Abstract
Aims
Fibroblast growth factor 21 (FGF21) is closely linked with metabolic disorders including diabetes. Evidences suggested that FGF21 may play a protective role against diabetic kidney disease (DKD). However, the relationship between genetic variants in the FGF21 gene region and DKD remains unknown. In our study, we aimed to investigate the association of genetic variations in this gene region with DKD and DKD-related clinical traits.
Materials and Methods
We recruited 1340 Han Chinese participants with type 2 diabetes, including 596 DKD patients and 744 patients who was diagnosed with diabetes for more than 5 years but did not progress to DKD. Three single-nucleotide polymorphisms were selected (rs2071699, rs838136, and rs499765) and genotyped. The association between these SNPs and DKD as well as DKD-related quantitative traits was analyzed.
Results
We did not find any significant association between these SNPs and susceptibility to DKD in the present study. However, a significant association with estimated glomerular filtration rate (eGFR) was detected: in the non-DKD group, rs838136 was significantly associated with eGFR under an additive model (β = 0.013 ± 0.006, P = 0.0295, β was calculated for log10eGFR) as well as a recessive model (P = 0.0385) and rs499765 was associated with eGFR under a dominant model (P = 0.0411) and in the DKD group, rs499765 showed a trend toward association with eGFR under an additive model (β = −0.022 ± 0.012, P = 0.0820, β was calculated for log10eGFR) and showed a significant association with eGFR under a dominant model (P = 0.0182).
Conclusions
Our findings indicated that genetic variations adjacent to FGF21 were associated with eGFR in Chinese diabetic patients.
1. Introduction
Diabetes has become one of the most common chronic diseases in the world, and the prevalence of the disease has experienced an exploding increase in recent years, especially in the Chinese population [1]. As an important microvascular complication of diabetes, diabetic kidney disease (DKD) is the most frequent cause of end-stage renal disease (EASD) in different countries [2]. A genetic basis has been suggested for this disease according to the evidence of familial clustering of DKD [3] and higher risks of DKD in certain racial groups [4]. Thus, investigation of the genetic profile of DKD could reveal the underlying mechanism of the disease.
FGF21 is an endocrine member of the fibroblast growth factor (FGF) gene family, predominantly expressed in the liver. It exerts beneficial metabolic effects in animal models, such as reducing blood glucose and triglyceride levels, increasing insulin sensitivity, suppressing hepatic glucose production, increasing expenditure, and losing weight [5–8]. So it is recognized as a novel metabolic regulator, and its therapeutic potential has also been suggested for the treatment of metabolic disorders including diabetes and obesity.
Recently, studies have suggested that FGF21 may play a role in the pathogenesis of DKD. Kim et al. revealed that daily administration of FGF21 in diabetic db/db mice markedly decreased urinary albumin excretion and mesangial expansion, suppressed profibrotic molecule synthesis, and improved renal lipid metabolism and oxidative stress injury [9]. Cheng et al. found that fenofibrate prevents the development of diabetic nephropathy in mice with type 1 diabetes via upregulating FGF21 and stimulating PI3K/Akt/GSK-3β/Fyn-mediated activation of the Nrf2 pathway [10]. Furthermore, a recent study compared the therapeutic effect of rhFGF21 and PEG-rhFGF21 on diabetic nephropathy in diet-induced obesity (DIO) mice, and the results showed that PEG-rhFGF21 was more efficacious in ameliorating functional and morphological abnormalities induced by diabetic nephropathy in db/db and DIO mice [11]. However, whether genetic variants in the FGF21 gene region relate to DKD is still unknown. So we aimed to evaluate the association of genetic variants in the FGF21 gene region with DKD and its related quantitative traits in this study.
2. Materials and Methods
2.1. Subjects
1340 unrelated Han Chinese participants were recruited from the Shanghai Diabetes Institute Inpatient Database of Shanghai Jiao Tong University Affiliated Sixth People's Hospital, including 596 DKD patients (DKD group) and 744 patients who was diagnosed with diabetes for more than 5 years but did not progress to DKD (non-DKD group). All subjects were diagnosed with type 2 diabetes according to the 1999 WHO criteria (https://www.who.int/entity/diabetes/currentpublications/en). Patients with estimated glomerular filtration rate (eGFR) < 90 mL/min per 1.73 m2, albuminuria ≥ 30 mg/24 h, or ACR (albumin-to-creatinine ratio) ≥ 30 μg/mg were diagnosed with DKD.
