Abstract
The complete genome sequence of Bacillus subtilis strain DM2 isolated from petroleum-contaminated soil on the Tibetan Plateau was determined. The genome of strain DM2 consists of a circular chromosome of 4,238,631 bp for 4458 protein-coding genes and a plasmid of 84,240 bp coding for 103 genes. Thirty-four genomic islands coding for 330 proteins and 5 prophages are found in the genome. The DDH value shows that strain DM2 belongs to B. subtilis subsp. subtilis subspecies, but significant variations of the genome are also present. Comparative analysis showed that the genome of strain DM2 encodes some strain-specific proteins in comparison with B. subtilis subsp. subtilis str. 168, such as carboxymuconolactone decarboxylase family protein, gfo/Idh/MocA family oxidoreductases, GlsB/YeaQ/YmgE family stress response membrane protein, HlyC/CorC family transporters, LLM class flavin-dependent oxidoreductase, and LPXTG cell wall anchor domain-containing protein. Most of the common strain-specific proteins in DM2 and MJ01 strains, or proteins unique to DM2 strain, are involved in the pathways related to stress response, signaling, and hydrocarbon degradation. Furthermore, the strain DM2 genome contains 122 genes coding for developed two-component systems and 138 genes coding for ABC transporter systems. The prominent features of the strain DM2 genome reflect the evolutionary fitness of this strain to harsh conditions and hydrocarbon utilization.
1. Introduction
Petroleum exploitation and utilization have caused a widespread distribution of hydrocarbons in the environment. Petroleum comprises alkanes, aromatic hydrocarbons, and nonhydrocarbon compounds. These compounds pose a threat to the ecosystem and human health [1]. Many microbes have been described by their ability to degrade petroleum hydrocarbons and have been used in the bioremediation of petroleum-contaminated environment. However, the microbes inhabiting petroleum-contaminated environments at low-temperature and higher-altitude biotope remain to be studied and exploited.
Bacillus subtilis, an extensively studied gram-positive model bacterial species in the Bacillus genus, has been isolated from a variety of distinct environments, such as diverse soils and waters [2, 3], fermented foods [4], marine sand [5], rumen and intestinal tract [6], and plant endophytic bacteria [7, 8]. The versatile physiological functions of this bacterium have been explored for industrial production [9], bioremediation of polluted environment [10–12], plant growth promotion and pathogen control [7], and even for use as probiotics for humans [13]. The diverse habitats of these strains reflect versatile metabolic pathways and a robust capacity of environmental adaptation of a widely distributed species [3, 14]. Recently, comparative genomics have shown the significant genetic variations among B. subtilis strains that inhabit diverse environments [3, 14–17]. Thus, the ecophysiological diversity of this species provides an ideal model for revealing its genetic and molecular basis of successful environmental adaptability [14].
Recently, several Bacillus strains had been isolated from petroleum-contaminated soils and were explored to degrade hydrocarbon compounds, such as B. subtilis A1 [18], Bacillus sp. M3 [19], and Bacillus sp. Q2 [20]. In addition, genome sequences of two B. subtilis strains are available. B. subtilis strain B-1, which was isolated from an oil field, can form a thick biofilm with an extracellular matrix consisting mainly of gamma-polyglutamate [21]. The B-1 genome displays 50% sequence homology with that of the model laboratory strain B. subtilis 168. Another B. subtilis strain, MJ01, was isolated from oil-contaminated soil and evaluated as a new biosurfactant-producing strain [12]. Digital DNA-DNA hybridization showed the most similarity (94.7%) with the genome of B. subtilis subsp. spizizenii TU-B-10. In this study, we isolated a new B. subtilis strain from petroleum-contaminated soil on the Tibetan Plateau in China. To further understand its genetic traits for hydrocarbon degradation and adaptability to low-temperature environment, we analyzed the whole genome sequence and compared it with the genomes of other B. subtilis strains representing distinct ecotypes or physiological traits. Our aim was to reveal the ecological fitness associated with microbial survival strategies that are relevant to petroleum-contaminated and low-temperature soil environments.
2. Materials and Methods
2.1. Strain Isolation and Measurement of Petroleum Degradation
B. subtilis strain DM2 was isolated from oil field soils in the town of Huatugou, which is located in the northwest of Qinghai province of China (90.71°E, 38.29°N, 2907 m). The strain was isolated and cultured in MM medium (3.5 g/L MgCl2, 1.0 g/L NH4NO3, 0.35 g/L KCl, 0.05 g/L CaCl2, 1.0 g/L KH2PO4, 1.0 g/L K2HPO4, 0.01 g/L FeCl3, 0.08 g/L KBr, 1 × 10−4 g/L ZnSO4·7H2O, and 24 mg/L SrCl2·6H2O, pH 7.5), with 2% (v/v) petroleum as a sole carbon source. To assess petroleum degradation, cells were inoculated in 100 mL liquid MM medium with 2% (v/v) petroleum and cultured on a rotary shaker at 20°C and 150 rpm. After 96 h of fermentation, the residual petroleum in the medium was extracted using petroleum ether. The extraction was subsequently evaporated in a rotary evaporator at 40°C and the amount of residual oil was measured using the gravimetric method described by Latha and Kalaivani [22], i.e., amount of petroleum degraded = weight of petroleum added in the medium − weight of residual oil, and the degradation rate was consequently calculated.
2.2. DNA Extraction and 16S rRNA Gene Sequencing
For DNA extraction, the strain was inoculated in liquid LB medium at 25°C and incubated at 150 rpm on a rotary shaker for 60 h. Genomic DNA was extracted using a Bacterial Genomic DNA Extraction Kit (AxyPrep, Corning Inc., NY, USA) according to its instructions. The 16S rRNA gene sequence was amplified using the primers 27F 5′-AGAGTTTGATCCTGGCTCAG and 1492R 5′-TACCTTGTTACGACTT [23]; the sequence was aligned with the NCBI database, and the 16S rRNA gene sequence obtained in this study was deposited into NCBI under accession number MK014304.
