Abstract
Ebola virus is the causative agent of Ebola virus disease, a severe, often fatal illness in humans. So far, there are no US Food and Drug Administration (FDA)-approved therapeutics directed against Ebola virus. Here, we selected the host factor Niemann-Pick C1 (NPC1), which has been shown to be essential for Ebola virus entry into host cytoplasm, as a therapeutic target for suppression by locked nucleic acid-modified antisense oligonucleotides. Screening of antisense oligonucleotides in human and murine cell lines led to identification of candidates with up to 94% knockdown efficiency and 50% inhibitory concentration (IC50) values in the submicromolar range. Selected candidate oligonucleotides led to efficient NPC1 protein knockdown in vitro without alteration of cell viability. Furthermore, they did not have immune stimulatory activity in cell-based assays. Treatment of Ebola-virus-infected HeLa cells with the most promising candidates resulted in significant (>99%) virus titer reduction, indicating that antisense oligonucleotides against NPC1 are a promising therapeutic approach for treatment of Ebola virus infection.
Keywords: Ebola virus, antisense oligonucleotide, NPC1, LNA
Introduction
Ebola virus (EBOV), as well as four other filoviruses—Bundibugyo virus (BDBV), Sudan virus (SUDV), Marburg virus (MARV), and Ravn virus (RAVV)—are causative agents of severe disease in humans, such as severe hemorrhagic fever, and are often associated with high morbidity and mortality rates.1, 2, 3 These viruses belong to the family Filoviridae of non-segmented negative-strand RNA viruses and are biosafety level 4 pathogens transmitted by contact with body fluids, fomites, and droplets from infected patients. Filoviruses are considered a significant threat to public health and global security because of their pandemic potential and the risk of being used as a bioweapon.1, 4, 5, 6 Therefore, accelerated efforts in the development of therapeutics is a key objective in the filovirus research community, especially since the 2013–2016 EBOV disease (EVD) epidemic in Western Africa. No vaccines or therapeutic agents with final US Food and Drug Administration (FDA) approval are currently available, and supportive care remains the standard for Ebola virus disease treatment. However, to reduce EBOV spread and the pandemic risk of the current outbreak in Democratic Republic of the Congo (750 confirmed cases and 449 confirmed deaths, as of February 9, 2019) (https://www.who.int/ebola/situation-reports/drc-2018/en/) use of rVSV-ZEBOV Ebola vaccine, as well as antiviral drugs and antibodies against EBOV, have been temporarily approved (https://www.who.int/ebola/drc-2018/faq-vaccine/en/, https://www.who.int/ebola/drc-2018/treatments-approved-for-compassionate-use/en/).
Filovirus particles have a uniform diameter of 80 nm and variable lengths. A single transmembrane glycoprotein (GP), consisting of two subunits, GP1 and -2, is inserted into the virus envelope as a trimeric complex. GP mediates cell attachment and endocytosis by binding to attachment proteins of the host cell.7, 8 In late endosomes, the host cysteine proteases cathepsin-B and -L cleave and remove large C-terminal regions of the GP1 subunit,8, 9 thereby unmasking a binding site for the host factor Niemann-Pick C1 (NPC1). This cholesterol transport protein has been shown to be an essential host factor10, 11 and endosomal entry receptor for filoviruses.12, 13 In cooperation with Niemann-Pick C2 (NPC2), NPC1 is an endosomal transmembrane protein that mediates transport of luminal cholesterol across the endosomal and lysosomal membrane for dispersal to other cellular compartments.14, 15 Loss-of-function mutations in NPC1 or NPC2 cause a rare and often fatal hereditary neurovisceral disorder in humans.16, 17 Over time, patients with NPC disease accumulate cholesterol and glycosphingolipids in various tissues and organs, leading to neurological dysfunction and organ failure. Herbert et al.18 demonstrated that Npc1-deficient mice are completely protected from EBOV infection and free of replicating virus. These results strongly implicate the NPC1 protein as a direct mediator of filovirus infection in vivo. It has been reported that NPC1 inhibition by U18666A, an amphipathic steroid, as well as the EBOV-specific antiviral compound 3.47 significantly inhibit filovirus replication by interfering with viral entry10, 11 in vitro and in vivo.18 However, both compounds have not yet reached clinical trials.
Besides small molecules and therapeutic antibodies, oligonucleotide-based gene expression inhibitors have developed into fully accepted therapeutics. The majority of compounds progressing through clinical trials19, 20, 21 are either antisense oligonucleotides (ASOs), comprising single-stranded DNA-like molecules that recruit endogenous RNase H for target mRNA degradation or small-interfering RNAs (siRNAs) that work through the RNA-induced silencing complex (RISC). ASOs typically have a length of 12–21 nucleotides (nt). In our study, nucleotides were joined via phosphorothioate (PTO) linkages. The phosphorothioate linkage substitutes a sulfur atom for a non-bridging oxygen. This modification renders the internucleotide bond resistant to nuclease degradation and enhances plasma-protein binding while retaining the ability to direct RNase H activity in the cell.22, 23 In addition, ribose moieties in the flanks of the oligonucleotide are modified by an extra bridge connecting the 2′ oxygen and 4′ carbon. This modification locks the conformation of the ribose, conferring high affinity to the RNA. Therefore, this modification is termed the “locked nucleic acid (LNA) modification.”24, 25
Here, we demonstrate the potency of LNA-containing ASO (LNA-ASO) molecules targeting mRNA of the host factor NPC1, which is essential for filovirus replication. We show that NPC1-specific ASOs efficiently reduce viral replication in cultured cells without affecting cell viability or inducing immune-stimulatory responses.
Results
Selection of ASOs Targeting Host Factor Niemann-Pick C1
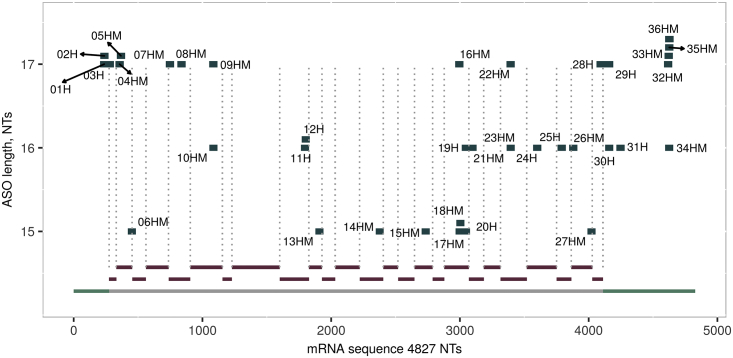
NPC1-specific 15-, 16- and 17-mer ASOs were selected based on human NCBI reference sequence (accession number GenBank: NM_000271.4) (Table S1). The main criterion for sequence selection was selectivity, to avoid undesired off-target effects. Several sequences were completely cross-reactive to murine Npc1, several had one or more mismatches. ASO length, LNA modification pattern, and localization of ASO binding sequence on human NPC1 mRNA are depicted in Tables 1 and S1 and Figure 1.
Table 1.
Sequence and Modification of the ASOs 05HM, 28H, and Neg1
| Name | mRNA Binding Sequence | Position | Length | Sequence |
|---|---|---|---|---|
| 05HM | GGAGAGTGTGGAATTGC | 359 | 17 | +G*+C*+A*A*T*T*C*C*A*C*A*C*T*C*+T*+C*+C |
| 28H | AGCGCGAACGGCTTCTA | 4,086 | 17 | +T*+A*+G*A*A*G*C*C*G*T*T*C*G*C*+G*+C*+T |
| Neg1 | N/A | N/A | 18 | +C*+G*+T*T*T*A*G*G*C*T*A*T*G*T*A*+C*+T*+T |
Depicted are name of ASO, mRNA binding sequence, position on mRNA, ASO length as well as ASO sequence and modification: LNA (+) and/or phosphorothioate (*). Human-specific ASOs (H) as well as cross-reactive ASOs targeting both, human and murine NPC1 (HM), were selected.
Figure 1.
ASO Distribution on Human NPC1 mRNA
All ASOs are depicted according to their location on the human NPC1 mRNA along the x axis. Distinct exons (red) and UTRs (green) are shown in the bottom part of the figure. The lengths of the ASOs are indicated on the y axis.
NPC1 ASOs Efficiently Reduce NPC1 mRNA Expression in Human and Murine Cell Lines
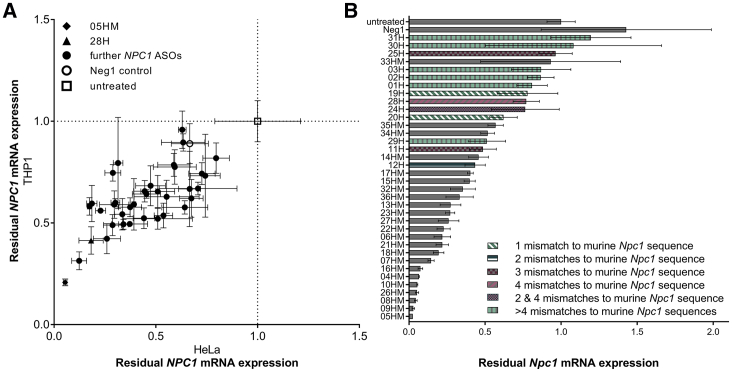
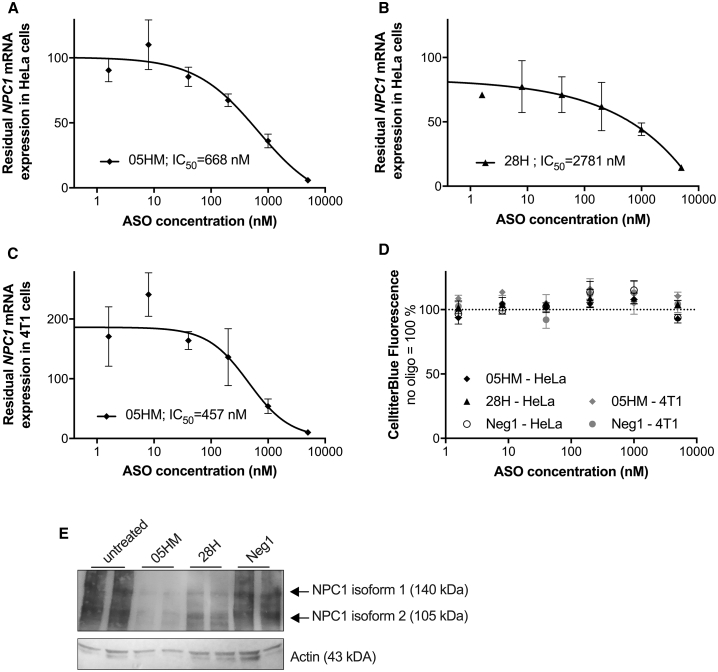
The in vitro activity of the 36 NPC1-specific ASOs was evaluated in two human and one murine cell line endogenously expressing NPC1 mRNA. After treating these cells with LNA-ASO without using a transfection reagent,26 the level of NPC1 mRNA was measured after 3 days of treatment. Human HeLa and THP-1 cells were used as cell lines for screening, as both cell lines are susceptible to EBOV infection. NPC1-specific ASOs led to reduced NPC1 mRNA expression levels in both human cell lines with correlating efficacies (Figure 2A). As expected, cross-reactive ASOs having full complementarity to both human and murine NPC1 mRNA were more efficient in murine 4T1 cells than ASOs that are human-specific and have mismatches to the murine target (Figure 2B). Therefore, an increased number of mismatches of the human-specific ASOs to the murine Npc1 sequence resulted in decreased efficacy in murine 4T1 cells (Figure 2B). In all three cell lines, the human-mouse cross-reactive ASO 05HM was the most efficient candidate with 95% (HeLa), 79% (THP-1), and 98% (4T1) NPC1 mRNA knockdown, while the human-specific ASOs 28H and 29H were among the most potent ASOs in human cells, but had poor activity in murine cells (Figure 2). To test dose-dependence of effects, HeLa and 4T1 cells were exposed to increasing concentrations of ASO 05HM and 28H. Endogenous mRNA levels were evaluated after 3 days of treatment with ASOs, and the 50% inhibitory concentration (IC50) for the inhibition of NPC1 expression was determined (Figures 3A–3C). As already indicated by the aforementioned screening results, ASO 05HM (IC50 = 668 nM) was more potent in the HeLa cells than was ASO 28H (IC50 = 2,781 nM; Figures 3A and 3B). In the murine cell line 4T1, the cross-reactive ASO 05HM was even more effective (IC50 = 457 nM; Figure 3C). Notably, treatment with ASOs did not affect cell viability at any concentration (Figure 3D). Using immunoblot analysis, knockdown efficacy on protein level was evaluated and confirmed in HeLa cells, treated twice for 3 days with ASO 05HM and 28H (Figure 3E). Both ASOs clearly reduced NPC1 protein levels compared with untreated cells or with cells treated with control Neg1 that is not complementary to any human or murine RNA (Figure 3E).27 Again, treatment of HeLa cells with ASO 05HM resulted in a more pronounced NPC1 knockdown than incubation with ASO 28H (Figure 3E).
