of cellulase and pectinase for extraction of quercetin from onion skins [50].
Fig. 8.
HPLC Chromatograms of onion extract upon hydrolysis by (a) bare cellulase (0.5%), (b) Ez-C5-IOMNPs, (c) Ez-C6-IOMNPs (d) Ez-C14-IOMNPs (e) Ez-C22-IOMNPs.
Fig. 9.
HPLC Chromatograms of onion extract upon hydrolysis by (a) Ez-C5-IOMNPs, (b) Ez-C5-SNPs (c) Ez-C5-S-IOMNPs (d) Ez-C5-C-IOMNPs.
Table 2.
Calculated and observed mass of polyphenols extracted from onion skins.
| Compound (Polyphenols) | Retention Time of Standards (min) | Calculated Mass | Observed Mass |
|---|---|---|---|
| Quercetin | 13.8 | 302.238 | 303.0779 |
| Quercetin -3 -Glucoside | 5.4; 6.9; 8.5 | 464.38 | 465.1506 |
| Quercetin-3,4′-Glucoside | 3.5 | 626.52 | 627.1548 |
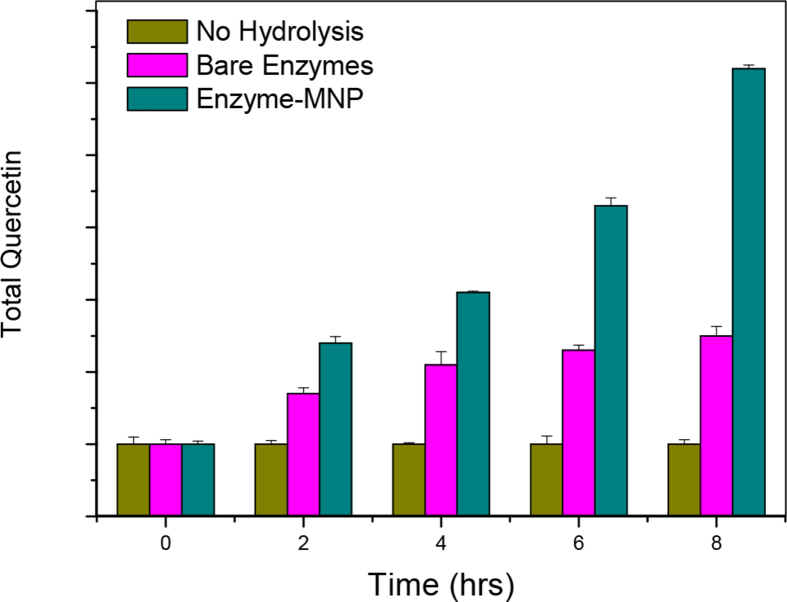
Further, the release of quercetin from onion skins was monitored over time after treatment with cellulase-MNPs. In samples that were treated with free cellulase the release of quercetin was attenuated between 4-6 hrs (see Fig. 10). In contrast, use of cellulase-MNP conjugates resulted in substantial release of quercetin beyond 8 hrs of treatment of the onion skins.
Fig. 10.
Release of Quercetin from onion skins over time in samples treated with no enzymes, bare enzymes and MNP-enzymes.
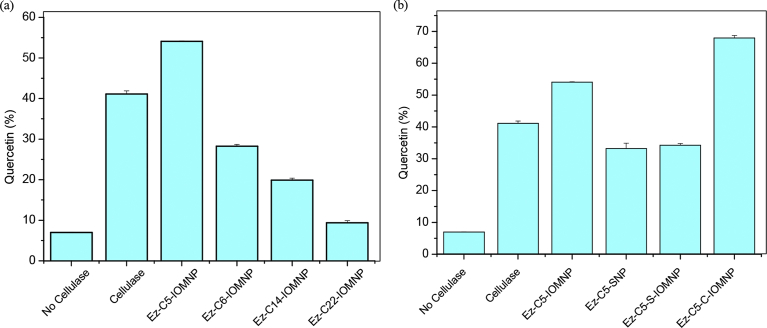
A comparison of IOMNP-immobilized cellulase bearing different linker lengths with respect to the hydrolysis of quercetin glycosides clearly reveals the superiority of the APTES- conjugated nanobiocatalysts (see Fig. 11a). In fact, the hydrolysis of mono- and diglucosides of quercetin to their aglycosidic form can be easily correlated to catalytic activity of the nanoconstructs as determined by the DNS assay (see Figs. 11b and 7b). Similarly, when comparing different nanoparticles bearing the same linker length, chitosan-coated IOMNPs is most effective in the conversion of quercetin glycosides to free quercetin (see Fig. 11b). The efficiency of cellulase-nanoconstructs towards hydrolysis of quercetin glycosides mirrors the catalytic efficiency as measured by the DNS assay. The results obtained on onion skins are consistent with behaviour of cellulase-nanoconstructs on standard quercetin diglucoside and monoglucoside. Our results indicate that APTES-glutaraldehyde is the optimum chemical linker and chitosan-coated iron oxide the best nanomaterial for cellulase immobilization.