2.3. Genomic DNA Sequencing, Assembly, and Annotation
The PacBio genomic DNA library was prepared using TruSeq Nano DNA LT Library Preparation Kits (Illumina Inc., San Diego, CA, USA) after purification of the strain DNA and examination using a Nanodrop 2500. The DNA library sequencing was performed on a PacBio RS II platform using Illumina MiSeq at Majorbio Inc. (Shanghai, China). After quality control of the raw reads generated from sequencing, the resulting clean reads were assembled de novo using Newbler (version 2.8) and Hierarchical Genome Assembly Process (HGAP) version 3.0. The protein-coding genes, tRNA genes, and rRNA genes within the genomic sequence assembled were predicted using Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/), tRNAscan-SE v1.3.1, and Barrnap 0.4.2, respectively. The tandem repeat and interspersed repeat sequences were predicted using RepeatMasker and TRF software, respectively. The predicted protein-coding genes were subjected to BLASTn against the Nr, string (v9.05), and GO databases using BLAST2.2.28+. The COG (Clusters of Orthologous Groups of proteins) annotation of the predicted genes was obtained by BLASTp search against the string database (http://string-db.org/), and the functional protein clustering was performed according to the COG results. The predicted genes were further compared by blast against KEGG (Kyoto Encyclopedia of Genes and Genomes) database to gain their KOs and pathways. Genomic Island (GI) in the genome was predicted using IslandViewer 4 (https://www.pathogenomics.sfu.ca/islandviewer/) and PHAST software (version 1.5). The complete genome sequences generated in the present study were deposited in GenBank under the accession numbers CP030937 and CP030938.
2.4. Phylogenetic Analysis of the Strain
The protein sequences of 24 housekeeping genes, including CTP synthase, DNA primase, DNA-directed RNA polymerase beta-subunit, LSU ribosomal protein L3p, LSU ribosomal protein L4p, LSU ribosomal protein L5p, LSU ribosomal protein L6p, LSU ribosomal protein L7/L12, LSU ribosomal protein L11p, LSU ribosomal protein L13p, LSU ribosomal protein L16p, LSU ribosomal protein L20p, LSU ribosomal protein L27p, phosphoglycerate kinase, ribosome recycling factor, SSU ribosomal protein S2p, SSU ribosomal protein S3p, SSU ribosomal protein S5p, SSU ribosomal protein S9p, SSU ribosomal protein S10p, SSU ribosomal protein S11p, SSU ribosomal protein S13p, transcription termination protein NusA, and translation elongation factor Ts, from certain Bacillaceae members were downloaded from GenBank [13]. The protein sequences extracted from GenBank and the present isolate were aligned using MEGA 7.0, and a phylogenetic tree was consequently produced based on neighbor-joining method.
2.5. Comparative Genomics
To discern the characteristic of DM2 genome, the genomes of six Bacillus strains, i.e., B. subtilis subsp. subtilis str. 168, B. subtilis PY79, B. subtilis TO-A JPC, B. subtilis MJ01, B. subtilis B-1, B. subtilis TO-A JPC, and B. subtilis UD1022, which were isolated from different biotopes with their genome sequences deposited in GenBank, were retrieved from NCBI. The genome of strain DM2 was submitted to the Integrated Microbial Genomes (IMG) database (https://img.jgi.doe.gov/) for comparative genome analysis.
3. Results and Discussion
3.1. Isolation and Identification of a Petroleum-Degrading Strain DM2
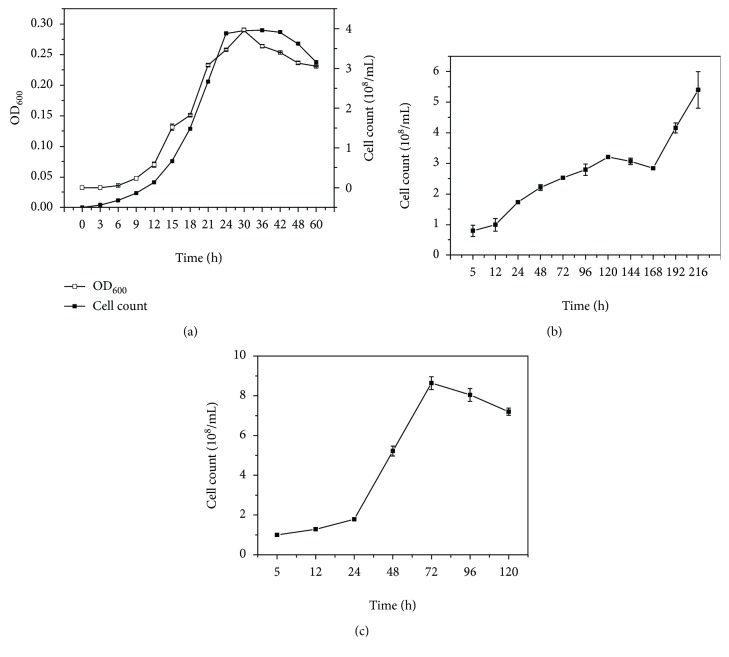
Strain DM2 was isolated from the soil of an oil field located in a cryogenic region at an altitude of 2909 m using MM medium with petroleum as the sole carbon source. The strain grew well in liquid LB medium and reached its maximum growth rate after 24 h of shaking culture (Figure 1(a)). The strain could also grow in the oligotrophic liquid MM medium containing 2% (v/v) of the mixture of alkanes (C12 : C14 : C15 = 1 : 1 : 1) as the sole carbon source (Figure 1(b)). However, when 2% petroleum was added to MM medium as the sole carbon source, the strain exhibited better growth than with the alkane mixture as the carbon source (Figure 1(c)), suggesting a low degradation capacity for middle-chain alkanes. Further experiments indicated that, when the strain incubated in liquid MM medium containing 2% petroleum as the carbon source at 20°C for 96 h, 53.92%±4.74 of petroleum in medium was degraded suggesting its strong petroleum-degrading ability at the culture conditions. The 16S rRNA gene sequence of strain DM2 showed 99% similarity with Bacillus subtilis. Thus, the isolate was identified as B. subtilis DM2.
Figure 1.
The growth features of strain DM2 in different medium. (a) Growth curve in LB medium. (b) Growth curve in MM medium with the mixture of alkanes (C12 : C14 : C15 = 1 : 1 : 1) as the sole carbon source. (c) Growth curve in MM medium with 2% petroleum as the sole carbon source.