Figure 2.
Screening of NPC1-Specific LNA-ASOs in Human and Murine Cell Lines
(A and B) Human HeLa and THP1 cells (A) as well as murine cell line 4T1 (B) were treated with 10 μM of ASOs. After 3 days cell lysates were used to determine NPC1 and HPRT1 mRNA levels. (A) Shown is the correlation of residual NPC1 mRNA expression (means and SD of triplicate wells) after treatment in HeLa (x axis) versus THP1 cells (y axis). Values were normalized to HPRT1 and relative to the untreated control (set as 1; empty square). ASO 05HM is indicated as a filled diamond, ASO 28H as a filled triangle, Neg1 as an empty circle, and other NPC1-specific ASOs as filled circles. (B) Residual Npc1 mRNA expression in 4T1 cells after treatment with respective NPC1-specific ASOs or negative control Neg1. Values were normalized to Hprt1 and are shown relative to the untreated control (set as 1). Error bars show SD (triplicate wells). The number of mismatches to murine Npc1 sequence of human specific ASOs (H) are shown in different colors and patterns.
Figure 3.
IC50 Determination and Protein Knockdown Efficacy of ASOs 05HM and 28H
(A–D) HeLa and 4T1 cells were used to generate dose-response curves by treating them with different concentrations of ASO 05HM (A and C) and 28H (B), respectively. On day 3, cell viability was determined using a CellTiter Blue Assay Kit (D). Then, cells were lysed and NPC1 and HPRT1 mRNA levels were determined. Values were normalized to the housekeeping gene HPRT1 and are shown relative to untreated cells (set as 100). IC50 values were calculated using Prism 6 (GraphPad Software). Data are means and SD (triplicate wells). (E) HeLa cells were treated twice for 3 days with 10 μM ASO 05HM, 28H, negative control oligonucleotide Neg1 or were left untreated. On day 6, cells were lysed, and lysates were analyzed for NPC1 and Actin protein expression using SDS-PAGE and immunoblot analysis in duplicate wells.
Taken together, these data demonstrate the potential of the NPC1-specific LNA-ASOs for efficient knockdown of NPC1 gene expression in human and murine cell lines.
Selected NPC1-Specific LNA-ASOs Do Not Cause Adverse Effects In Vitro
Immune activation leading to cytokine release is characteristic of therapeutic oligonucleotides, either as an unwanted side effect or intended pharmacology. This immune activation is mediated by pattern recognition receptors, such as the Toll-like receptors (TLRs). Binding of immune stimulatory ligands, e.g., bacterial DNA or immune stimulatory oligonucleotides, with or without nonmethylated CpG dinucleotides,28 results in TLR activation. As immune activation can lead to a severe, possibly life-threatening, condition of excessive cytokine release,29 the selected LNA-ASOs 05HM and 28H were analyzed for their potential to activate TLR9 and to induce cytokine release in human cells, enabling a safety assessment for future clinical studies.
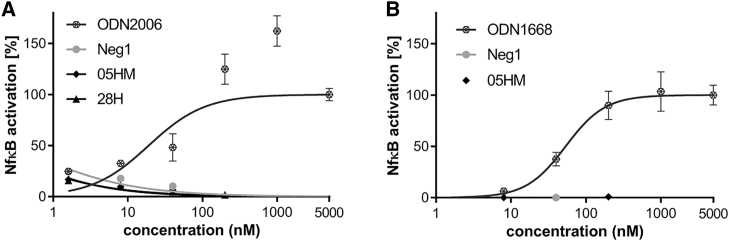
To assess the potential of NPC1-specific ASOs to activate TLR9-mediated signaling, an HEK-Blue hTLR9 SEAP reporter assay was used to measure activation of nuclear factor-kappa light-chain enhancer of activated B cells (NF-кB) induced by TLR9. In contrast to the human TLR9 agonist CpG ODN2006, neither ASO 05HM nor ASO 28H triggered human NF-кB activation (Figure 4A). Murine Nf-кb was also not induced by treatment with cross-reactive ASO 05HM tested in stably transfected HEK-mTlr9_Nf-кb-LUC cells (Figure 4B), whereas the murine Tlr9 agonist ODN1668 induced a considerable dose-dependent response.
Figure 4.
NPC1-Specific ASOs 05HM and 28H Did Not Activate TLR9 Signaling
(A) HEK-Blue hTLR9 cells were treated with ODN2006 or LNA-ASOs 05HM and 28H, respectively, with the indicated concentrations. After 20 h SEAP reporter activity was measured at 620 nm, using a microplate reader. (B) HEK cells expressing a mouse Tlr9 Nf-кb luciferase reporter plasmid were treated with the indicated concentrations of ODN1668 or LNA-ASO 05HM. After 20 h, the cells were treated with ONE-Glo EX reagent, and luminescence was measured at 560 nm. Values were normalized to untreated cells and are means with SD (triplicate wells).
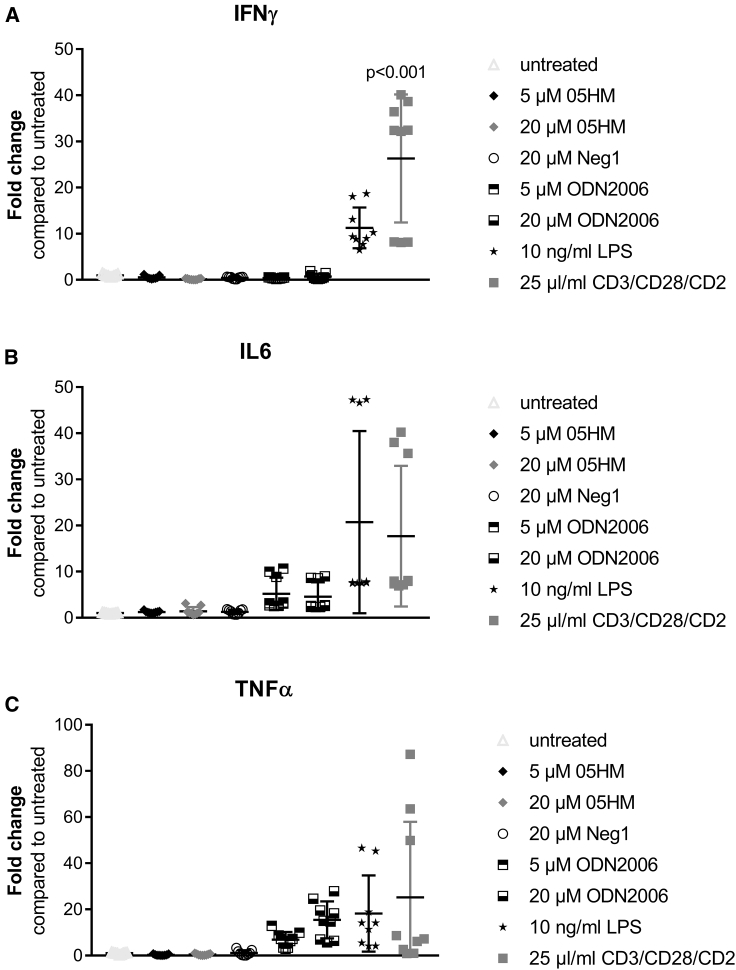
TLRs are expressed by numerous cells of the immune system, such as B lymphocytes, monocytes, natural killer (NK) cells, keratinocytes, melanocytes, and plasmacytoid dendritic cells (pDCs). The peripheral blood mononuclear cell (PBMC)-based assay is well established for determination of immune activation by different drugs, including TLR ligands and oligonucleotides.28, 30, 31 Here, PBMC isolated from leukocytes preparations of three different donors were used. In contrast to pattern recognition receptor agonists, ODN2006, lipopolysaccharide (LPS), or immune stimulatory CD3/CD28/CD2, the ASO 05HM failed to trigger a cytokine response (Figure 5).
Figure 5.
NPC1-Specific ASO 05HM Did Not Stimulate Cytokine Release from Treated PBMCs
(A–C) PBMCs from three different donors were treated with oligonucleotides and immune stimulatory agents (each condition in triplicate wells), with the indicated concentrations. The third day after treatment, supernatants were harvested and used for determination of cytokine release using ELISA: (A) IFNγ, (B) IL6, and (C) TNFα. Values were displayed as fold changes compared to untreated. Means and SDs were also indicated. ANOVA test was used to test for significant differences and p values were determined using Dunnett’s test in GraphPad Prism 7.04 Software.
These data support that the most potent ASOs, 05HM and 28H, do not trigger a host innate immune response.
NPC1-Specific LNA-ASOs Efficiently Inhibit EBOV Replication In Vitro
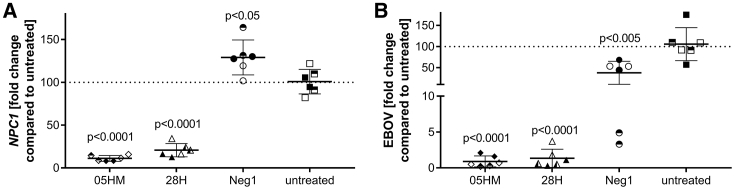
LNA-ASOs 05HM and 28H were then tested for antiviral activity during EBOV infection. To ensure efficient downregulation of endosomal NPC1 protein (Figure 3E), HeLa cells were treated twice with ASO 05HM and 28H for 3 days. Then, cells were infected with EBOV at an MOI of 0.01 and incubated for 24 h in medium containing the respective ASOs. Compared to untreated cells, NPC1 mRNA was significantly reduced (by about 89% [05HM] and 80% [28H]; Figure 6A), whereas EBOV virus genome copies were decreased by about 99% (05HM) and 98% (28H), 1 day after infection, as quantified by qPCR (Figure 6B). Treatment with the control oligonucleotides Neg1 (Figure 6A; Figure S2A), Neg1B, or S5 (Figure S2A) did not reduce NPC1 mRNA levels. However, EBOV replication was also decreased (by about 62% [Neg1, Figure 6B] and 70% [Neg1B and S5; Figure S2B]), most likely due to the backbone-mediated effects of phosphorothioate-modified oligonucleotides (Figure 6B; Figure S2A).