2.2. Clinical Measurements
Anthropometric and biochemical traits related to diabetes were collected in detail for each participant, including height, weight, blood pressure, fasting glucose, 2-hour glucose, glycated hemoglobin A1c (HbA1c), serum lipid profile, liver function, and renal function. The albuminuria level was measured using the samples from 24-hour urine collection, which was repeated in three consecutive days, and then the mean values were used for further analysis. eGFR was calculated by using a Chinese population-specific formula derived from the Modification of Diet in Renal Disease (MDRD) equation [12].
2.3. SNP Selection, Genotyping, and Quality Control Analysis
We chose three SNPs flanking the FGF21 gene on chromosome 19 (all within 8 kb) including rs2071699, rs838136, and rs499765. All these SNPs were genotyped using the MassARRAY Compact Analyzer (Sequenom, San Diego, CA, USA), with matrix-assisted laser desorption ionization time-of-flight mass spectroscopy detecting primer extension of multiplex products. All SNPs met the quality control thresholds with a genotyping call rate over 90%. The minor allele frequencies for rs2071699, rs499765, and rs838136 were 28.9%, 41.6%, and 39.3%, respectively.
2.4. Statistical Analysis
Allele frequencies were determined by gene counting. The Hardy-Weinberg equilibrium tests were then performed for all of the SNPs in the DKD group and the non-DKD group separately. The χ2 test was conducted to compare the allele frequencies of each SNP between the two groups, and multivariable logistic regression analysis was used to examine the differences of genotype distribution between the two groups, with adjustment for confounding factors. A generalized linear model was applied to test the effects of each SNP on quantitative traits, adjusting for the covariates. eGFR and 24-hour urinary albumin excretion rate (AER) were log10 transformed before linear regression because of their skewed distribution. All these analyses were conducted using SAS software (version 8.0; SAS Institute, Cary, NC, USA), with a two-tailed P value of <0.05 considered statistically significant.
3. Results
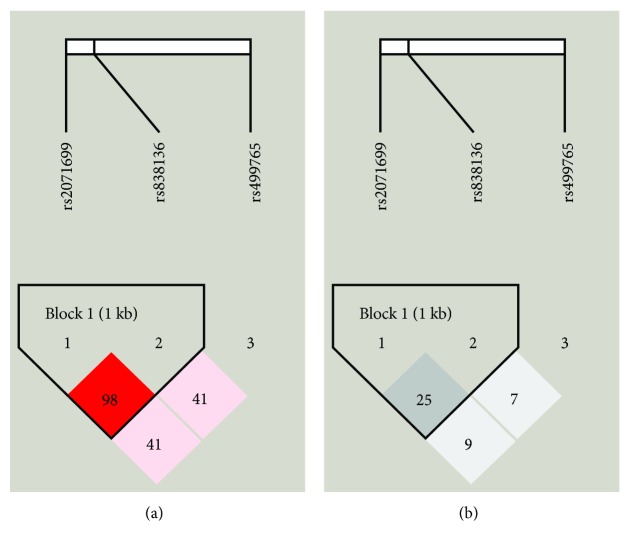
All of the SNPs conformed to the Hardy-Weinberg equilibrium (Table 1). The linkage disequilibrium pattern of these SNPs is shown in Figure 1. rs2071699 and rs838136 constructed a haplotype block (block 1) in this region with |D′|=0.98 and r2 = 0.25. The clinical characteristics of both groups are presented in Table 2. Significant differences were detected between the DKD group and non-DKD group with respect to gender, duration of diabetes, BMI, systolic blood pressure (SBP), diastolic blood pressure (DBP), HbA1c, AER, eGFR, uric acid, creatinine, blood urea nitrogen, total cholesterol, and total triglycerides (P < 0.05, Table 2).
Table 1.
Hardy-Weinberg equilibrium tests.
| Group | n | rs2071699 | rs499765 | rs838136 | |||
|---|---|---|---|---|---|---|---|
| CC/CT/TT | P value | GG/GC/CC | P value | AA/AG/GG | P value | ||
| DKD group | 596 | 290/239/42 | 0.4479 | 201/263/104 | 0.2716 | 199/282/93 | 0.6791 |
| Non-DKD group | 744 | 357/306/58 | 0.5000 | 244/349/124 | 0.9668 | 282/320/113 | 0.1650 |
Figure 1.