3.2. The Genome Organization of B. subtilis DM2
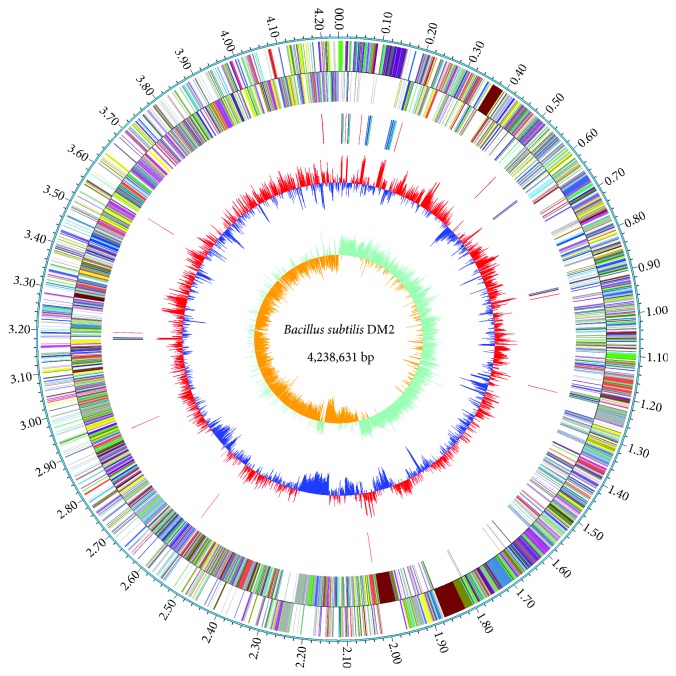
The genome of strain DM2 consists of a circular chromosome of 4,238,631 bp with G+C content of 43.52% and a plasmid of 84,240 bp with G+C content of 35.08%. The detailed information on the genome is summarized in Table 1 and Figure 2. To further discern the characteristics of the genome, we downloaded the genomic data of six B. subtilis strains from the NCBI database and comparatively analyzed their genomes (Table 2). Of those, B. subtilis subsp. subtilis str. 168 is a subspecies and a model strain of B. subtilis. B. subtilis PY79 is one of the most widely used laboratory strains [24]. B. subtilis B-1 is a petroleum-degrading and biofilm-producing strain isolated from the oil field biofilms [21]. B. subtilis MJ01 is also a petroleum-degrading strain isolated from oil-contaminated soil [12]. B. subtilis TO-A JPC is a probiotic strain isolated from a probiotic drug Vibact® [13]. B. subtilis UD1022 is a plant growth-promoting strain isolated from the plant rhizosphere soil [7]. Among them, DM2 has the largest genome and the highest number of predicted genes and protein-coding genes. The previous studies hypothesized that there is a correlation between microbial genome size and their environment adaptability [25, 26]. Whether such distinct genetic traits of strain DM2 is related to its successful adaptation to its habitat needs further study.
Table 1.
Genome organization of B. subtilis strain DM2.
| Genome organization | Chromosome | Plasmid |
|---|---|---|
| Genome | 1 | 1 |
| Genome size (bp) | 4,238,631 | 84,240 |
| G+C content (%) | 43.52 | 35.08 |
| Gene number | 4458 | 103 |
| Total gene length (bp) | 3,754,593 | 73,761 |
| Gene average length(bp) | 842.21 | 716.13 |
| Gene density (kb) | 1.05 | 1.22 |
| GC content in gene region (%) | 44.18 | 35.75 |
| Gene/genome (%) | 88.58 | 87.56 |
| Intergenetic region length (bp) | 484,038 | 10,479 |
| GC content in intergenetic region (%) | 38.34 | 30.36 |
| Tandem repeat number | 22 | - |
| Tandem repeat length (bp) | 6612 | - |
Figure 2.
Circular chromosome genome of B. subtilis DM2. Circle 1 (from outside to inside) represents the genome size. Circle 2 represents the protein-coding genes in the positive strand of chromosome. Circle 3 represents the protein-coding genes in the negative strand of chromosome. Circle 4 represents rRNA and tRNA genes. Circle 5 represents G+C content, <average content (blue-colored), ≥average content (red-colored). Circle 6 represents the GC skew (G-C)/(G+C).
Table 2.
Genomic characteristics of several Bacillus subtilis strains.
| Genome organization | B. subtilis strains | ||||||
|---|---|---|---|---|---|---|---|
| DM2 | PY79 | 168 | UD1022 | B-1 | MJ01 | TO-A JPC | |
| Isolated habitat | Oil field soil | Prototrophic laboratory strain | Hay infusion | Rhizosphere soil | Oil field biofilm | Oil-polluted soil | Probiotic drug |
| Genome size (Mb) | 4.24 | 4.03 | 4.22 | 4.03 | 3.94 | 4.11 | 4.09 |
| GC content (%) | 43.5 | 43.8 | 43.5 | 43.9 | 46.5 | 43.9 | 43.8 |
| Total genes | 4527 | 4187 | 4419 | 4129 | 3868 | 4223 | 4236 |
| Protein-coding genes | 4458 | 4138 | 4255 | 3933 | 3706 | 3999 | 4058 |
| rRNA number | 30 | 30 | 30 | 30 | 19 | 30 | 30 |
| tRNA number | 88 | 85 | 86 | 86 | 59 | 86 | 86 |
| Other RNA number | 5 | 1 | 5 | 1 | 1 | 5 | 5 |
| Pseudogene number | 93 | 2 | 43 | 79 | 83 | 103 | 57 |
| NCBI accession | CP030937 | CP006881 | CP010052 | CP011534 | CP009684 | CP018173 | CP011882 |
3.3. Functional Protein Classification
The genome of strain DM2 encodes 4458 proteins, of which 712 are hypothetical proteins. The predicted protein sequences were aligned against the COG database using BLASTp. A total of 3163 proteins were annotated to at least one COG category. The top protein categories are amino acid transport and metabolism, carbohydrate transport and metabolism, general function prediction only, transcription, function unknown, translation, ribosomal structure and biogenesis, coenzyme transport and metabolism, cell wall/membrane/envelope biogenesis, inorganic ion transport and metabolism, signal transduction mechanisms, and energy production and conversion (Table S1). The protein numbers under categories of extracellular structures, lipid transport and metabolism, secondary metabolite biosynthesis, transport and catabolism, posttranslational modification, protein turnover, chaperones, replication, recombination and repair, and signal transduction mechanisms increase over those of the model strain B. subtilis subsp. subtilis str. 168 (Table S1). These categories include a large number of stress response and environmental adaptation proteins, implying that strain DM2 has a strong adaptive capacity to environments.