Fig. 11.
Comparison of the percentage Quercetin present in onion extract after (a) hydrolysis by nanobiocatalysts with different linkers and (b) hydrolysis by nanobiocatalysts with different compositions.
3.5. Reusability of nano-biocatalysts
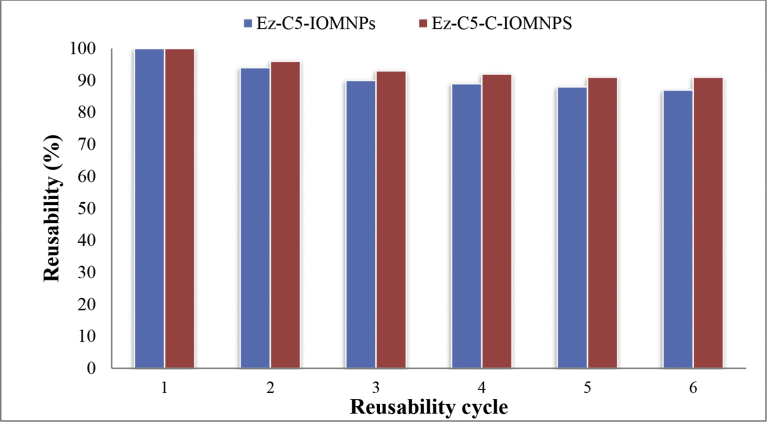
Recyclability of Ez-C5-IOMNPs and Ez-C5-C-IOMNPs nanobiocatalysts was assessed on onion extract. Once, the hydrolysis of onion skins had been performed, the nanobiocatalysts were magnetically decanted from reaction mixture and used for next cycle of hydrolysis. Catalytic activity of reusable magnetic nanobiocatalysts for the first cycle was considered 100%. Nanobiocatalysts were reused for five cycles with 87% and 91% catalytic activity being recovered in fifth cycle, respectively, for Ez-C5-IOMNPs and Ez-C5-C-IOMNPs (Fig. 12).
Fig. 12.
Reusability of Ez-C5-IOMNPs and Ez-C5-C-IOMNPs towards onion skin hydrolysis.
4. Conclusions
We have investigated the nanoparticle and chemical linkage in different cellulase nanobiocatalysts in an attempt to optimize these components for effective hydrolytic activity of the enzyme. Chitosan-coated iron oxide nanoparticles with APTES-conjugated cellulase are found to possess the best enzyme loading and catalytic activity. The optimized nanoparticle identity and linker length are highlighted through application on onion skins. The distribution of aglycosidic and glycosidic forms of trapped polyphenol quercetin bear a distinctive correlation with the activity of nanobiocatalysts. These results are important for the practical application of immobilized cellulase and suggest an alternative strategy for measurement of cellulase activity.
Declarations
Author contribution statement
Sanjay Kumar: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Vinod Morya: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Joshna Gadhavi, Anjani Vishnoi, Jaskaran Singh: Analyzed and interpreted the data.