Linkage disequilibrium analyses for SNPs tested in the FGF21 gene region. (a) Shades of pink indicate the strength of pairwise linkage disequilibrium based on |D′|, and numbers represent |D′| which is expressed as a percentage. (b) Shades of grey indicate the strength of pairwise linkage disequilibrium based on r2, and numbers represent r2 which is expressed as a percentage.
Table 2.
Clinical characteristics of the study samples.
| Parameters | Total | Non-DKD group | DKD group | P value |
|---|---|---|---|---|
| n | 1340 | 744 | 596 | |
| Male/female | 637/703 | 304/440 | 333/263 | <0.0001 |
| Age (years) | 64 (55, 72) | 64 (56, 72) | 64 (54, 73) | 0.4578 |
| Duration of diabetes (years) | 10 (7, 14) | 10 (8, 14) | 9 (3, 14) | <0.0001 |
| BMI (kg/m2) | 24.3 (21.9, 26.7) | 23.7 (21.6, 26.2) | 24.9 (22.3, 27.2) | <0.0001 |
| SBP (mmHg) | 135 (120, 150) | 130 (120, 150) | 140 (130, 150) | <0.0001 |
| DBP (mmHg) | 80 (75, 90) | 80 (70, 85) | 80 (80, 90) | <0.0001 |
| HbA1c (%) | 8.7 (7.3, 10.3) | 8.3 (7.1, 10.0) | 9.1 (7.5, 10.9) | <0.0001 |
| AER (mg/24 h) | 21.0 (8.4, 77.2) | 9.2 (6.4, 14.5) | 89.3 (46.3, 288.8) | <0.0001 |
| eGFR (mL/min/1.73 m2) | 116.5 (94.4, 140.7) | 120.7 (102.8, 141.8) | 108.9 (81.2, 136.3) | <0.0001 |
| Uric acid (μmol/L) | 309 (250, 371) | 293 (241, 351) | 330 (271, 397) | <0.0001 |
| Creatinine (μmol/L) | 68 (56, 83) | 64 (54, 75) | 74 (59, 96) | <0.0001 |
| Blood urea nitrogen (mmol/L) | 5.8 (4.9, 7.1) | 5.7 (4.8, 6.8) | 6.1 (4.9, 7.8) | <0.0001 |
| TC (mmol/L) | 4.6 (4.1, 5.4) | 4.6 (4.0, 5.2) | 4.7 (4.1, 5.5) | 0.0003 |
| TG (mmol/L) | 1.4 (1.0, 2.1) | 1.3 (0.9, 1.9) | 1.6 (1.0, 2.3) | <0.0001 |
Data are shown as median (interquartile range) or mean ± SD. P value refers to the comparison of parameters between the DKD group and the non-DKD group. P values < 0.05 are shown in bold. DKD: diabetic kidney disease; BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; AER: 24-hour urinary albumin excretion rate; eGFR: estimated glomerular filtration rate; TC: total cholesterol; TG: total triglycerides.
We first evaluated the association between genetic variants and DKD susceptibility. As shown in Table 3, the allele frequencies of the three SNPs did not differ between the DKD group and the control group (P > 0.05). We also compared the genotype distribution of these SNPs between the two groups using logistic regression analysis with adjustment for age, gender, BMI, duration of diabetes, HbA1c, SBP, and DBP. However, no significant difference was found either (P > 0.05). Furthermore, there is no difference of haplotype distribution between the DKD and non-DKD groups (Table 4).
Table 3.
Associations between SNPs and DKD susceptibility in type 2 diabetic patients.
| SNP | Major/minor allele | Major allele frequency | P allele | OR (95% CI) | Genotype count | P genotype | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|
| DKD | Non-DKD | DKD AA/Aa/aa† |
Non-DKD AA/Aa/aa† |
||||||
| rs2071699 | C/T | 0.717 | 0.707 | 0.5845 | 1.049 (0.884, 1.245) | 290/239/42 | 357/306/58 | 0.5511 | 1.062 (0.871, 1.296) |
| rs499765 | G/C | 0.585 | 0.584 | 0.9306 | 1.007 (0.860, 1.179) | 201/263/104 | 244/349/124 | 0.7234 | 0.968 (0.811, 1.157) |
| rs838136 | A/G | 0.592 | 0.618 | 0.1818 | 0.897 (0.766, 1.052) | 199/282/93 | 282/320/113 | 0.4146 | 0.928 (0.776, 1.110) |
†AA = major allele homozygous; Aa = heterozygous; aa = minor allele homozygous. Pallele refers to the P value for the comparison of the allele frequency between the DKD group and the control group. Pgenotype refers to the P value for the comparison of genotype distribution between the DKD group and the control group, which was adjusted for age, gender, BMI, duration of diabetes, systolic blood pressure, diastolic blood pressure, and HbA1c. The ORs with 95% CIs shown are for the major alleles of the SNPs.