3.4. Whole-Genome Alignments Reveal Heterogeneity within Strains of B. subtilis
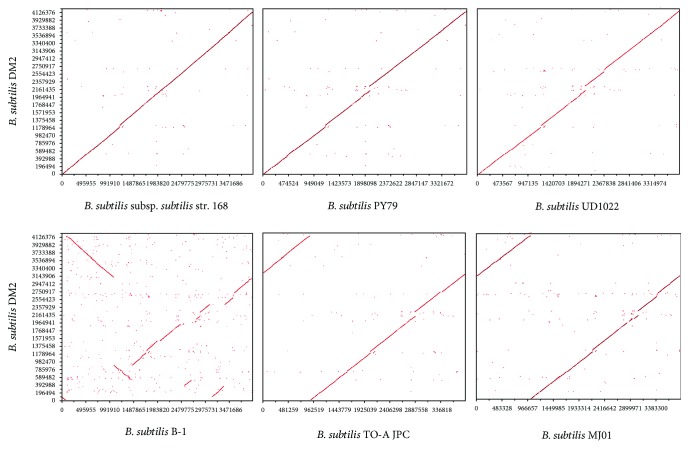
In general, the genetic distance and gene similarity between two organisms can be determined by DNA-DNA hybridization (DDH). Recently, the Genome Blast Distance Phylogeny (GBDP) approach was improved for in silico genome-to-genome comparison [27, 28]. The principle of this approach is, firstly, two genomes are aligned using BLAST to generate a set of high-scoring segment pairs, and secondly, a single genome-to-genome distance value is calculated from the total number of identical base pairs by a specific distance formula [28]. The DDH values between the whole genomes of strain DM2 and other B. subtilis strains, which have publicly available complete genomes, were calculated using the genome-genome distance calculator (GGDC) server at http://ggdc.dsmz.de [28]. Because the length of high-scoring pairs was used for calculation instead of the genome length, the Formula II values were used as the analysis standards. Strain DM2 was the closest to B. subtilis PY79 with 89% DDH value followed by B. subtilis NCIB 3610 and B. subtilis subsp. subtilis str. 168, both with 88.6% DDH value, suggesting that strain DM2 belongs to B. subtilis subsp. subtilis subspecies. However, DDH values < 70%, which is a threshold for species delimitation in Archaea and Bacteria [28], for strain DM2 and B. subtilis subsp. stercoris, B. subtilis subsp. spizizenii, B. subtilis subsp. inaquosorum, and B. subtilis subsp. spizizenii suggest significant genome variations among these B. subtilis strains (Table 3). Pairwise genome alignments show that the genomic organization of strain DM2 has high similarity with B. subtilis subsp. subtilis str. 168, B. subtilis PY79, and B. subtilis UD1022. No rearrangement is evident, but only a few chromosomal deletions between 1178 and 1375 Kbps are observed in the chromosome. However, chromosomal inversions are observed among strains DM2, B. subtilis MJ01, and B. subtilis TO-A JPC. Synteny analysis showed various genome rearrangements between strains DM2 and B-1 with numerous genomic insertions, deletions, and inversions (Figure 3). These results indicate that the core genomes of the Bacillus subtilis strains are conserved.
Table 3.
DNA-DNA hybridization values as calculated by GGDC server.
| Query genome | Reference genome | Formula I | Formula II | Formula III |
|---|---|---|---|---|
| B. subtilis DM2 | B. subtilis PY79 | 92.4 | 89 | 94.2 |
| B. subtilis DM2 | B. subtilis subsp. subtilis NCIB 3610 | 95.1 | 88.6 | 96.1 |
| B. subtilis DM2 | B. subtilis subsp. subtilis str. 168 | 93.5 | 88.6 | 94.9 |
| B. subtilis DM2 | B. subtilis subsp. subtilis CU1050 | 92.9 | 88.6 | 94.5 |
| B. subtilis DM2 | B. subtilis subsp. subtilis str. AG1839 | 92.8 | 88.5 | 94.4 |
| B. subtilis DM2 | B. subtilis QB928 | 92.8 | 88.4 | 94.4 |
| B. subtilis DM2 | B. subtilis BSn5 | 92.1 | 88.4 | 93.9 |
| B. subtilis DM2 | B. subtilis subsp. subtilis str. OH 131.1 | 92.2 | 87.9 | 93.9 |
| B. subtilis DM2 | B. subtilis UD1022 | 91.4 | 85.3 | 92.9 |
| B. subtilis DM2 | B. subtilis subsp. subtilis RO-NN-1 | 89.5 | 83.3 | 91.2 |
| B. subtilis DM2 | B. subtilis subsp. stercoris JS | 88.4 | 62.9 | 86.4 |
| B. subtilis DM2 | B. subtilis subsp. stercoris MD-5 | 87.9 | 62.7 | 85.9 |
| B. subtilis DM2 | B. subtilis MJ01 | 84.5 | 51.2 | 79.7 |
| B. subtilis DM2 | B. subtilis subsp. spizizenii TU-B-10 | 83.9 | 50.5 | 79 |
| B. subtilis DM2 | B. subtilis subsp. inaquosorum KCTC 13429 | 81.1 | 50.5 | 76.6 |
| B. subtilis DM2 | B. subtilis subsp. inaquosorum DE111 | 80 | 50.3 | 75.7 |
| B. subtilis DM2 | B. subtilis subsp. spizizenii W23 | 83 | 49.5 | 77.9 |
| B. subtilis DM2 | B. vallismortis NBIF-001 | 33.1 | 21 | 28.9 |
| B. subtilis DM2 | B. velezensis FZB42 | 33 | 20.9 | 28.7 |
| B. subtilis DM2 | B. subtilis J-5 | 32.4 | 20.9 | 28.4 |
| B. subtilis DM2 | B. subtilis B-1 | 32 | 20 | 28 |
Figure 3.