Bhaskar Datta: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
The authors acknowledge Ministry of Human Resources and Development (MHRD) and Department of Science and Technology (DST) of Govt. of India for financial support through IMPRINT grant no. 6349.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Galante Y.M., De Conti A., Montverdi R. In: Trichoderma & Gliocladium - Enzymes, Biological Control and Commercial Applications. Harman G.F., Kubicek C.P., editors. Taylor & Francis; London: 1998. pp. 327–342. [Google Scholar]
- 2.Grassin C., Coutel Y. In: Enzymes in Food Technology. Whitehurst R.J., van Oort M., editors. Wiley-Blackwell; Oxford, UK: 2009. [Google Scholar]
- 3.Khoshnevisan K., Bordbar A.-K., Zare D., Davoodi D., Noruzi M., Barkhi M., Tabatabaei M. Immobilization of cellulase enzyme on superparamagnetic nanoparticles anddetermination of its activity and stability. Chem. Eng. J. 2011;171:669–673. [Google Scholar]
- 4.Jordan J., Challa S.S.R.K., Theegala C. Preparation and characterization of cellulase-bound magnetite nanoparticles. J. Mol. Catal. B Enzym. 2011;68:139–146. [Google Scholar]
- 5.Kumar S., Sharma P., Ratrey P., Datta B. Reusable nanobiocatalysts for the efficient extraction of pigments from orange peel. J. Food Sci. Technol. 2016;53:3013–3019. doi: 10.1007/s13197-016-2272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao X., Guo G., Huang J., Du Z., Huang Z., Ma L., Li P., Gu L. J. Chem. Technol. Biotechnol. 2006;81:189–195. [Google Scholar]
- 7.Wu L., Yuan X., Sheng J. J. Membr. Sci. 2005;250:167–173. [Google Scholar]
- 8.Tebeka I.R.M., Silva A.G.L., Petri D.F.S. Langmuir. 2009;25:1582–1587. doi: 10.1021/la802882s. [DOI] [PubMed] [Google Scholar]
- 9.Saville B.A., Khavkine M., Seetharam G., Marandi B., Zuo Y.L. Appl. Biochem. Biotechnol. 2004;113 – 116:251–259. doi: 10.1385/abab:113:1-3:251. [DOI] [PubMed] [Google Scholar]
- 10.Li C.Z., Yoshimoto M., Fukunaga K., Nakao K. Bioresour. Technol. 2007;98:1366–1372. doi: 10.1016/j.biortech.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Hirsh S.L., Bilek M.M.M., Nosworthy N.J., Kondyurin A., dos Remedios C.G., McKenzie D.R. Langmuir. 2010;26:14380–14388. doi: 10.1021/la1019845. [DOI] [PubMed] [Google Scholar]
- 12.Bilek M.M.M., McKenzie D.R. Biophys. Rev. 2010;2:55–65. doi: 10.1007/s12551-010-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kammerer D.R., Claus A., Schieber A., Carle R. J. Food Sci. 2005;70:C157–C163. [Google Scholar]
- 14.Maier T., Goeppert A., Kammerer D.R., Schieber A., Carle R. Eur. Food Res. Technol. 2008;227:267–275. [Google Scholar]
- 15.Wilkins M.R., Widmer W.W., Grohmann K., Cameron R.G. Bioresour. Technol. 2007;98:1596–1601. doi: 10.1016/j.biortech.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Fabre G., Bayach I., Berka K., Paloncyova M., Starok M., Rossi C., Duroux J.-L., Otyepka M., Trouillas P. Chem. Commun. 2015;51:7713–7716. doi: 10.1039/c5cc00636h. [DOI] [PubMed] [Google Scholar]
- 17.Bjorklund G., Chirumbolo S. Nutrition. 2017;33:311–321. doi: 10.1016/j.nut.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 18.del Rio D., Rodriguez-Mateos A., Spencer J.P.E., Tognolini M., Borges G., Crozier A. Antioxidants Redox Signal. 2013;18:1818–1892. doi: 10.1089/ars.2012.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katarzyna G., Duthie G.G., Stewart D., Leslie S.J., Megson I.L. Br. J. Pharmacol. 2017;174:1209–1225. doi: 10.1111/bph.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saura-Calixto F., Serrano J., Goni I. J. Agric. Food Chem. 2007;55:9443–9449. doi: 10.1021/jf072146o. [DOI] [PubMed] [Google Scholar]
- 21.Manach C., Scalbert A., Morand C., Remsy C., Jimenez L. Am. J. Clin. Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 22.Bouayed J., Deuser H., Hoffman L., Bohn T. Food Chem. 2012;131:1466–1472. [Google Scholar]
- 23.Perez-Gregorio R.M., Garcia-Falcon M.S., Simal-Gandara J., Rodrigues A.S., Almeida D.P.F. J. Food Compos. Anal. 2010;23:592–598. [Google Scholar]