Figure 6.
NPC1-Specific ASOs 05HM and 28H Specifically Inhibit EBOV Replication
(A and B) HeLa cells pretreated with the respective ASO were infected with EBOV at an MOI of 0.01. At 1 day after infection (p.i.), NPC1 (A) and EBOV (B) levels were quantified by RT-qPCR and normalized to the internal control α-tubulin. Shown is the fold change compared to untreated control (set at 100), which was calculated using the 2−ΔΔCt method. Error bars show SD (n = 3, each in duplicate). Duplicates are labeled with identical symbol shapes. ANOVA was used to test for significant differences and p values were determined using Dunnett’s test, in GraphPad Prism 7.04 Software. See also Figure S2.
Treatment with phosphorothioate-modified non-targeting oligonucleotides reduced EBOV replication by 62-70% in vitro. Furthermore, oligonucleotides specifically targeting NPC1, an essential receptor of EBOV cell entry, led to a more potent inhibition of EBOV replication—up to 99%.
Discussion
EBOV causes a severe, often fatal illness in humans, is transmitted to people from wild animals, and spreads in the human population through human-to-human transmission.32 Case fatality rates of Ebola virus disease have varied from 25% to 90% in past outbreaks, with an average of around 50%.33, 34 There are two distinct paths for potential EBOV treatments: post-exposure prophylaxis and treatment of symptomatic patients. Both have different challenges, but a common strategy may be to limit virus replication, to allow the adaptive and innate immune systems time to fight the infection.35, 36 A limitation for ASO-based antiviral strategies directly targeting the virus may be the RNase H1-dependent mode of action of gapmer ASOs. Filoviruses replicate in the host cytoplasm and do not require the nucleus.37 However, the cellular compartment of ASO-mediated RNA degradation is a controversial issue within the ASO community.38, 39, 40, 41 We and others showed that ASOs targeting EBOV mRNA could efficiently block viral transcription or replication in cell-free42 and reporter gene assays (Figures S1A and S1B). In reporter-based assays, transcription of the reporter genes takes place in the nucleus. However, EBOV-specific ASOs were not capable of inhibiting viral propagation in Huh7 cells, with or without the use of a transfection reagent (Figures S1C and S1D). These results indicate, that the predominant location of RNase H1-mediated target RNA degradation is the nucleus that is, as previously mentioned, bypassed during EBOV replication. The use of ASO approaches aimed at sterically blocking viral translation in the cytoplasm could avoid this issue. However, translation-blocking approaches are much less efficient compared to RNase-H1-dependent mechanisms. Whereas one translation-blocking ASO-molecule is needed to repress translation of one target mRNA molecule, one RNase H1-depending ASO could mediate degradation of many individual RNA molecules. Therefore, much higher doses would be necessary for a translation-blocking approach in the cytoplasm. The antiviral efficacy of translation-blocking approaches was suggested by Chery et al.42 indirectly by use of reporter-based assays. However, direct evidence for antiviral effects such as copy number determination of an EBOV isolate after ASO treatment is missing. A fundamental disadvantage of approaches targeting EBOV directly is the error-prone viral polymerase of RNA viruses that enables incorporation of the mutations that facilitate resistance against antiviral drugs. Furthermore, antiviral approaches have not been capable of overcoming strain-specific differences, so far. Although the efficacy of antibody cocktails, such as ZMapp, for EBOV in nonhuman primates is clear, a future challenge will be to identify similar treatments for other filoviruses.43
For the aforementioned reasons, targeting an essential human host factor for viral replication may be one of the most promising approaches for fighting EBOV. An essential host factor for filovirus replication in vitro and in vivo is the cholesterol transport protein NPC1. This endosomal entry receptor has been shown to mediate cytoplasmic release of viral ribonucleoproteins,10, 13 making it an ideal target for blocking filovirus replication.
Various small molecule therapeutics against host factor NPC1 have been described, some with demonstrated efficacy against filovirus replication in mice.43 However, those molecules targeting NPC1 did not reach clinical trials, so far, probably because of the described toxic effects44 or the need for further mechanistic characterization, as well as improvement of compounds.43 The limited success of the available NPC1 inhibitors in protecting mice from EBOV challenge highlights the need for new molecules or approaches to target NPC1 in vivo.18
A unique advantage of ASOs targeting essential host factors for filovirus replication, particularly in the context of emergency prophylactic or post-exposure therapeutics, is their target specificity, cost-effectiveness, and fast generation. Mode of action, uptake, biodistribution, pharmacokinetics, and safety issues have been extensively studied.24, 26, 45, 46, 47, 48, 49, 50
In the present study, a total of 36 ASOs was designed for the initial screen broadly covering the NPC1 mRNA sequence. Fifteen-, 16-, and 17-mer ASOs were tested, and two ASOs, comprising 17 nt, were selected for further experiments. As recommended by the Oligonucleotide Safety Working Group,51 an extensive in silico approach was used to avoid suppression of off-target genes. The bioinformatic analysis revealed no perfect match to any exonic or intronic off-target sequence. Even allowing one mismatch, still no off-target hit was detected in the NCBI RefSeq data base. This off-target characteristic is well in the range of the characteristic of other ASOs recently published in the field.42, 52 This should also decrease risk of generation of ASO-generated RNA fragments, which, as Dieckmann et al.45 speculated, could cause toxicity.
ASOs have been repeatedly described to stimulate immune activation.22, 53, 54, 55, 56 Reasons for immune stimulatory activities were reported to be nonmethylated CpG dinucleotides within the oligonucleotide sequence, as well as the stabilizing phosphorothioate backbone.57, 58, 59 Studies using Tlr9-deficient mice demonstrated that this Tlr subtype is essential for the effects that are mediated by bacterial DNA or CpG oligonucleotides.60 Since expression, ligand preference, and function of pattern-recognition receptors is highly species specific,61 cytokine release in humans is hard to predict in animal studies. The human TLR9 is expressed in B cells and pDCs.61, 62, 63 Both cell types are stimulated by CpG oligonucleotides to upregulate cell surface costimulatory molecules and to secrete a variety of cytokines.64, 65 These effects can lead to indirect activation of other cell populations, such as monocytes and NK or T cells.65 The in vitro TLR9 assays as well as the PBMC ex vivo test used in this study are therefore helpful in deselecting ASOs with immune stimulatory potential early in the screening and compound characterization process, to prevent unexpected harmful effects in clinical development. In contrast to the CpG oligonucleotides ODN2006 and ODN1668, which were used as positive controls, both of the selected NPC1-targeting ASOs 05HM and 28H activated neither human nor murine TLR9 (Figure 4), nor did ASO 05HM stimulate cytokine release in treated PBMCs (Figure 5). These results are in line with the findings of Vollmer and colleagues,28 who demonstrated that LNA modification of ASOs significantly decreases the immune stimulatory effects of ASOs. Our bioinformatics analysis enabled the selection of cross-reactive ASOs that target human as well as murine NPC1 mRNA for screening in human and murine cell lines. Again, the benefit of our rational ASO design was confirmed, as cross-reactive ASOs were effective in both species in vitro (Figure 2). As expected, a decreased knockdown of murine Npc1 by human-specific ASOs with no 100% complementarity to the murine mRNA was observed in murine cells. Thereby, a higher number of mismatches to the murine Npc1 sequence was associated with decreased activity in murine cells (Figure 2B). In Npc1−/− mice, it was previously demonstrated that the Npc1 protein acts as a direct mediator for EBOV infection in vivo.18 Since ASO 05HM targets human and murine NPC1, it is also a suitable tool for testing in vivo efficacy against different filovirus strains in mice.
NPC1 ASO screening in two different human cell lines resulted in strong correlation of ASO activity, which enabled reliable selection of candidate ASOs. Consistently, ASO 05HM was the most effective candidate during single-dose screens in one murine and two human and cell lines (Figure 2), during IC50 determination (Figures 3A–3C), protein knockdown (Figure 3E), and the infection assay in HeLa cells (Figure 6). All experiments were performed by using a method called “gymnosis” without using a transfection reagent.26 The pattern of gene silencing of in vitro gymnotically delivered oligonucleotides correlates particularly well with in vivo silencing and is therefore of particular significance for drug discovery.26 Recently, Chery and colleagues42 also reported on efficient knockdown of NPC1 by use of target-specific ASOs. However, these ASOs were delivered into cells using lipofection and may have divergent characteristics in vivo.
Macrophages, monocytes, and dendritic cells are the primary target cells during acute EBOV infection, and several organs, such as liver, kidneys, and spleen, show high viral loads during the course of infection,66, 67, 68, 69 These cell types and organs are also good targets for phosphorothioate-modified ASOs which have a preferred biodistribution to kidney, liver, and immune cells and are not capable of crossing the blood-brain barrier.46 Therefore they cannot affect the transport of cholesterol in neurons within the CNS, which could cause a Niemann-Pick disease-like phenotype.
Treatment with NPC1-specific ASO resulted in significantly decreased EBOV replication (>99%; Figure 6B). Similar results were obtained by Chery and colleagues42 by use of a VSVluc-EboV GP reporter virus. Reduced luciferase expression up to 80% was detected after transfection of HeLa cells with an NPC1-specific ASO. However, neither was the immune stimulatory potential of the selected ASO tested nor was a negative control ASO included to test for potential unspecific effects. Therefore, whether the observed decrease in reporter virus replication is a backbone-mediated or a specific effect caused by NPC1 knockdown cannot be distinguised.42 In the present study, the control oligonucleotides clearly diminished the amount of EBOV genome copies 24 h after infection, though to a lesser extent (Figure 6B; Figure S2B). In addition to their sequence-specific functionality, single-stranded oligonucleotides are polymers with polyanionic characteristics that are largely conserved, regardless of their nucleotide sequence. Phosphorothioation of oligonucleotides confers increased hydrophobicity and has been shown to specifically mediate antiviral activity in a manner independent of the increased nuclease stability present with this modification.70, 71 Phosphorothioates with broad-spectrum activity against viral and other infectious diseases are also called “nucleic acid polymers” (NAPs).72 The structure-function relationship of the antiviral activity of NAPs, as well as their molecular mechanism of action, was first elucidated in a study describing the specific antiviral effects of NAPs during the entry of HIV-1.71 In this study, the entry inhibition effect of NAPs was shown to be independent of sequence, but dependent on size.71 A large amphipathic protein domain in the viral gp41 glycoprotein is required for interaction with NAPs and conserved in class I fusion GPs from many other viruses susceptible to antiviral polymers. Following on the findings in the initial study in HIV-1, NAPs were subsequently shown to have the same sequence-independent and phosphorothioate as well as length-dependent antiviral effects in other viruses with class I fusion GPs, including herpesviruses, cytomegalovirus, influenza virus, and lymphocytic choriomeningitis virus.73, 74, 75, 76, 77 Meanwhile, the company Replicor develops antiviral drug candidates against hepatitis B and D viruses based on the mechanism by which NAPs inhibit viral propagation.