Ortholog dot plot of B. subtilis DM2 genome vs. B. subtilis PY79, B. subtilis subsp. subtilis str. 168, B. subtilis UD1022, B. subtilis B-1, B. subtilis TO-A JPC, and B. subtilis MJ01 genomes. Each dot represents a reciprocal best hit by BLASTp.
3.5. Phylogenetic Analysis of B. subtilis Strain DM2
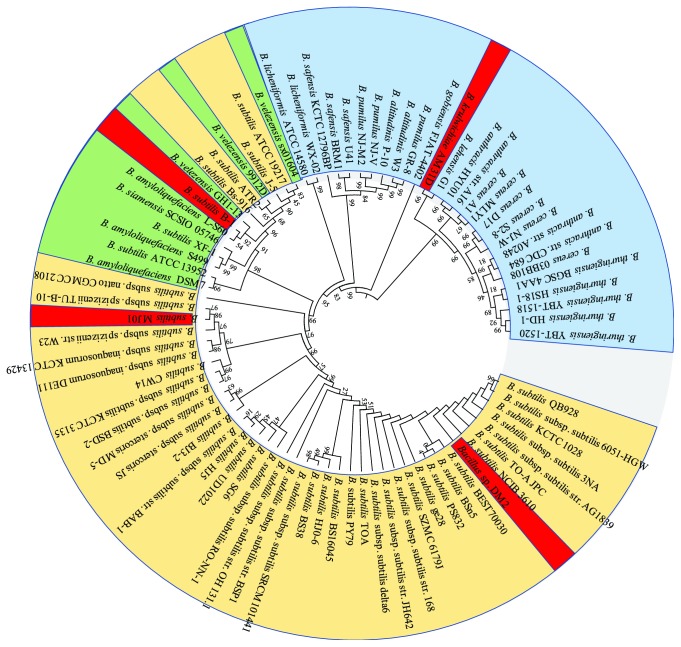
To understand the phylogenetic relationship of strain DM2, the protein sequences of 24 housekeeping genes of the members of B. subtilis and other Bacillus species were aligned using MEGAx. The neighbor-joining phylogenetic tree shows multiple clades (Figure 4). Although the DDH value indicates the closest similarity between the strains DM2 and B. subtilis subsp. subtilis str. 168, strain DM2 belongs to a separate clade in the phylogenetic tree compared with the other petroleum-degrading strains such as B. subtilis MJ01, B. subtilis B-1, and B. krulwichiae, which are only distantly related. Furthermore, an intermix of strains B. velezensis and B. amyloliquefaciens within the clades of B. subtilis implies that these strains have the closest phylogenetic relationship. In addition to B. krulwichiae, B. pumilus, and B. safensis, which have been reported as isolated from oil-contaminated environments, to date [29–31], most of the oil-degrading Bacillus strains belong to B. subtilis, suggesting that B. subtilis possesses the functional diversity and adaptive capacity to various environments.
Figure 4.
Neighbor-joining phylogenetic tree based on the protein sequences of 24 housekeeping genes of the Bacillaceae family members. The members of B. subtilis (orange-colored), outgroups of B. subtilis (blue-colored), the members of B. velezensis and B. amyloliquefaciens (green-colored), and the isolates from the crude-oil environment (red-colored) are included.
3.6. Core Proteome Analysis of Strain DM2
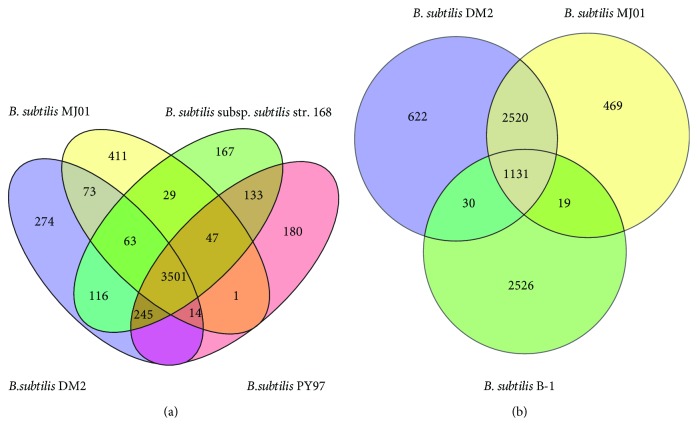
The orthologous proteins of four B. subtilis strains, which have the closest phylogeny or functional similarity, were aligned using Proteinortho V2.3 Perl script (Figure 5(a)). A total of 3501 proteins formed the core set of proteins of the four strains. Strain DM2 shares 3925, 3777, and 3651 common orthologous proteins with strains B. subtilis 168, PY79, and MJ01, respectively. To further unravel the differences in orthologous proteins among the Bacillus strains isolated from oil-contaminated environments, the orthologous proteins of strains DM2, MJ01, and B-1 were aligned. The result shows that 1131 orthologous proteins are shared by three strains; 3651 orthologous proteins are shared by strains DM2 and MJ01, but only 1161 orthologous proteins are shared by strains DM2 and B-1 (Figure 5(b)). This result indicates that there are great differences between strains DM2 and B-1, although both are the B. subtilis members capable of degrading petroleum. The 1131 orthologous proteins shared with the three strains were further aligned using BLASTp to identify the conserved function genes. The analysis shows that, apart from housekeeping genes, the genes responsible for sporulation/spore germination proteins, chaperones, membrane transport proteins, and transcriptional regulators are the functionally conserved Bacillus genes. Moreover, the genes encoding ring-cleaving dioxygenase, fatty acid desaturase, cytochrome P450, oxidoreductase, and FAD-binding oxidoreductase are highly conserved genes in these three petroleum-degrading strains, providing the molecular basis for petroleum biodegradation.
Figure 5.
Comparative analysis of protein sets among four B. subtilis strains selected. (a) Orthologous set of proteins in B. subtilis DM2, B. subtilis subsp. subtilis str. 168, B. subtilis PY79, and B. subtilis MJ01. (b) Orthologous set of proteins in B. subtilis DM2, B. subtilis MJ01, and B. subtilis B-1.