- 24.Park H.J., McConnell J.T., Boddohi S., Kipper M.J., Johnson P.A. Colloids Surfaces B Biointerfaces. 2011;83:198–203. doi: 10.1016/j.colsurfb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Goonasekera C.S., Jack K.S., Cooper-White J.J., Grondahl L. J. Mater. Chem. B. 2013;1:5842–5852. doi: 10.1039/c3tb21110j. [DOI] [PubMed] [Google Scholar]
- 26.Wang X.-D., Shen Z.-X., Sang T., Cheng X.-B., Li M.-F., Chen L.-Y., Wang Z.-S. J. Colloid Interface Sci. 2010;341:23–29. doi: 10.1016/j.jcis.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Seenuvasan M., Malar C.G., Preethi S., Balaji N., Iyyapan J., Kumar M.A., Kumar K.S. Mater. Sci. Eng. C Mater. Biol. Appl. 2013;33:2273–2279. doi: 10.1016/j.msec.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 28.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Miller G.L. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 30.Zhua A., Yuan L., Liao T. Int. J. Pharm. 2008;350:361–368. doi: 10.1016/j.ijpharm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y.-K., Lui C.N.P., Lin T.-H., Chang C., Chou P.-T., Yung K.K.L., Tsang S.C.E. Faraday Discuss. 2014;175:13–26. doi: 10.1039/c4fd00132j. [DOI] [PubMed] [Google Scholar]
- 32.Song J., Su P., Yang Y., Wang T., Yang Y. J. Mater. Chem. B. 2016;2016:5873–5882. doi: 10.1039/c6tb01857b. [DOI] [PubMed] [Google Scholar]
- 33.Kuo C.H., Liu Y.C., Chang C.M., Chen J.H., Chang C., Shieh C.J. Carbohydr. Polym. 2011;87:2538–2545. [Google Scholar]
- 34.Olariu C.I., Yiu H.H.P., Bouffier L., Nedjadi T., Costello E., Williams S.R., Halloran C.M., Rosseinsky M.J. J. Mater. Chem. 2011;21:12650–12659. [Google Scholar]
- 35.Bordbar A.-K., Rastegari A.A., Amiri R., Ranjbaksh E., Abbasi M., Khosropour A.R. Biotechnol. Res. Int. 2014;7 doi: 10.1155/2014/705068. 705068/705061-705066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng B., Hong R.Y., Wang L.S., Guo L., Li H.-Z., Ding J., Zheng Y., Wei D.G. Colloid. Surf. Physicochem. Eng. Asp. 2008;328:52–59. [Google Scholar]
- 37.Zhang Y., Kohler N., Zhang M. Biomaterials. 2002;23:1553–1561. doi: 10.1016/s0142-9612(01)00267-8. [DOI] [PubMed] [Google Scholar]
- 38.Bruni S., Cariati F., Casu M., Lai A., Musinu A., Piccaluga G., Solinas S. Nanostruct. Mater. 1999;11:573–586. [Google Scholar]
- 39.Cheng K., Peng S., Xu C., Sun S. J. Am. Chem. Soc. 2009;131:10637–10644. doi: 10.1021/ja903300f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen X., Ye L. Macromolecules. 2011;44:5631–5637. doi: 10.1021/ma200837n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antsiferova Y., Sotnikova N., Parfenyuk E. BioMed. Res. Int. 2013;11 doi: 10.1155/2013/924362. 924362/924361 - 924311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rammohan N., Holbrook R.J., Rotz M.W., MacRenaris K.W., Preslar A.T., Carney C.E., Reichova V., Meade T.J. Bioconjug. Chem. 2017;28:153–160. doi: 10.1021/acs.bioconjchem.6b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan B., Huang X. Chirality. 2008;20:265–277. doi: 10.1002/chir.20418. [DOI] [PubMed] [Google Scholar]
- 44.Singh V., Kaul S., Singla P., Kumar V., Sandhir R., Chung J.H., Garg P., Singhal N.K. Int. J. Biol. Macromol. 2018;115:590–599. doi: 10.1016/j.ijbiomac.2018.04.097. [DOI] [PubMed] [Google Scholar]
- 45.Dyawanapelly S., Jagtap D.D., Dandekar P., Ghosh G., Jain R. Colloids Surfaces B Biointerfaces. 2017;154:408–420. doi: 10.1016/j.colsurfb.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 46.Chen T., Yang W., Guo Y., Yuan R., Xu L., Yan Y. Enzym. Microb. Technol. 2014;63:50–57. doi: 10.1016/j.enzmictec.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Ashtari K., Khajeh K., Fasihi J., Ashtari P., Ramazani A., Vali H. Int. J. Biol. Macromol. 2012;50:1063–1069. doi: 10.1016/j.ijbiomac.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 48.Zang L., Qiu J., Wu X., Zhang W., Sakai E., Wei Y. Ind. Eng. Chem. Res. 2014;53:3448–3454. [Google Scholar]
- 49.Wach A., Pyrzynska K., Biesaga M. Food Chem. 2007;100:699–704. [Google Scholar]
- 50.Choi I.S., Cho E.J., Moon J.-H., Bae H.-J. Food Chem. 2015;188:537–542. doi: 10.1016/j.foodchem.2015.05.028. [DOI] [PubMed] [Google Scholar]