Interestingly, EBOV GP, which is required for virus entry into the host cell, is also a class I fusion protein.78 Keeping this fact in mind, it may be reasonable that, to some extent, phosphorothioated oligonucleotides inhibit EBOV replication, independent of sequence.
These findings could explain the 62-70% reduction of viral replication detected with control phosphorothioates that did not reduce NPC1 mRNA levels (Figures 6A and 6B; Figures S2A and S2B). However, the specific oligonucleotides 05HM and 28H significantly reduced the EBOV titer after 24 h of infection by 97%–99% compared to non-treated cells (Figure 6B; Figure S2B). This result strengthens our hypothesis that RNase H1-mediated targeting of an essential host factor for virus entry has an additional effect of solely backbone-mediated reduction of EBOV entry, thereby vastly improving therapeutic antiviral potential.
Taken together, the findings in this study demonstrate that knockdown of intracellular receptor NPC1 by target-specific ASOs is a promising approach for treatment of EBOV infection. Our selected human-mouse cross-reactive NPC1-specific ASO may be used in future studies to further investigate the efficacy of Npc1-specific ASO against EBOV in mouse models.
Materials and Methods
Selection of NPC1-Specific ASOs
The human mRNA sequence of NPC1, as defined by NCBI accession number GenBank: NM_000271.4, was taken as a basis to design the ASOs. For this sequence (4,827 bp) the possible 15-, 16-, and 17-mer sequence fragments were extracted. All 14,442 fragments were tested for specificity against the complete NCBI RefSeq (https://www.ncbi.nlm.nih.gov/refseq/) and ENSEMBL (http://www.ensembl.org/) databases, by using PatMaN.79 Possible matches, including up to three mismatches against any mRNA (RefSeq) or any intron (ENSEMBL), were collected to enable a filtering of those fragments showing the highest specificity. As a cross-reactivity to mice was additionally desired, the sequences were tested against all mouse sequences of both databases, as well.
The main criterion for the selection process was the absence of a perfect match to any human or mice off-target mRNA or intron sequence. In addition, only a limited number of off-target matches allowing up to three mismatches were tolerated. As recommended by the distributor (https://www.qiagen.com/de/shop/pcr/primer-sets/custom-lna-oligonucleotides/?akamai-feo=off&clear=true#productdetails), sequences containing a triple C or triple G pattern were avoided. The melting temperatures of the reverse complements of the remaining sequences were determined in silico, considering the effects of LNA modifications. The typical LNA-gapmer format of three modifications at each side80, 81 was optimized, if there were strong deviations from the internally defined range of an appropriate melting temperature. In the first screen, 23 sequences showing a perfect match to human and mice NPC1 mRNA and 13 sequences specific only to the human mRNA were used to design ASOs containing phosphorothioate bonds and the optimal LNA pattern.
For screening, IC50 determination and analysis of protein knockdown, 40 nmol of ASOs were synthesized by Eurogentec (Cologne, Germany). After selection of two of the most effective candidates, 5 mg of the ASOs 05HM and 28H were purchased from Exiqon (Copenhagen, Denmark) for in vitro infection assays. Control oligonucleotides Neg1 and S5 were synthesized by Exiqon (Copenhagen, Denmark), and negative control oligonucleotide Neg1b was purchased from Axolabs (Kulmbach, Germany). ASOs were purified by reverse-phase high-performance liquid chromatography (HPLC) and subsequently lyophilized. After receiving the ASOs, they were solubilized in diethyl-pyrocarbonate (DEPC)-treated H2O to a concentration of 1 mM.
Screening of NPC1-Specific ASOs
HeLa (ACC 57; DSMZ, Braunschweig, Germany;), THP-1 (ACC 16; DSMZ), and 4T1 (CRL-2539; ATCC-LGC Standards, Wesel, Germany) cells were cultured in DMEM GlutaMAX (32430027; Gibco, Life Technologies, Darmstadt, Germany) supplemented with 10% fetal calf serum (FCS; 10270106; Gibco), 1 mM sodium pyruvate (11360070; Gibco), and 1× antibiotic-antimycotic (15240062; Gibco).
The cells were detached using trypsin (25200056; Gibco), seeded into 96-well plates at 6,000 (HeLa), 20,000 (THP-1), and 5,000 (4T1) cells per well and treated with the NPC1 ASO compounds on day 0. Treatments were done at a final concentration of 10 μM with triplicate experiments for each compound. A control oligonucleotide, Neg1, was used at a final concentration of 10 μM with triplicate experiments. On day 3, cells were lysed, and mRNA levels were measured according to the manufacturers’ instructions, using QuantiGene Singleplex Gene Expression Assay (QS0011; Life Technologies, Darmstadt, Germany) and the following probe sets: human probe sets specific for NPC1 (SA-10502; Life Technologies) and the housekeeping gene HPRT1 (SA-10030; Life Technologies) or probe sets specific for murine Npc1 (SB-12805; Life Technologies) and Hprt1 (SB-15463; Life Technologies).
Residual NPC1 mRNA expression levels were calculated by comparing the NPC1 values normalized to HPRT1 in the ASO-treated samples with that measured in the untreated control.
IC50 Determination
HeLa and 4T1 cells were used to generate dose-response curves by treating them with different concentrations (5,000 nM, 1,000 nM, 200 nM, 40 nM, 8 nM, and 1.6 nM) of ASO 05HM (4T1; HeLa) and 28H (HeLa). On day 3 after treatment, cell viability was determined using the CellTiter-Blue Assay (G8081; Promega, Mannheim, Germany) according to the manufacturer’s instructions. Afterwards, the cells were lysed, and mRNA levels were measured according to the manufacturers’ instructions, using the QuantiGene Singleplex Gene Expression Assay (QS0011; Life Technologies) and the following probe sets: human probe sets specific for NPC1 (SA-10502; Life Technologies) and the housekeeping gene HPRT1 (SA-10030; Life Technologies) or probe sets specific for murine Npc1 (SB-12805; Life Technologies) and Hprt1 (SB-15463; Life Technologies). Values were normalized to the housekeeping gene HPRT1 and cell control (untreated cells). IC50 values were calculated using Prism 6 (GraphPad Software).
Immunoblot Analysis
HeLa cells were seeded into 12-well plates at 75,000 cells per well and treated twice with 10 μM ASO 05HM and 28H for 3 days each. On day 6, cells were lysed using 100 μL RIPA buffer (89900; Thermo Scientific, Life Technologies) per well supplemented with Halt Protease Inhibitor Cocktail (1861278; Thermo Scientific). Samples were prepared for SDS-PAGE using 4× Laemmli sample buffer (161-0747; Bio-Rad Laboratories, München, Germany) and proteins were separated using Mini-Protean TGX™ Precast Gels (456-8025; Bio-Rad Laboratories), Precision Plus Protein Western C Standard (161-0376; Bio-Rad Laboratories), and 1× Tris/glycine/SDS (TGS; 161-0732; Bio-Rad Laboratories) according to the manufacturer’s instructions. Polyvinylidene fluoride (PVDF) membrane (162-0177; Bio-Rad Laboratories) was activated in 100% ethanol (5054.4; Carl Roth, Karlsruhe, Germany) and equilibrated in 1× Tris/glycine (TG) buffer (161-0734; Bio-Rad Laboratories). Separated proteins were transferred to the membrane using OmniPAGE mini vertical systems and Semi-dry Blotter (Cleaver Scientific, Warwickshire, United Kingdom) and 10× TG buffer. Protein detection was performed using the Amplified Opti-4CN Substrate Kit (1708238; Bio-Rad Laboratories), according to the manufacturer’s instructions and the following antibody dilutions: NPC1 antibody (1:500, ab55706; Abcam, Cambridge, United Kingdom), Actin antibody (1:500, VMA00048; Bio-Rad AbD Serotec, Puchheim, Germany), goat anti-mouse-horse radish peroxidase (GAM-HRP, 1:1,000, STAR207P; Bio-Rad AbD Serotec).
TLR9 Reporter Assay
HEK-Blue hTLR9 cells, which were generated by co-transfection of the hTLR9 gene and an optimized SEAP reporter gene into HEK293 cells, were obtained from InvivoGen (hkb-htlr9; Toulouse, France). The SEAP reporter gene was placed under the control of the interferon-beta (IFNβ) minimal promoter fused to five NF-кB and activator protein 1 (AP-1) binding sites. Stimulation with a TLR9 ligand activates NF-кB and AP-1, which induce the production of SEAP. Cells were cultivated in DMEM GlutaMAX (32430027; Gibco, Life Technologies) supplemented with 10% FCS (10270106; Gibco), 1 mM sodium pyruvate (11360070; Gibco), 1× antibiotic-antimycotic (15240062; Gibco), 10 μg/mL Blasticidin (ant-bl-05; InvivoGen) and 100 μg/mL Zeocin (ant-zn-1; InvivoGen). Cells were seeded at 25,000 cells per well in 96-well plates and incubated for 20 h at 37°C and 5% CO2. Then, they were treated with ODN2006 (tlrl-2006; InvivoGen) or LNA-ASOs 05HM and 28H, with 5-fold serial dilutions, starting at a concentration of 5,000–1.6 nM. As a control, cells were treated with cell culture medium without addition of oligonucleotides. Each condition was performed in triplicate. Twenty hours after cell treatment, 100 mL Quanti-Blue (QB) Solution (rep-qbs; InvivoGen) was prepared by adding 1 mL of QB reagent and 1 mL of QB buffer to 98 mL of sterile water in a sterile glass bottle or flask. QB Solution was gently mixed and incubated for 10 min at room temperature before it was added to the sample. HEK-Blue-hTLR9 cell supernatants (20 μL per well) were harvested into a fresh 96-well plate and 180 μL QB solution was added to each well. Samples were incubated for 2 h at 37°C, and SEAP activity was determined by measurement of the optical density at 620 nm with a microplate reader.
Stably transfected HEK cells expressing a mouse Tlr9 Nf-кb luciferase reporter plasmid, kindly provided by Prof. Holger Garn, University of Marburg, were used for the murine Tlr9 reporter gene assay. Stimulation with mouse Tlr9 ligands activate Nf-кb which induces the expression of firefly luciferase (Photinus pyralis). HEK-mTlr9_ Nf-кb-LUC cells (25,000/well) were plated in a white-walled, 96-well tissue-culture plate in DMEM GlutaMAX (32430027; Gibco, supplemented as described above). Twenty hours after cell seeding, the cells were treated with 5-fold serial dilutions of ODN1668 (tlrl-1668; InvivoGen) or LNA-ASO 05HM, starting at a concentration of 5,000–1.6 nM and incubated for 20 h at 37°C in the incubator. As a control, cells were treated with cell culture medium without addition of oligonucleotides. Each condition was performed in triplicate. After the incubation period, the plates were centrifuged for 5 min at 500 g, and the cell supernatants were removed. ONE-Glo EX reagent (50 μL, E8110; Promega) was added to each well, and cells were lysed according to the manufacturer’s instructions. Luminescence was immediately measured at 560 nm.