3.7. Characterization of Proteins Encoded by Strain DM2 Genome
The above analyses indicate that strain DM2 has the most protein-coding genes among the B. subtilis strains compared in this selected panel. Comparison analysis of the identified protein coding regions of strain DM2, model strain 168, and another oil-degrading strain MJ01 shows that strain DM2 shares several common proteins with strain MJ01, including DNA-binding response regulators, EamA family transporters, NAD(P)-dependent oxidoreductases, two-component sensor histidine kinases, two-component system response regulators, and methyl-accepting chemotaxis proteins. Most of these proteins may function in response to stresses and signal transduction. Some of them are also involved in hydrocarbon degradation, such as NAD(P)-dependent alcohol dehydrogenases. Furthermore, some proteins are DM2-specific, such as carboxymuconolactone decarboxylase family, gfo/Idh/MocA family oxidoreductases, GlsB/YeaQ/YmgE family stress response membrane proteins, HlyC/CorC family transporters, LLM class flavin-dependent oxidoreductase, and LPXTG cell wall anchor domain-containing proteins (Table S2). However, most of the abovementioned proteins are absent from strain 168. We conclude that the strain-specific proteins further imply the vigorous adaptability of strain DM2 to the harsh biotope.
3.8. Horizontal Gene Transfers in the Genome of Strain DM2
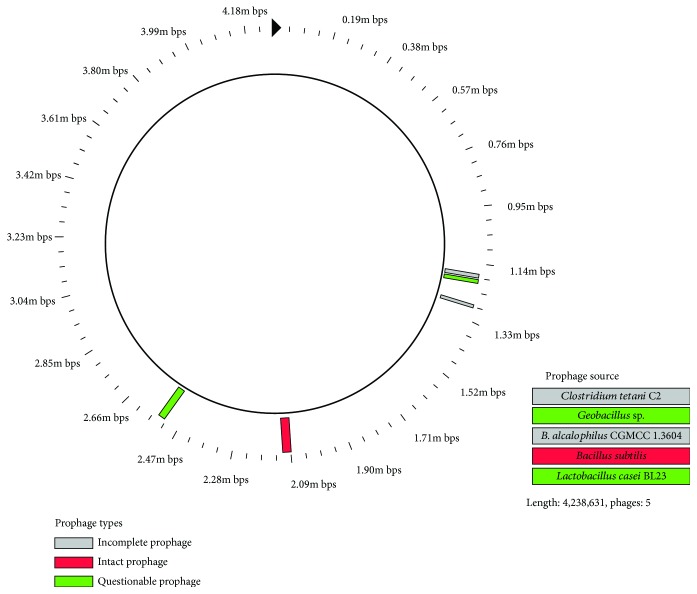
The analysis of nonorthologous proteins reveals that the genome contains many horizontal gene transfers. A total of 34 gene islands (GIs) are found in the genome, which consist of 4000 bp-100,000 bp DNA in size (Table 4). Of the total 510 genes, 330 genes are annotated as hypothetical proteins with unknown function, 37 are annotated as recombinase- and phage-related proteins, most of which are the phage-specific site integrases. Most of the genes in GI are associated with metabolism (22), transcriptional regulation (33), signal transduction, and membrane transport (10). Notably, several genes, including glycosyl transferase family A, fatty acid desaturase, short chain dehydrogenase family protein, stress response protein, and cold-shock protein, which are involved in glycosyl transfer, lipid metabolism, and stress response, are found in the GIs. The harboring of these genes in GIs suggests that horizontal gene transfer provides additional clues about metabolic diversity [26] and confers several functional genes to strain DM2 to cope with the harsh environment and to promote petroleum degradation [32]. In addition, a total of 5 prophages are found in the genome of strain DM2, which comprise an intact (123 kb), two incomplete (30 kb), and two questionable (28 kb and 61 kb) prophages (Figure 6). The protein sequences of prophages were further searched in the Nr database using BLASTp, and the result indicates that only an intact prophage protein initiated from B. subtilis, whereas the remaining protein sequences evolved from the outgroup prophages of Bacillus. The presence of prophages in the genome reflects phage-related genetic modifications and is well-known to regulate bacterial population density. Therefore, the gene transfer, which occurred in strain DM2, plays critical roles in the acquisition of the resistance genes and adaptation to harsh environments [33].
Table 4.
Features of the gene islands found in the genome of B. subtilis DM2.
| GI | Size (bp) | Genes | Hypothetical proteins | Integrase/phage portal protein |
|---|---|---|---|---|
| 1 | 10,848 | 9 | 3 | 1 |
| 2 | 4247 | 3 | 1 | 0 |
| 3 | 10,269 | 4 | 3 | 0 |
| 4 | 11,092 | 4 | 4 | 0 |
| 5 | 7217 | 7 | 4 | 0 |
| 6 | 8419 | 8 | 5 | 1 |
| 7 | 26,611 | 26 | 13 | 1 |
| 8 | 4522 | 5 | 4 | 0 |
| 9 | 5365 | 5 | 2 | 0 |
| 10 | 4179 | 7 | 2 | 2 |
| 11 | 4778 | 11 | 5 | 1 |
| 12 | 5696 | 7 | 4 | 0 |
| 13 | 18,793 | 27 | 18 | 2 |
| 14 | 6295 | 3 | 2 | 0 |
| 15 | 7427 | 11 | 7 | 0 |
| 16 | 122,642 | 164 | 122 | 5 |
| 17 | 4157 | 7 | 5 | 0 |
| 18 | 6329 | 14 | 10 | 0 |
| 19 | 24,186 | 41 | 32 | 2 |
| 20 | 4642 | 9 | 8 | 0 |
| 21 | 8129 | 7 | 7 | 0 |
| 22 | 5209 | 9 | 8 | 1 |
| 23 | 14,435 | 7 | 6 | 0 |
| 24 | 6531 | 9 | 4 | 1 |
| 25 | 40,710 | 53 | 23 | 19 |
| 26 | 4006 | 8 | 6 | 1 |
| 27 | 6429 | 8 | 2 | 0 |
| 28 | 4134 | 2 | 2 | 0 |
| 29 | 5506 | 7 | 1 | 0 |
| 30 | 4168 | 2 | 0 | 0 |
| 31 | 4087 | 6 | 5 | 0 |
| 32 | 7752 | 7 | 4 | 0 |
| 33 | 6928 | 7 | 4 | 0 |
| 34 | 4389 | 6 | 4 | 0 |
| Total | 420,127 | 510 | 330 | 37 |
Figure 6.