Table 4.
Haplotype analysis of block 1 in the study population.
| Haplotype frequency | P value | ||
|---|---|---|---|
| DKD group | Non-DKD group | ||
| CG | 0.405 | 0.380 | 0.1986 |
| CA | 0.312 | 0.326 | 0.4413 |
| TA | 0.280 | 0.291 | 0.5386 |
We then investigated the effects of SNPs on the quantitative traits related to DKD, including eGFR and AER. In the non-DKD group, rs838136 was significantly associated with eGFR under an additive model (β = 0.013 ± 0.006, P = 0.0295, Table 5) as well as a recessive model (P = 0.0385, Table 5) and rs499765 was associated with eGFR under a dominant model (P = 0.0411, Table 5). In the DKD group, rs499765 showed a trend toward association with eGFR under an additive model (β = −0.022 ± 0.012, P = 0.0820, Table 5) and showed a significant association with eGFR under a dominant model (P = 0.0182, Table 5). As for AER, no evidence of association was found for the SNPs in either group (Table 6).
Table 5.
Associations between the SNPs and eGFR (mL/min/1.73 m2) in the study subjects.
| Group | SNP | Major/minor allele | AA† | Aa† | aa† | Additive model | Dominant model | Recessive model | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE)‡ | P value | β (SE) | P value | β (SE) | P value | ||||||
| DKD group | rs2071699 | C/T | 109.6 (82.3, 136.3) | 106.0 (81.3, 134.0) | 120.6 (86.5, 144.5) | -0.006 (0.014) | 0.6515 | -0.001 (0.018) | 0.9337 | -0.032 (0.035) | 0.3517 |
| rs499765 | G/C | 105.6 (70.8, 134.7) | 112.0 (85.8, 137.4) | 109.9 (81.3, 135.2) | -0.022 (0.012) | 0.0820 | -0.044 (0.019) | 0.0182 | -0.007 (0.023) | 0.7539 | |
| rs838136 | A/G | 106.5 (81.5, 138.6) | 109.1 (81.5, 133.1) | 113.1 (89.4, 144.5) | -0.018 (0.013) | 0.1758 | -0.016 (0.019) | 0.3937 | -0.035 (0.024) | 0.1534 | |
|
| |||||||||||
| Non-DKD group | rs2071699 | C/T | 120.3 (101.5, 141.5) | 119.3 (102.9, 138.9) | 122.5 (109.5, 147.6) | -0.006 (0.007) | 0.3575 | -0.009 (0.009) | 0.3047 | -0.004 (0.016) | 0.7964 |
| rs499765 | G/C | 117.5 (102.1, 136.9) | 123.4 (103.5, 148.0) | 116.8 (103.5, 135.6) | -0.010 (0.006) | 0.1159 | -0.019 (0.009) | 0.0411 | -0.004 (0.012) | 0.7560 | |
| rs838136 | A/G | 121.7 (104.3, 146.4) | 120.7 (102.2, 141.1) | 114.9 (96.8, 136.6) | 0.013 (0.006) | 0.0295 | 0.014 (0.009) | 0.1108 | 0.024 (0.012) | 0.0385 | |
Data are shown as geometric means (SE). P values were adjusted for age, gender, BMI, duration of diabetes, systolic blood pressure, diastolic blood pressure, and HbA1c. P values < 0.05 are shown in bold. †AA = major allele homozygous; Aa = heterozygous; aa = minor allele homozygous. ‡β (SE) was calculated for log10eGFR and refers to major alleles. eGFR: estimated glomerular filtration rate.
Table 6.