Cytokine Release
PBMCs were isolated from leukapheresis products donated by three healthy individuals (Klinikum rechts der Isar, TU München, ethics commission reference: 329/16 S), by density gradient centrifugation. The leukapheresis product was diluted 1:10 with PBS (10010-023; Gibco) and carefully loaded onto 15 mL Biocoll separating solution (L6115; Biochrom, Berlin, Germany) in 50 mL Falcon tubes. Density gradient centrifugation was performed at 800 g for 20 min at room temperature with the brake turned off. Afterward, the mononuclear cell layer was collected carefully and transferred into a new 50 mL Falcon tube, and PBS was added to a final volume of 50 mL. After centrifugation at 500 g for 5 min at room temperature (brake turned on) supernatants were discarded, and cell pellets were pooled in PBS in a total volume of 50 mL. Cells were counted, divided into aliquots, and stored on liquid nitrogen for further use. PBMCs (400,000) were seeded in RPMI-1640 medium (72400-047; Gibco) supplemented with 10% FCS, 1 mM sodium pyruvate, 1× antibiotic-antimycotic, and 50 U/mL Benzonase (70746-3; Merck, Darmstadt, Germany) per well of a 96-well plate. Afterwards, the cells were treated with RPMI containing either oligonucleotides (5 or 20 μM ASO 05HM or 20 μM negative control Neg1) or immune stimulatory agents: 20 μM ODN2006 (tlrl-2006; InvivoGen), 10 ng/mL LPS (L4391; Sigma Aldrich, Taufkirchen, Germany), or 25 μL/mL CD3/CD28/CD2 (10970; StemCell Technologies, Inc., Grenoble, France). Each condition was performed in triplicate. Treated cells were then rested for 72 h at 37°C and 5% CO2. The third day after treatment, 96-well plates were subjected to centrifugation at 2,000 rpm for 10 min at 4°C. Cell supernatants were collected in 96-well plates. For the measurement of IFNγ secretion, 50 μL of cell supernatant was diluted with 50 μL ELISA/ELISPOT diluent. IFNγ measurement was performed with IFNγ Human Uncoated ELISA Kit from eBioscience (88-7316-88; Thermo Fisher Scientific) according to the manufacturer’s protocol. For the measurement of IL6 and TNFα secretion, 50 μL of cell supernatant was diluted with 50 μL Assay Diluent A. IL-6 and TNFα measurements were performed with IL-6 and TNFα ELISA Kits, respectively, from BioLegend (Koblenz, Germany [IL6; 430505] and [TNFα; 430201]), according to the manufacturer’s protocols.
EBOV Infection Assay
All work with infectious EBOV was performed in compliance with national regulations at the BSL4 Laboratory of the Institute of Virology, Philipps-University, Marburg.
HeLa cells were pretreated in duplicate two times for 3 days by adding medium containing 10 μM of ASOs 05HM and 28H and negative control oligonucleotides Neg1, Neg1B, or S5 or medium only (mock). At 1 h prior to infection, ASOs were removed. Cells were then infected with EBOV Mayinga (NCBI accession number GenBank: AF086833.2) at an MOI of 0.01 for 3 h in the absence of ASOs. Subsequently, the cells were washed to remove unbound input virus and incubated in the presence of 10 μM of the respective ASO. At 24 h after infection, cellular RNA was isolated using the RNeasy Kit (74106; QIAGEN, Hilden, Germany). For virus quantification, EBOV L- or GP-specific primer sets and probes were used for qRT-PCR analysis.82 Knockdown of NPC1 was analyzed via qRT-PCR with NPC1 specific primers (VHPS-6283; Real-Time Primers, Elkins Park, PA) using the QuantiTect SYBR Green RT-PCR Kit (204143; QIAGEN). Ct values were normalized to the internal control α-tubulin (2−ΔCt), and the fold change over mock was calculated using the 2−ΔΔCt method.83
Statistical Analysis
GraphPad Prism 7.04 Software was used for statistical calculations. The efficacy of the ASOs was assessed by one-way ANOVA followed by Dunnett’s multiple-comparison test, to contrast the treatment groups (including control ASOs) with mock control. Differences were considered statistically significant when p < 0.05.
Author Contributions
The study design was developed by A.S., E.D., M.K., M.H., M.E., S.B., and F.J. The bioinformatic design of the ASOs was created by S.M. The experiments were executed by A.S., E.D., M.K., M.H., T.T., and R.K. Data were evaluated by A.S., M.K., M.H., and E.D. The manuscript was written by A.S., E.D., S.M., and F.J.
Conflicts of Interest
A.S., S.M., M.H., T.T., R.K. and F.J. are/were employees of Secarna Pharmaceuticals & Co. KG (Planegg, Germany). The rest of the authors declare no competing interests.
Acknowledgments
This work was supported by “Projektträger Bayern” (Bayerisches Forschungsprogramm “Bio- und Gentechnologie” (BayBIO); grant FKZ BIO-1505-0004). We thank Holger Garn (Marburg, Germany) for kindly providing us with the HEK cells expressing a mouse TLR9 NF-кB luciferase reporter plasmid. We also thank Lisa Hinterwimmer and Monika Schell for excellent technical support.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.04.018.
Supplemental Information
References
- 1.Feldmann H., Jones S., Klenk H.-D., Schnittler H.-J. Ebola virus: from discovery to vaccine. Nat. Rev. Immunol. 2003;3:677–685. doi: 10.1038/nri1154. [DOI] [PubMed] [Google Scholar]; Feldmann, H., Jones, S., Klenk, H.-D., and Schnittler, H.-J. (2003). Ebola virus: from discovery to vaccine. Nat. Rev. Immunol. 3, 677-685. [DOI] [PubMed]
- 2.Geisbert T.W., Jahrling P.B. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10(12, Suppl):S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]; Geisbert, T.W., and Jahrling, P.B. (2004). Exotic emerging viral diseases: progress and challenges. Nat. Med. 10(12, Suppl) S110-S121. [DOI] [PubMed]
- 3.Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kuhn, J.H., Becker, S., Ebihara, H., Geisbert, T.W., Johnson, K.M., Kawaoka, Y., Lipkin, W.I., Negredo, A.I., Netesov, S.V., Nichol, S.T., et al. (2010). Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 155, 2083-2103. [DOI] [PMC free article] [PubMed]
- 4.Working Group on Civilian Biodefense Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]; Working Group on Civilian Biodefense (2002). Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287, 2391-2405. [DOI] [PubMed]
- 5.Jaax N., Jahrling P., Geisbert T., Geisbert J., Steele K., McKee K., Nagley D., Johnson E., Jaax G., Peters C. Transmission of Ebola virus (Zaire strain) to uninfected control monkeys in a biocontainment laboratory. Lancet. 1995;346:1669–1671. doi: 10.1016/s0140-6736(95)92841-3. [DOI] [PubMed] [Google Scholar]; Jaax, N., Jahrling, P., Geisbert, T., Geisbert, J., Steele, K., McKee, K., Nagley, D., Johnson, E., Jaax, G., and Peters, C. (1995). Transmission of Ebola virus (Zaire strain) to uninfected control monkeys in a biocontainment laboratory. Lancet 346, 1669-1671. [DOI] [PubMed]
- 6.Johnson E., Jaax N., White J., Jahrling P. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int. J. Exp. Pathol. 1995;76:227–236. [PMC free article] [PubMed] [Google Scholar]; Johnson, E., Jaax, N., White, J., and Jahrling, P. (1995). Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int. J. Exp. Pathol. 76, 227-236. [PMC free article] [PubMed]
- 7.Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, J.E., Fusco, M.L., Hessell, A.J., Oswald, W.B., Burton, D.R., and Saphire, E.O. (2008). Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177-182. [DOI] [PMC free article] [PubMed]
- 8.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]; White, J.M., Delos, S.E., Brecher, M., and Schornberg, K. (2008). Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189-219. [DOI] [PMC free article] [PubMed]
- 9.Falasca L., Agrati C., Petrosillo N., Di Caro A., Capobianchi M.R., Ippolito G., Piacentini M. Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death Differ. 2015;22:1250–1259. doi: 10.1038/cdd.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]; Falasca, L., Agrati, C., Petrosillo, N., Di Caro, A., Capobianchi, M.R., Ippolito, G., and Piacentini, M. (2015). Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death Differ. 22, 1250-1259. [DOI] [PMC free article] [PubMed]
- 10.Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carette, J.E., Raaben, M., Wong, A.C., Herbert, A.S., Obernosterer, G., Mulherkar, N., Kuehne, A.I., Kranzusch, P.J., Griffin, A.M., Ruthel, G., et al. (2011). Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340-343. [DOI] [PMC free article] [PubMed]
- 11.Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cote, M., Misasi, J., Ren, T., Bruchez, A., Lee, K., Filone, C.M., Hensley, L., Li, Q., Ory, D., Chandran, K., and Cunningham, J. (2011). Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344-348. [DOI] [PMC free article] [PubMed]
- 12.Krishnan A., Miller E.H., Herbert A.S., Ng M., Ndungo E., Whelan S.P., Dye J.M., Chandran K. Niemann-Pick C1 (NPC1)/NPC1-like1 chimeras define sequences critical for NPC1's function as a flovirus entry receptor. Viruses. 2012;4:2471–2484. doi: 10.3390/v4112471. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krishnan, A., Miller, E.H., Herbert, A.S., Ng, M., Ndungo, E., Whelan, S.P., Dye, J.M., and Chandran, K. (2012). Niemann-Pick C1 (NPC1)/NPC1-like1 chimeras define sequences critical for NPC1's function as a flovirus entry receptor. Viruses 4, 2471-2484. [DOI] [PMC free article] [PubMed]
- 13.Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A., Ndungo E., Sandesara R.G., Carette J.E., Kuehne A.I. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miller, E.H., Obernosterer, G., Raaben, M., Herbert, A.S., Deffieu, M.S., Krishnan, A., Ndungo, E., Sandesara, R.G., Carette, J.E., Kuehne, A.I., et al. (2012). Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31, 1947-1960. [DOI] [PMC free article] [PubMed]
- 14.Cruz J.C., Sugii S., Yu C., Chang T.-Y. Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J. Biol. Chem. 2000;275:4013–4021. doi: 10.1074/jbc.275.6.4013. [DOI] [PubMed] [Google Scholar]; Cruz, J.C., Sugii, S., Yu, C., and Chang, T.-Y. (2000). Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J. Biol. Chem. 275, 4013-4021. [DOI] [PubMed]
- 15.Infante R.E., Wang M.L., Radhakrishnan A., Kwon H.J., Brown M.S., Goldstein J.L. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Infante, R.E., Wang, M.L., Radhakrishnan, A., Kwon, H.J., Brown, M.S., and Goldstein, J.L. (2008). NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA 105, 15287-15292. [DOI] [PMC free article] [PubMed]