Characteristics and position of the prophage found in the Bacillus subtilis DM2 genome. The color in the circles represents the completeness of the prophage, incomplete prophage (gray-colored), intact prophage (red-colored), and potential prophage (green-colored).
3.9. KEGG Pathway Enrichment Reveals the Metabolic Character of Strain DM2
3.9.1. Metabolism
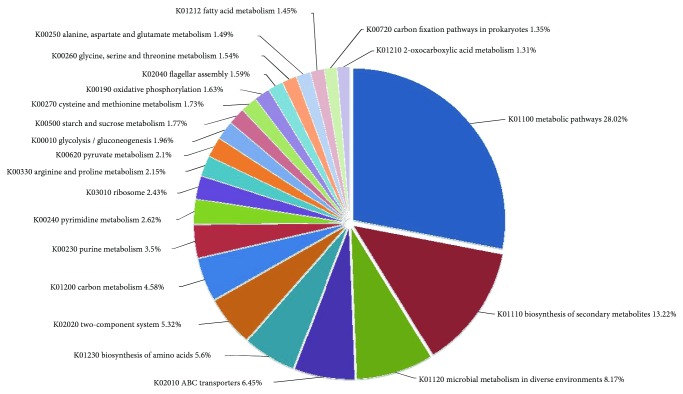
A total of 600 genes are enriched in the metabolic and synthetic pathways, including whole genes associated with glycolysis, TCA cycle, and pentose phosphate pathway, but are lacking the key gene coding for 2-dehydro-3-deoxy-phosphogluconate in Entner-Doudoroff pathway. Strain DM2 can use sucrose, fructose, galactose, rhamnose, mannose, and C5-branched dibasic acid as substrates. However, the key genes involved in pathways of fucose, allose, and sorbitol are lacking. The genes involved in polysaccharide metabolism, such as dextranase and amylase genes, are found in strain DM2 genome. All the genes required for the anabolic pathways of amino acids, purines, and pyrimidine synthesis are present in the genome. A total of 18 genes are involved in nitrogen metabolic pathway, of which 7 genes encode nitrite reductase (nirB, nirC, nirD, narG, narH, narI, and narJ) that catalyze nitrate to ammonia. In addition, 2 genes encode nitronate monooxygenases that catalyze nitroalkane to nitrite. The lack of a gene coding for nitrogenase suggests the absence of nitrogen fixation. All key enzymes involved in synthesis of cysteine from sulfate are found in the genome, but the gene coding for sulfate transport system substrate-binding protein (Cysp) is absent. Thus, the presence of genes coding for sulfonate transport protein (ssuA) and alkanesulfonate monooxygenase (ssuD) in the DM2 genome implies that the strain uses alkanesulfonate rather than sulfate as a sulfur supply (Figure 7).
Figure 7.
Top KEGG pathways enriched in strain DM2 genome and the percentage of genes under each pathway.
3.9.2. Osmoprotectant Transport Systems
The osmoprotectant transport system (Opu) in the genome of the strain comprises two opuA (orf3675 and orf3680), four opuBD (orf3672, orf3674, orf3677, and orf3679), and two opuC (orf3673 and orf3678) genes. The genes that are involved in the absorption and synthesis of glutamate, which acts as osmoprotectant, include three gltA (orf0968, orf2624, and orf3162), a gltD (orf2036), a gltB (orf2037), a gltC (orf2038), and two gltP/T (orf0240 and orf1048) genes.
3.9.3. Pathways for Degradation of Petroleum Hydrocarbons and Xenobiotics
Strain DM2 genome harbors a total of 37 genes that may be responsible for hydrocarbon degradation. Of those, 11 genes encode dioxygenases, 13 genes encode monooxygenases, 8 genes encode cytochrome P450 enzymes, and single genes encode fatty acid desaturase, dihydropteridine reductase, and NADH-dependent butanol dehydrogenase. Among them, catechol-2,3-dioxygenase, biphenyl-2,3-dioxygenase, 4-hydroxyphenylacetate 3-monooxygenase, cytochrome P450 CYP102A2_3, and ring-cleaving dioxygenase are the key aromatic degradation enzymes. Some monooxygenases, cytochrome P450 enzymes, NADH-dependent butanol dehydrogenases, fatty acid beta-hydroxylases, and fatty acid desaturases are involved in the degradation pathways of alkanes and alkenes (Table 5). The enzyme fatty acid desaturase is also an important member that plays roles in the adaptability to low temperature [13]. Interestingly, a ubiquitous gene coding for alkane monooxygenase (AlkB) is not found in the genome of strain DM2. Additionally, another gene coding for bacterial luciferase (LadA) involved in the degradation of long-chain hydrocarbon [34], which is harbored in the genomes of strains 168 and MJ01, is not found in DM2 genome.
Table 5.
List of proteins involve in degradation of petroleum compounds.