Associations between the SNPs and AER (mg/24 h) in the study subjects.
| SNP | Major/minor allele | AA† | Aa† | aa† | β (SE)‡ | P value | |
|---|---|---|---|---|---|---|---|
| DKD group | rs2071699 | C/T | 100.6 (48.3, 318.7) | 82.2 (45.1, 283.4) | 78.5 (37.1, 262.0) | 0.030 (0.039) | 0.4389 |
| rs499765 | G/C | 90.0 (52.4, 334.3) | 89.1 (42.7, 289.0) | 89.8 (44.6, 267.5) | 0.030 (0.034) | 0.3782 | |
| rs838136 | A/G | 77.3 (43.0, 282.3) | 103.2 (50.1, 292.6) | 90.1 (46.1, 227.4) | -0.028 (0.035) | 0.4327 | |
|
| |||||||
| Non-DKD group | rs2071699 | C/T | 9.5 (6.5, 14.7) | 8.7 (6.0, 13.8) | 9.4 (6.8, 13.6) | 0.014 (0.014) | 0.3311 |
| rs499765 | G/C | 9.7 (6.1, 14.1) | 9.0 (6.4, 14.7) | 8.5 (6.5, 14.1) | 0.0006 (0.013) | 0.9632 | |
| rs838136 | A/G | 8.9 (6.2, 13.8) | 8.8 (6.2, 14.9) | 10.0 (6.9, 14.6) | -0.007 (0.013) | 0.5667 | |
Data are shown as geometric means (SE). P values were adjusted for age, gender, BMI, duration of diabetes, systolic blood pressure, diastolic blood pressure, and HbA1c. †AA = major allele homozygous; Aa = heterozygous; aa = minor allele homozygous. ‡β (SE) was calculated for log10eGFR and refers to major alleles. AER: 24-hour urinary albumin excretion rate.
4. Discussion
As a potent metabolic regulator, FGF21 has been closely related to obesity-related disorders, such as hyperglycemia, dyslipidemia, insulin resistance, and hepatosteatosis [5–8]. There were also supporting evidences that suggested that FGF21 may play a protective role against DKD [9–11], through both improvement of systemic metabolic alterations and antifibrotic effects [9]. We attempted to examine the association between genetic variants in the FGF21 gene region and DKD in a Chinese population in the current study. However, we did not detect any significant association with the susceptibility to DKD for this gene region. Haplotype analysis did not show any significant result either. Since our sample size was relatively small, the association between the FGF21 gene region and DKD should be further explored in the Chinese population with a larger sample size.
eGFR and albuminuria are the main phenotypes of DKD [13]. A few studies have suggested that serum FGF21 levels were linked with eGFR as well as albuminuria in diabetic patients. A prospective study demonstrated an independent association between elevated serum FGF21 levels and eGFR decline; thus, the circulating FGF21 level was proposed as a predictor for progressive kidney disease in subjects with type 2 diabetes and normoalbuminuria [14]. This is in accordance with the results revealed by our study that rs838136 and rs499765 were significantly associated with eGFR after adjusting for confounding factors. Individuals who carried more G alleles of rs838136 showed lower eGFR, which means that the G allele of rs838136 may be a risk factor for eGFR decline in the non-DKD group. Individuals with the GG genotype of rs499765 showed lower eGFR than those with the CC+CG genotype. rs838136 and rs499765 are both located in the flanking region of the FGF21 gene, and they may affect the transcriptional regulation, resulting in abnormal splicing, or the translational dynamics of FGF21. The mechanism underlying the link between the FGF21 gene region and eGFR is still unknown and should be further investigated by functional studies. Evidences also showed the independent association of FGF21 levels with urinary albumin excretion (UAE) or albuminuria in type 2 diabetic patients [15, 16]. However, no evidence of association with AER was found for variants in the FGF21 gene region in our study.
Several limitations should be noted in the current study. First, the sample size is relatively small and only Southern Chinese population was included, which limits the power of our study. Secondly, we did not measure the circulating FGF21 levels in this study. Further investigations with a larger sample size as well as functional studies are needed to elucidate how FGF21 is involved in renal function in patients with type 2 diabetes.
In conclusion, our data indicated that genetic variants adjacent to FGF21 were associated with eGFR in subjects with type 2 diabetes. Further genetic and functional studies are necessary to replicate the association and illuminate the underlying mechanisms.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (grant number 81600659), the Open Research Project of Shanghai Key Laboratory of Diabetes Mellitus (grant number SHKLD-KF-1605), and Wenzhou Public Welfare Science and Technology Project (grant numbers Y20140683 and Y20170047). We appreciate the involvement of all the doctors, researchers, and patients in this study.