- 16.Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]; Carstea, E.D., Morris, J.A., Coleman, K.G., Loftus, S.K., Zhang, D., Cummings, C., Gu, J., Rosenfeld, M.A., Pavan, W.J., Krizman, D.B., et al. (1997). Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228-231. [DOI] [PubMed]
- 17.Naureckiene S., Sleat D.E., Lackland H., Fensom A., Vanier M.T., Wattiaux R., Jadot M., Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]; Naureckiene, S., Sleat, D.E., Lackland, H., Fensom, A., Vanier, M.T., Wattiaux, R., Jadot, M., and Lobel, P. (2000). Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290, 2298-2301. [DOI] [PubMed]
- 18.Herbert A.S., Davidson C., Kuehne A.I., Bakken R., Braigen S.Z., Gunn K.E., Whelan S.P., Brummelkamp T.R., Twenhafel N.A., Chandran K. Niemann-pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio. 2015;6 doi: 10.1128/mBio.00565-15. e00565–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Herbert, A.S., Davidson, C., Kuehne, A.I., Bakken, R., Braigen, S.Z., Gunn, K.E., Whelan, S.P., Brummelkamp, T.R., Twenhafel, N.A., Chandran, K., et al. (2015). Niemann-pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio 6, e00565-15. [DOI] [PMC free article] [PubMed]
- 19.Viney N.J., van Capelleveen J.C., Geary R.S., Xia S., Tami J.A., Yu R.Z., Marcovina S.M., Hughes S.G., Graham M.J., Crooke R.M. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet. 2016;388:2239–2253. doi: 10.1016/S0140-6736(16)31009-1. [DOI] [PubMed] [Google Scholar]; Viney, N.J., van Capelleveen, J.C., Geary, R.S., Xia, S., Tami, J.A., Yu, R.Z., Marcovina, S.M., Hughes, S.G., Graham, M.J., Crooke, R.M. (2016). Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 388, 2239-2253. [DOI] [PubMed]
- 20.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]; Janssen, H.L.A., Reesink, H.W., Lawitz, E.J., Zeuzem, S., Rodriguez-Torres, M., Patel, K., van der Meer, A.J., Patick, A.K., Chen, A., Zhou, Y., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685-1694. [DOI] [PubMed]
- 21.Schluep T., Lickliter J., Hamilton J., Lewis D.L., Lai C.-L., Lau J.Y., Locarnini S.A., Gish R.G., Given B.D. Safety, Tolerability, and Pharmacokinetics of ARC-520 Injection, an RNA Interference-Based Therapeutic for the Treatment of Chronic Hepatitis B Virus Infection, in Healthy Volunteers. Clin. Pharmacol. Drug Dev. 2017;6:350–362. doi: 10.1002/cpdd.318. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schluep, T., Lickliter, J., Hamilton, J., Lewis, D.L., Lai, C.-L., Lau, J.Y., Locarnini, S.A., Gish, R.G., and Given, B.D. (2017). Safety, Tolerability, and Pharmacokinetics of ARC-520 Injection, an RNA Interference-Based Therapeutic for the Treatment of Chronic Hepatitis B Virus Infection, in Healthy Volunteers. Clin. Pharmacol. Drug Dev. 6, 350-362. [DOI] [PMC free article] [PubMed]
- 22.Dias N., Stein C.A. Potential roles of antisense oligonucleotides in cancer therapy. The example of Bcl-2 antisense oligonucleotides. Eur. J. Pharm. Biopharm. 2002;54:263–269. doi: 10.1016/s0939-6411(02)00060-7. [DOI] [PubMed] [Google Scholar]; Dias, N., and Stein, C.A. (2002). Potential roles of antisense oligonucleotides in cancer therapy. The example of Bcl-2 antisense oligonucleotides. Eur. J. Pharm. Biopharm. 54, 263-269. [DOI] [PubMed]
- 23.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]; Eckstein, F. (2014). Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 24, 374-387. [DOI] [PubMed]
- 24.Hagedorn P.H., Persson R., Funder E.D., Albæk N., Diemer S.L., Hansen D.J., Møller M.R., Papargyri N., Christiansen H., Hansen B.R. Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov. Today. 2018;23:101–114. doi: 10.1016/j.drudis.2017.09.018. [DOI] [PubMed] [Google Scholar]; Hagedorn, P.H., Persson, R., Funder, E.D., Albæk, N., Diemer, S.L., Hansen, D.J., Moller, M.R., Papargyri, N., Christiansen, H., Hansen, B.R., et al. (2018). Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov. Today 23, 101-114. [DOI] [PubMed]
- 25.Frieden M., Ørum H. Locked nucleic acid holds promise in the treatment of cancer. Curr. Pharm. Des. 2008;14:1138–1142. doi: 10.2174/138161208784246234. [DOI] [PubMed] [Google Scholar]; Frieden, M., and Orum, H. (2008). Locked nucleic acid holds promise in the treatment of cancer. Curr. Pharm. Des. 14, 1138-1142. [DOI] [PubMed]
- 26.Stein C.A., Hansen J.B., Lai J., Wu S., Voskresenskiy A., Høg A., Worm J., Hedtjärn M., Souleimanian N., Miller P. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stein, C.A., Hansen, J.B., Lai, J., Wu, S., Voskresenskiy, A., Hog, A., Worm, J., Hedtjarn, M., Souleimanian, N., Miller, P., et al. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 38, e3. [DOI] [PMC free article] [PubMed]
- 27.Jaschinski F., Korhonen H., Janicot M. Design and Selection of Antisense Oligonucleotides Targeting Transforming Growth Factor Beta (TGF-β) Isoform mRNAs for the Treatment of Solid Tumors. Methods Mol. Biol. 2015;1317:137–151. doi: 10.1007/978-1-4939-2727-2_9. [DOI] [PubMed] [Google Scholar]; Jaschinski, F., Korhonen, H., and Janicot, M. (2015). Design and Selection of Antisense Oligonucleotides Targeting Transforming Growth Factor Beta (TGF-β) Isoform mRNAs for the Treatment of Solid Tumors. Methods Mol. Biol. 1317, 137-151. [DOI] [PubMed]
- 28.Vollmer J., Jepsen J.S., Uhlmann E., Schetter C., Jurk M., Wader T., Wüllner M., Krieg A.M. Modulation of CpG Oligodeoxynucleotide-Mediated Immune Stimulation by Locked Nucleic Acid. Oligonucleotides. 2004;14:23–31. doi: 10.1089/154545704322988021. [DOI] [PubMed] [Google Scholar]; Vollmer, J., Jepsen, J.S., Uhlmann, E., Schetter, C., Jurk, M., Wader, T., Wullner, M., and Krieg, A.M. (2004). Modulation of CpG Oligodeoxynucleotide-Mediated Immune Stimulation by Locked Nucleic Acid. Oligonucleotides 14, 23-31. [DOI] [PubMed]
- 29.Suntharalingam G., Perry M.R., Ward S., Brett S.J., Castello-Cortes A., Brunner M.D., Panoskaltsis N. Cytokine Storm in a Phase 1 Trial of the Anti-CD28 Monoclonal Antibody TGN1412. N. Engl. J. Med. 2006;355:1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]; Suntharalingam, G., Perry, M.R., Ward, S., Brett, S.J., Castello-Cortes, A., Brunner, M.D., and Panoskaltsis, N. (2006). Cytokine Storm in a Phase 1 Trial of the Anti-CD28 Monoclonal Antibody TGN1412. N. Engl. J. Med. 355, 1018-1028. [DOI] [PubMed]
- 30.Coch C., Lück C., Schwickart A., Putschli B., Renn M., Höller T., Barchet W., Hartmann G., Schlee M. A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS ONE. 2013;8:e71057. doi: 10.1371/journal.pone.0071057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Coch, C., Luck, C., Schwickart, A., Putschli, B., Renn, M., Holler, T., Barchet, W., Hartmann, G., and Schlee, M. (2013). A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS ONE 8, e71057. [DOI] [PMC free article] [PubMed]
- 31.Ren T., Wen Z.-K., Liu Z.-M., Qian C., Liang Y.-J., Jin M.-L., Cai Y.Y., Xu L. Targeting toll-like receptor 9 with CpG oligodeoxynucleotides enhances anti-tumor responses of peripheral blood mononuclear cells from human lung cancer patients. Cancer Invest. 2008;26:448–455. doi: 10.1080/07357900701681608. [DOI] [PubMed] [Google Scholar]; Ren, T., Wen, Z.-K., Liu, Z.-M., Qian, C., Liang, Y.-J., Jin, M.-L., Cai, Y.Y., and Xu, L. (2008). Targeting toll-like receptor 9 with CpG oligodeoxynucleotides enhances anti-tumor responses of peripheral blood mononuclear cells from human lung cancer patients. Cancer Invest. 26, 448-455. [DOI] [PubMed]
- 32.Vetter P., Fischer W.A., 2nd, Schibler M., Jacobs M., Bausch D.G., Kaiser L. Ebola Virus Shedding and Transmission: Review of Current Evidence. J. Infect. Dis. 2016;214(Suppl 3):S177–S184. doi: 10.1093/infdis/jiw254. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vetter, P., Fischer, W.A., 2nd, Schibler, M., Jacobs, M., Bausch, D.G., and Kaiser, L. (2016). Ebola Virus Shedding and Transmission: Review of Current Evidence. J. Infect. Dis. 214 (Suppl 3), S177-S184. [DOI] [PMC free article] [PubMed]
- 33.Easton V., McPhillie M., Garcia-Dorival I., Barr J.N., Edwards T.A., Foster R., Fishwick C., Harris M. Identification of a small molecule inhibitor of Ebola virus genome replication and transcription using in silico screening. Antiviral Res. 2018;156:46–54. doi: 10.1016/j.antiviral.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Easton, V., McPhillie, M., Garcia-Dorival, I., Barr, J.N., Edwards, T.A., Foster, R., Fishwick, C., and Harris, M. (2018). Identification of a small molecule inhibitor of Ebola virus genome replication and transcription using in silico screening. Antiviral Res. 156, 46-54. [DOI] [PMC free article] [PubMed]
- 34.Kadanali A. Vol. 2. North. Clin. Istanbul; 2015. . pp. 81–86. (An overview of Ebola virus disease). [DOI] [PMC free article] [PubMed] [Google Scholar]; Kadanali, A. (2015). An overview of Ebola virus disease. North. Clin. Istanbul. 2, 81-86. [DOI] [PMC free article] [PubMed]
- 35.Bray M., Paragas J. Experimental therapy of filovirus infections. Antiviral Res. 2002;54:1–17. doi: 10.1016/s0166-3542(02)00005-0. [DOI] [PubMed] [Google Scholar]; Bray, M., and Paragas, J. (2002). Experimental therapy of filovirus infections. Antiviral Res. 54, 1-17. [DOI] [PubMed]
- 36.Feldmann H., Jones S.M., Schnittler H.-J., Geisbert T. Therapy and prophylaxis of Ebola virus infections. Curr. Opin. Investig. Drugs. 2005;6:823–830. [PubMed] [Google Scholar]; Feldmann, H., Jones, S.M., Schnittler, H.-J., and Geisbert, T. (2005). Therapy and prophylaxis of Ebola virus infections. Curr. Opin. Investig. Drugs 6, 823-830. [PubMed]
- 37.Mühlberger E. Filovirus replication and transcription. Future Virol. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]; Muhlberger, E. (2007). Filovirus replication and transcription. Future Virol. 2, 205-215. [DOI] [PMC free article] [PubMed]
- 38.Kitson J.D.A., Kamola P.J., Kane L. Hybridization-Dependent Effects: The Prediction, Evaluation, and Consequences of Unintended Target Hybridization. In: Ferrari N., Seguin R., editors. Oligonucleotide-Based Drugs and Therapeutics. John Wiley & Sons; 2018. pp. 191–225. [Google Scholar]; Kitson, J.D.A., Kamola, P.J., and Kane, L. (2018). Hybridization-Dependent Effects: The Prediction, Evaluation, and Consequences of Unintended Target Hybridization. In Oligonucleotide-Based Drugs and Therapeutics, N. Ferrari and R. Seguin, eds. (John Wiley & Sons), pp. 191-225.
- 39.Crooke S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017;27:70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]; Crooke, S.T. (2017). Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 27, 70-77. [DOI] [PMC free article] [PubMed]
- 40.Castanotto D., Lin M., Kowolik C., Wang L., Ren X.-Q., Soifer H.S., Koch T., Hansen B.R., Oerum H., Armstrong B. A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 2015;43:9350–9361. doi: 10.1093/nar/gkv964. [DOI] [PMC free article] [PubMed] [Google Scholar]; Castanotto, D., Lin, M., Kowolik, C., Wang, L., Ren, X.-Q., Soifer, H.S., Koch, T., Hansen, B.R., Oerum, H., Armstrong, B., et al. (2015). A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 43, 9350-9361. [DOI] [PMC free article] [PubMed]
- 41.Tse M.T. Nuclear RNA more susceptible to knockdown: Antisense therapeutics. Nat. Rev. Drug Discov. 2012;11:674. doi: 10.1038/nrd3825. [DOI] [PubMed] [Google Scholar]; Tse, M.T. (2012). Nuclear RNA more susceptible to knockdown: Antisense therapeutics. Nat. Rev. Drug Discov. 11, 674. [DOI] [PubMed]
- 42.Chery J., Petri A., Wagschal A., Lim S.-Y., Cunningham J., Vasudevan S., Kauppinen S., Näär A.M. Development of Locked Nucleic Acid Antisense Oligonucleotides Targeting Ebola Viral Proteins and Host Factor Niemann-Pick C1. Nucleic Acid Ther. 2018;28:273–284. doi: 10.1089/nat.2018.0722. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chery, J., Petri, A., Wagschal, A., Lim, S.-Y., Cunningham, J., Vasudevan, S., Kauppinen, S., and Naar, A. M. (2018). Development of Locked Nucleic Acid Antisense Oligonucleotides Targeting Ebola Viral Proteins and Host Factor Niemann-Pick C1. Nucleic Acid Ther. 28, 273-284. [DOI] [PMC free article] [PubMed]
- 43.Nyakatura E.K., Frei J.C., Lai J.R. Chemical and Structural Aspects of Ebola Virus Entry Inhibitors. ACS Infect. Dis. 2015;1:42–52. doi: 10.1021/id500025n. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nyakatura, E.K., Frei, J.C., and Lai, J.R. (2015). Chemical and Structural Aspects of Ebola Virus Entry Inhibitors. ACS Infect. Dis. 1, 42-52. [DOI] [PMC free article] [PubMed]
- 44.Amritraj A., Wang Y., Revett T.J., Vergote D., Westaway D., Kar S. Role of cathepsin D in U18666A-induced neuronal cell death: potential implication in Niemann-Pick type C disease pathogenesis. J. Biol. Chem. 2013;288:3136–3152. doi: 10.1074/jbc.M112.412460. [DOI] [PMC free article] [PubMed] [Google Scholar]; Amritraj, A., Wang, Y., Revett, T.J., Vergote, D., Westaway, D., and Kar, S. (2013). Role of cathepsin D in U18666A-induced neuronal cell death: potential implication in Niemann-Pick type C disease pathogenesis. J. Biol. Chem. 288, 3136-3152. [DOI] [PMC free article] [PubMed]
- 45.Dieckmann A., Hagedorn P.H., Burki Y., Brügmann C., Berrera M., Ebeling M., Singer T., Schuler F. A Sensitive In Vitro Approach to Assess the Hybridization-Dependent Toxic Potential of High Affinity Gapmer Oligonucleotides. Mol. Ther. Nucleic Acids. 2018;10:45–54. doi: 10.1016/j.omtn.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dieckmann, A., Hagedorn, P.H., Burki, Y., Brugmann, C., Berrera, M., Ebeling, M., Singer, T., and Schuler, F. (2018). A Sensitive In Vitro Approach to Assess the Hybridization-Dependent Toxic Potential of High Affinity Gapmer Oligonucleotides. Mol. Ther. Nucleic Acids 10: 45-54. [DOI] [PMC free article] [PubMed]
- 46.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]; Geary, R.S., Norris, D., Yu, R., and Bennett, C.F. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 87, 46-51. [DOI] [PubMed]
- 47.Stein C.A., Castanotto D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stein, C.A., and Castanotto, D. (2017). FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 25, 1069-1075. [DOI] [PMC free article] [PubMed]
- 48.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shen, X., and Corey, D.R. (2018). Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584-1600. [DOI] [PMC free article] [PubMed]
- 49.Juliano R.L. Intracellular Trafficking and Endosomal Release of Oligonucleotides: What We Know and What We Don’t. Nucleic Acid Ther. 2018;28:166–177. doi: 10.1089/nat.2018.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]; Juliano, R.L. (2018). Intracellular Trafficking and Endosomal Release of Oligonucleotides: What We Know and What We Don’t. Nucleic Acid Ther. 28, 166-177. [DOI] [PMC free article] [PubMed]
- 50.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.-H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]; Crooke, S.T., Wang, S., Vickers, T.A., Shen, W., and Liang, X.-H. (2017). Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 35, 230-237. [DOI] [PubMed]
- 51.Lindow M., Vornlocher H.-P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]; Lindow, M., Vornlocher, H.-P., Riley, D., Kornbrust, D.J., Burchard, J., Whiteley, L.O., Kamens, J., Thompson, J.D., Nochur, S., Younis, H., et al. (2012). Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 30, 920-923. [DOI] [PubMed]
- 52.Javanbakht H., Mueller H., Walther J., Zhou X., Lopez A., Pattupara T., Blaising J., Pedersen L., Albæk N., Jackerott M. Liver-Targeted Anti-HBV Single-Stranded Oligonucleotides with Locked Nucleic Acid Potently Reduce HBV Gene Expression In Vivo. Mol. Ther. Nucleic Acids. 2018;11:441–454. doi: 10.1016/j.omtn.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Javanbakht, H., Mueller, H., Walther, J., Zhou, X., Lopez, A., Pattupara, T., Blaising, J., Pedersen, L., Albæk, N., Jackerott, M., et al. (2018). Liver-Targeted Anti-HBV Single-Stranded Oligonucleotides with Locked Nucleic Acid Potently Reduce HBV Gene Expression In Vivo. Mol. Ther. Nucleic Acids 11, 441-454. [DOI] [PMC free article] [PubMed]
- 53.Branda R.F., Moore A.L., Mathews L., McCormack J.J., Zon G. Immune stimulation by an antisense oligomer complementary to the rev gene of HIV-1. Biochem. Pharmacol. 1993;45:2037–2043. doi: 10.1016/0006-2952(93)90014-n. [DOI] [PubMed] [Google Scholar]; Branda, R.F., Moore, A.L., Mathews, L., McCormack, J.J., and Zon, G. (1993). Immune stimulation by an antisense oligomer complementary to the rev gene of HIV-1. Biochem. Pharmacol. 45, 2037-2043. [DOI] [PubMed]
- 54.Pisetsky D.S., Reich C.F. Stimulation of murine lymphocyte proliferation by a phosphorothioate oligonucleotide with antisense activity for herpes simplex virus. Life Sci. 1994;54:101–107. doi: 10.1016/0024-3205(94)00780-2. [DOI] [PubMed] [Google Scholar]; Pisetsky, D.S., and Reich, C.F. (1994). Stimulation of murine lymphocyte proliferation by a phosphorothioate oligonucleotide with antisense activity for herpes simplex virus. Life Sci. 54, 101-107. [DOI] [PubMed]
- 55.Zhao Q., Temsamani J., Iadarola P.L., Jiang Z., Agrawal S. Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem. Pharmacol. 1996;51:173–182. doi: 10.1016/0006-2952(95)02177-9. [DOI] [PubMed] [Google Scholar]; Zhao, Q., Temsamani, J., Iadarola, P.L., Jiang, Z., and Agrawal, S. (1996). Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem. Pharmacol. 51, 173-182. [DOI] [PubMed]
- 56.Weiner G.J., Liu H.M., Wooldridge J.E., Dahle C.E., Krieg A.M. Immunostimulatory oligodeoxynucleotides containing the CpG motif are effective as immune adjuvants in tumor antigen immunization. Proc. Natl. Acad. Sci. USA. 1997;94:10833–10837. doi: 10.1073/pnas.94.20.10833. [DOI] [PMC free article] [PubMed] [Google Scholar]; Weiner, G.J., Liu, H.M., Wooldridge, J.E., Dahle, C.E., and Krieg, A.M. (1997). Immunostimulatory oligodeoxynucleotides containing the CpG motif are effective as immune adjuvants in tumor antigen immunization. Proc. Natl. Acad. Sci. USA 94, 10833-10837. [DOI] [PMC free article] [PubMed]
- 57.Sanjaya A., Elder J.R., Shah D.H. Identification of new CpG oligodeoxynucleotide motifs that induce expression of interleukin-1β and nitric oxide in avian macrophages. Vet. Immunol. Immunopathol. 2017;192:1–7. doi: 10.1016/j.vetimm.2017.08.005. [DOI] [PubMed] [Google Scholar]; Sanjaya, A., Elder, J.R., and Shah, D.H. (2017). Identification of new CpG oligodeoxynucleotide motifs that induce expression of interleukin-1β and nitric oxide in avian macrophages. Vet. Immunol. Immunopathol. 192, 1-7. [DOI] [PubMed]
- 58.Vollmer J., Weeratna R.D., Jurk M., Samulowitz U., McCluskie M.J., Payette P., Davis H.L., Schetter C., Krieg A.M. Oligodeoxynucleotides lacking CpG dinucleotides mediate Toll-like receptor 9 dependent T helper type 2 biased immune stimulation. Immunology. 2004;113:212–223. doi: 10.1111/j.1365-2567.2004.01962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vollmer, J., Weeratna, R.D., Jurk, M., Samulowitz, U., McCluskie, M.J., Payette, P., Davis, H.L., Schetter, C., and Krieg, A.M. (2004). Oligodeoxynucleotides lacking CpG dinucleotides mediate Toll-like receptor 9 dependent T helper type 2 biased immune stimulation. Immunology 113, 212-223. [DOI] [PMC free article] [PubMed]
- 59.Roberts T.L., Sweet M.J., Hume D.A., Stacey K.J. Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J. Immunol. 2005;174:605–608. doi: 10.4049/jimmunol.174.2.605. [DOI] [PubMed] [Google Scholar]; Roberts, T.L., Sweet, M.J., Hume, D.A., and Stacey, K.J. (2005). Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J. Immunol. 174, 605-608. [DOI] [PubMed]
- 60.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]; Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., and Akira, S. (2000). A Toll-like receptor recognizes bacterial DNA. Nature 408, 740-745. [DOI] [PubMed]
- 61.Barchet W., Wimmenauer V., Schlee M., Hartmann G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr. Opin. Immunol. 2008;20:389–395. doi: 10.1016/j.coi.2008.07.007. [DOI] [PubMed] [Google Scholar]; Barchet, W., Wimmenauer, V., Schlee, M., and Hartmann, G. (2008). Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr. Opin. Immunol. 20, 389-395. [DOI] [PubMed]
- 62.Bauer S., Kirschning C.J., Häcker H., Redecke V., Hausmann S., Akira S., Wagner H., Lipford G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bauer, S., Kirschning, C.J., Hacker, H., Redecke, V., Hausmann, S., Akira, S., Wagner, H., and Lipford, G.B. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 98, 9237-9242. [DOI] [PMC free article] [PubMed]