Contributor Information
Weiping Jia, Email: wpjia@sjtu.edu.cn.
Cheng Hu, Email: alfredhc@sjtu.edu.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding authors upon request.
Ethical Approval
The protocol for this research project has been approved by the Institutional Review Board of Shanghai Jiao Tong University Affiliated Sixth People's Hospital, and it conforms to the provisions of the Declaration of Helsinki.
Consent
All informed consent was obtained from the subjects.
Conflicts of Interest
The authors declared no conflict of interest.
References
- 1.Ma R. C. W. Epidemiology of diabetes and diabetic complications in China. Diabetologia. 2018;61(6):1249–1260. doi: 10.1007/s00125-018-4557-7. [DOI] [PubMed] [Google Scholar]
- 2.Lim A. Diabetic nephropathy – complications and treatment. International Journal of Nephrology and Renovascular Disease. 2014;7:361–381. doi: 10.2147/IJNRD.S40172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fava S., Azzopardi J., Hattersley A. T., Watkins P. J. Increased prevalence of proteinuria in diabetic sibs of proteinuric type 2 diabetic subjects. American Journal of Kidney Diseases. 2000;35(4):708–712. doi: 10.1016/S0272-6386(00)70019-9. [DOI] [PubMed] [Google Scholar]
- 4.Chandie Shaw P. K., Baboe F., van Es L. A., et al. South-Asian type 2 diabetic patients have higher incidence and faster progression of renal disease compared with Dutch-European diabetic patients. Diabetes Care. 2006;29(6):1383–1385. doi: 10.2337/dc06-0003. [DOI] [PubMed] [Google Scholar]
- 5.Kharitonenkov A., Shiyanova T. L., Koester A., et al. FGF-21 as a novel metabolic regulator. The Journal of Clinical Investigation. 2005;115(6):1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J., Lloyd D. J., Hale C., et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58(1):250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen B. M., Ding X., Morgan D. A., et al. FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metabolism. 2014;20(4):670–677. doi: 10.1016/j.cmet.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K. H., Jeong Y. T., Oh H., et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. National Medicine. 2013;19(1):83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 9.Kim H. W., Lee J. E., Cha J. J., et al. Fibroblast growth factor 21 improves insulin resistance and ameliorates renal injury in db/db mice. Endocrinology. 2013;154(9):3366–3376. doi: 10.1210/en.2012-2276. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y., Zhang J., Guo W., et al. Up-regulation of Nrf2 is involved in FGF21-mediated fenofibrate protection against type 1 diabetic nephropathy. Free Radical Biology and Medicine. 2016;93:94–109. doi: 10.1016/j.freeradbiomed.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L., Wang H., Xie J., Chen Z., Li X., Niu J. Potent long-acting rhFGF21 analog for treatment of diabetic nephropathy in db/db and DIO mice. BMC Biotechnology. 2017;17(1):p. 58. doi: 10.1186/s12896-017-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y. C., Zuo L., Chen J. H., et al. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. Journal of the American Society of Nephrology. 2006;17(10):2937–2944. doi: 10.1681/ASN.2006040368. [DOI] [PubMed] [Google Scholar]
- 13.Gross J. L., de Azevedo M. J., Silveiro S. P., Canani L. H., Caramori M. L., Zelmanovitz T. Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care. 2004;28(1):164–176. doi: 10.2337/diacare.28.1.164. [DOI] [PubMed] [Google Scholar]
- 14.Lee C. H., Hui E. Y. L., Woo Y. C., et al. Circulating fibroblast growth factor 21 levels predict progressive kidney disease in subjects with type 2 diabetes and normoalbuminuria. The Journal of Clinical Endocrinology & Metabolism. 2015;100(4):1368–1375. doi: 10.1210/jc.2014-3465. [DOI] [PubMed] [Google Scholar]
- 15.Jian W. X., Peng W. H., Jin J., et al. Association between serum fibroblast growth factor 21 and diabetic nephropathy. Metabolism. 2012;61(6):853–859. doi: 10.1016/j.metabol.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Esteghamati A., Khandan A., Momeni A., et al. Circulating levels of fibroblast growth factor 21 in early-stage diabetic kidney disease. Irish Journal of Medical Science. 2017;186(3):785–794. doi: 10.1007/s11845-017-1554-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding authors upon request.