- 63.Kadowaki N., Ho S., Antonenko S., Malefyt R.W., Kastelein R.A., Bazan F., Liu Y.J. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kadowaki, N., Ho, S., Antonenko, S., Malefyt, R.W., Kastelein, R.A., Bazan, F., and Liu, Y.J. (2001). Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 194, 863-869. [DOI] [PMC free article] [PubMed]
- 64.Krieg A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]; Krieg, A.M. (2002). CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20, 709-760. [DOI] [PubMed]
- 65.Uhlmann E., Vollmer J. Recent advances in the development of immunostimulatory oligonucleotides. Curr. Opin. Drug Discov. Devel. 2003;6:204–217. [PubMed] [Google Scholar]; Uhlmann, E., and Vollmer, J. (2003). Recent advances in the development of immunostimulatory oligonucleotides. Curr. Opin. Drug Discov. Devel. 6, 204-217. [PubMed]
- 66.Olejnik J., Forero A., Deflubé L.R., Hume A.J., Manhart W.A., Nishida A., Marzi A., Katze M.G., Ebihara H., Rasmussen A.L. Ebolaviruses Associated with Differential Pathogenicity Induce Distinct Host Responses in Human Macrophages. J. Virol. 2017;91 doi: 10.1128/JVI.00179-17. e00179–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Olejnik, J., Forero, A., Deflube, L.R., Hume, A.J., Manhart, W.A., Nishida, A., Marzi, A., Katze, M.G., Ebihara, H., Rasmussen, A.L., et al. (2017). Ebolaviruses Associated with Differential Pathogenicity Induce Distinct Host Responses in Human Macrophages. J. Virol. 91, e00179-17. [DOI] [PMC free article] [PubMed]
- 67.Geisbert T.W., Hensley L.E., Larsen T., Young H.A., Reed D.S., Geisbert J.B., Scott D.P., Kagan E., Jahrling P.B., Davis K.J. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Geisbert, T.W., Hensley, L.E., Larsen, T., Young, H.A., Reed, D.S., Geisbert, J.B., Scott, D.P., Kagan, E., Jahrling, P.B., and Davis, K.J. (2003). Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 163, 2347-2370. [DOI] [PMC free article] [PubMed]
- 68.Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. The Pathogenesis of Ebola Virus Disease. Annu. Rev. Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]; Baseler, L., Chertow, D.S., Johnson, K.M., Feldmann, H., and Morens, D.M. (2017). The Pathogenesis of Ebola Virus Disease. Annu. Rev. Pathol. 12, 387-418. [DOI] [PubMed]
- 69.Dahlmann F., Biedenkopf N., Babler A., Jahnen-Dechent W., Karsten C.B., Gnirß K., Schneider H., Wrensch F., O’Callaghan C.A., Bertram S. Analysis of Ebola Virus Entry Into Macrophages. J. Infect. Dis. 2015;212(Suppl 2):S247–S257. doi: 10.1093/infdis/jiv140. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dahlmann, F., Biedenkopf, N., Babler, A., Jahnen-Dechent, W., Karsten, C.B., Gnirß, K., Schneider, H., Wrensch, F., O’Callaghan, C.A., Bertram, S., et al. (2015). Analysis of Ebola Virus Entry Into Macrophages. J. Infect. Dis. 212 (Suppl 2), S247-S257. [DOI] [PMC free article] [PubMed]
- 70.Agrawal S., Tang J.Y., Brown D.M. Analytical study of phosphorothioate analogues of oligodeoxynucleotides using high-performance liquid chromatography. J. Chromatogr. A. 1990;509:396–399. doi: 10.1016/s0021-9673(01)93098-5. [DOI] [PubMed] [Google Scholar]; Agrawal, S., Tang, J.Y., and Brown, D.M. (1990). Analytical study of phosphorothioate analogues of oligodeoxynucleotides using high-performance liquid chromatography. J. Chromatogr. A 509, 396-399. [DOI] [PubMed]
- 71.Vaillant A., Juteau J.-M., Lu H., Liu S., Lackman-Smith C., Ptak R., Jiang S. Phosphorothioate oligonucleotides inhibit human immunodeficiency virus type 1 fusion by blocking gp41 core formation. Antimicrob. Agents Chemother. 2006;50:1393–1401. doi: 10.1128/AAC.50.4.1393-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vaillant, A., Juteau, J.-M., Lu, H., Liu, S., Lackman-Smith, C., Ptak, R., and Jiang, S. (2006). Phosphorothioate oligonucleotides inhibit human immunodeficiency virus type 1 fusion by blocking gp41 core formation. Antimicrob. Agents Chemother. 50, 1393-1401. [DOI] [PMC free article] [PubMed]
- 72.Vaillant A. Nucleic acid polymers: Broad spectrum antiviral activity, antiviral mechanisms and optimization for the treatment of hepatitis B and hepatitis D infection. Antiviral Res. 2016;133:32–40. doi: 10.1016/j.antiviral.2016.07.004. [DOI] [PubMed] [Google Scholar]; Vaillant, A. (2016). Nucleic acid polymers: Broad spectrum antiviral activity, antiviral mechanisms and optimization for the treatment of hepatitis B and hepatitis D infection. Antiviral Res. 133, 32-40. [DOI] [PubMed]
- 73.Abe T., Mizuta T., Hatta T., Miyano-Kurosaki N., Fujiwara M., Takai K., Shigeta S., Yokota T., Takaku H. Antisense therapy of influenza. Eur. J. Pharm. Sci. 2001;13:61–69. doi: 10.1016/s0928-0987(00)00208-6. [DOI] [PubMed] [Google Scholar]; Abe, T., Mizuta, T., Hatta, T., Miyano-Kurosaki, N., Fujiwara, M., Takai, K., Shigeta, S., Yokota, T., and Takaku, H. (2001). Antisense therapy of influenza. Eur. J. Pharm. Sci. 13, 61-69. [DOI] [PubMed]
- 74.Guzman E.M., Cheshenko N., Shende V., Keller M.J., Goyette N., Juteau J.-M., Boivin G., Vaillant A., Herold B.C. Amphipathic DNA polymers are candidate vaginal microbicides and block herpes simplex virus binding, entry and viral gene expression. Antivir. Ther. (Lond.) 2007;12:1147–1156. [PubMed] [Google Scholar]; Guzman, E.M., Cheshenko, N., Shende, V., Keller, M.J., Goyette, N., Juteau, J.-M., Boivin, G., Vaillant, A., and Herold, B.C. (2007). Amphipathic DNA polymers are candidate vaginal microbicides and block herpes simplex virus binding, entry and viral gene expression. Antivir. Ther. (Lond.) 12, 1147-1156. [PubMed]
- 75.Lee A.M., Rojek J.M., Gundersen A., Ströher U., Juteau J.-M., Vaillant A., Kunz S. Inhibition of cellular entry of lymphocytic choriomeningitis virus by amphipathic DNA polymers. Virology. 2008;372:107–117. doi: 10.1016/j.virol.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, A.M., Rojek, J.M., Gundersen, A., Stroher, U., Juteau, J.-M., Vaillant, A., and Kunz, S. (2008). Inhibition of cellular entry of lymphocytic choriomeningitis virus by amphipathic DNA polymers. Virology 372, 107-117. [DOI] [PMC free article] [PubMed]
- 76.Bernstein D.I., Goyette N., Cardin R., Kern E.R., Boivin G., Ireland J., Juteau J.M., Vaillant A. Amphipathic DNA polymers exhibit antiherpetic activity in vitro and in vivo. Antimicrob. Agents Chemother. 2008;52:2727–2733. doi: 10.1128/AAC.00279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bernstein, D.I., Goyette, N., Cardin, R., Kern, E.R., Boivin, G., Ireland, J., Juteau, J.M., and Vaillant, A. (2008). Amphipathic DNA polymers exhibit antiherpetic activity in vitro and in vivo. Antimicrob. Agents Chemother. 52, 2727-2733. [DOI] [PMC free article] [PubMed]
- 77.Cardin R.D., Bravo F.J., Sewell A.P., Cummins J., Flamand L., Juteau J.-M., Bernstein D.I., Vaillant A. Amphipathic DNA polymers exhibit antiviral activity against systemic murine Cytomegalovirus infection. Virol. J. 2009;6:214. doi: 10.1186/1743-422X-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cardin, R.D., Bravo, F.J., Sewell, A.P., Cummins, J., Flamand, L., Juteau, J.-M., Bernstein, D.I., and Vaillant, A. (2009). Amphipathic DNA polymers exhibit antiviral activity against systemic murine Cytomegalovirus infection. Virol. J. 6, 214. [DOI] [PMC free article] [PubMed]
- 78.Beniac D.R., Booth T.F. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci. Rep. 2017;7:46374. doi: 10.1038/srep46374. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beniac, D.R., and Booth, T.F. (2017). Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 A resolution. Sci. Rep. 7, 46374. [DOI] [PMC free article] [PubMed]
- 79.Prüfer K., Stenzel U., Dannemann M., Green R.E., Lachmann M., Kelso J. PatMaN: rapid alignment of short sequences to large databases. Bioinformatics. 2008;24:1530–1531. doi: 10.1093/bioinformatics/btn223. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prufer, K., Stenzel, U., Dannemann, M., Green, R.E., Lachmann, M., and Kelso, J. (2008). PatMaN: rapid alignment of short sequences to large databases. Bioinformatics 24, 1530-1531. [DOI] [PMC free article] [PubMed]
- 80.Tolstrup N., Nielsen P.S., Kolberg J.G., Frankel A.M., Vissing H., Kauppinen S. OligoDesign: Optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tolstrup, N., Nielsen, P.S., Kolberg, J.G., Frankel, A.M., Vissing, H., and Kauppinen, S. (2003). OligoDesign: Optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 31, 3758-3762. [DOI] [PMC free article] [PubMed]
- 81.Burdick A.D., Sciabola S., Mantena S.R., Hollingshead B.D., Stanton R., Warneke J.A., Zeng M., Martsen E., Medvedev A., Makarov S.S. Sequence motifs associated with hepatotoxicity of locked nucleic acid: modified antisense oligonucleotides. Nucleic Acids Res. 2014;42:4882–4891. doi: 10.1093/nar/gku142. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burdick, A.D., Sciabola, S., Mantena, S.R., Hollingshead, B.D., Stanton, R., Warneke, J.A., Zeng, M., Martsen, E., Medvedev, A., Makarov, S.S., et al. (2014). Sequence motifs associated with hepatotoxicity of locked nucleic acid: modified antisense oligonucleotides. Nucleic Acids Res. 42, 4882-4891. [DOI] [PMC free article] [PubMed]
- 82.Gibb T.R., Norwood D.A., Jr., Woollen N., Henchal E.A. Development and evaluation of a fluorogenic 5′ nuclease assay to detect and differentiate between Ebola virus subtypes Zaire and Sudan. J. Clin. Microbiol. 2001;39:4125–4130. doi: 10.1128/JCM.39.11.4125-4130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gibb, T.R., Norwood, D.A., Jr., Woollen, N., and Henchal, E.A. (2001). Development and evaluation of a fluorogenic 5′ nuclease assay to detect and differentiate between Ebola virus subtypes Zaire and Sudan. J. Clin. Microbiol. 39, 4125-4130. [DOI] [PMC free article] [PubMed]
- 83.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]; Schmittgen, T.D., and Livak, K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101-1108. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.