| Protein ID | Swissprot references | Swissprot closest homolog | Swissprot similarity % | KO | Gene name/enzyme |
|---|---|---|---|---|---|
| AXF32711 | spo31669.1 | Acireductone dioxygenase | 100 | K08967 | mtnD, mtnZ, ADI1 |
| AXF32173 | spp40402.2 | Alkanesulfonate monooxygenase | 100 | K04091 | ssuD |
| AXF32337 | spo07561.1 | Aromatic compound monooxygenase | 99 | None | _ |
| AXF34322 | spo34627.1 | Blue-light photoreceptor | 99 | None | _ |
| AXF32118 | spP54720.1 | Catechol-2,3-dioxygenase | 99 | None | _ |
| AXF32119 | spp54721.2 | Catechol-2,3-dioxygenase | 99 | K07104 | _ |
| AXF34401 | spo32085.1 | Cysteine dioxygenase | 97 | None | _ |
| AXF35528 | spp53554.1 | Cytochrome P450 | 98 | K16593 | bioI |
| AXF34015 | spo08336.1 | Cytochrome P450 CYP102A3 | 99 | K14338 | cypD_E, CYP102A2_3 |
| AXF31825 | spo34653.1 | Fatty acid desaturase | 82 | K10255 | FAD6, desA |
| AXF33257 | spo34653.1 | Fatty acid desaturase | 100 | K10255 | FAD6, desA |
| AXF31559 | spo31440.1 | Fatty acid peroxygenase | 100 | K15629 | CYP152A1 |
| AXF32654 | spp49852.1 | Flavohemoprotein | 100 | K05916 | hmp, YHB1 |
| AXF32298 | spp38049.1 | Heme-degrading monooxygenase | 99 | None | _ |
| AXF35412 | spo31534.2 | Heme-degrading monooxygenase | 100 | K07145 | isdG, isdI |
| AXF33211 | spc0SPC0.1 | Probable 4-hydroxyphenylacetate 3-monooxygenase | 100 | K00483 | hpaB |
| AXF34426 | spo05239.1 | Probable NADH-dependent butanol dehydrogenase 1 | 100 | K00100 | E1.1.1.- |
| AXF34427 | spo05240.1 | Probable NADH-dependent butanol dehydrogenase 2 | 100 | None | _ |
| AXF33973 | spo05413.1 | Probable nitronate monooxygenase | 99 | K00459 | ncd2, npd |
| AXF32025 | spo08394.1 | Putative cytochrome P450 CYP102A2 | 99 | K14338 | cypD_E, CYP102A2_3 |
| AXF32565 | spo34374.1 | Putative cytochrome P450 YjiB | 99 | K00517 | E1.14.-.- |
| AXF32015 | spo31535.1 | Putative lyase/dioxygenase | 100 | None | _ |
| AXF31633 | spo34504.2 | Putative monooxygenase | 99 | None | _ |
| AXF31730 | spp94425.1 | Putative monooxygenase | 100 | None | _ |
| AXF34230 | spo34846.1 | Putative monooxygenase | 99 | None | _ |
| AXF35096 | spp39606.1 | Putative monooxygenase | 99 | None | _ |
| AXF35236 | spp54950.1 | Putative monooxygenase | 100 | None | _ |
| AXF34689 | spo32254.1 | Putative monooxygenase | 100 | None | _ |
| AXF34228 | spo34974.1 | Putative monooxygenase MoxC | 99 | None | _ |
| AXF31862 | spp96693.1 | Putative ring-cleaving dioxygenase | 100 | K15975 | _ |
| AXF32639 | spo34689.1 | Putative ring-cleaving dioxygenase MhqA | 100 | K15975 | _ |
| AXF33297 | spo34543.1 | Putative ring-cleaving dioxygenase MhqE | 99 | K15975 | _ |
| AXF35280 | spp42106.2 | Quercetin 2,3-dioxygenase | 99 | K07155 | E1.13.11.24 |
The xenobiotic biodegradation and metabolism pathways consist of benzoate degradation, aminobenzoate degradation, chloroalkane and chloroalkene degradation, bisphenol degradation, azathioprine and 6-mercaptopurine degradation, fluorouracil degradation, and citalopram degradation (Table 6).
Table 6.
List of KO pathways for hydrocarbon and aromatic compound degradation.
| KO | Genes | Enzymes | Protein ID |
|---|---|---|---|
| K00121 | frmA, ADH, adhC | Alcohol dehydrogenase (EC: 1.1.1.1) | AXF33996 |
| K00128 | ALDH | Aldehyde dehydrogenase family protein (EC: 1.2.1.3) | AXF35274, AXF35172 |
| AXF35083, AXF33272 | |||
| AXF32034, AXF31595 | |||
| K00493 | CYP | Cytochrome P450 (EC: 1.14.14.1) | AXF32025 |
| K00626 | atoB | Acetyl-CoA C-acetyltransferase (EC: 2.3.1.9) | AXF32322, AXF33714 |
| K00074 | paaH | 3-Hydroxybutyryl-CoA dehydrogenase (EC: 1.1.1.157) | AXF33713 |
| K01670 | nmsA | Naphthyl-2-methylsuccinate synthase alpha subunit (EC: 4.1.99.-) | AXF34000 |
| K00632 | fadA/I | Acetyl-CoA acyltransferase (EC: 2.3.1.16) | AXF34569 |
| K01821 | praC | 4-Oxalocrotonate tautomerase (EC: 5.3.2.6) | AXF35041 |
| K01607 | pcaC | 4-Carboxymuconolactone decarboxylase (EC: 4.1.1.44) | AXF35133 |
| K01113 | phoD | Alkaline phosphatase D (EC: 3.1.3.1) | AXF31608 |
| K01077 | phoA | Alkaline phosphatase (EC: 3.1.3.1) | AXF31889 |
| AXF32230 | |||
| K01512 | acyP | Acylphosphatase (EC: 3.6.1.7) | AXF32061 |
| K00517 | CYP81F | Cytochrome P450 (EC: 1.14.14.-) | AXF32565 |
| K01034 | atoD | Acetate CoA/acetoacetate CoA-transferase alpha subunit (EC: 2.8.3.8) | AXF33316 |
| K01101 | - | 4-Nitrophenyl phosphatase (EC: 3.1.3.41) | AXF34513 |
| K00100 | bdhAB | Butanol dehydrogenase (EC: 1.1.1.-) | AXF34427 |
| K00148 | fdhA | Glutathione-independent formaldehyde dehydrogenase (EC: 1.2.1.46) | AXF35308 |
| K01428 | ureC | Urease subunit alpha (EC: 3.5.1.5) | AXF34944 |
| K01429 | ureB | Urease subunit beta (EC: 3.5.1.5) | AXF34945 |
| K01430 | ureA | Urease subunit gamma (EC: 3.5.1.5) | AXF34946 |
3.9.4. Stress Response and Signaling
The genomic analysis indicates that strain DM2 has developed systems of stress response and signal transduction. Many genes that are related to environmental stress response and signal transduction are found in the genome of strain DM2. A total of 122 genes encode two-component signal transduction proteins, including 19 histidine kinases, 21 response regulators, 11 sporulation and bacteria movement-related genes, and 8 diguanylate cyclases (Table 7). Of those, the histidine kinases act as the stimulus sensor and play a critical role in signal transduction [35]. These histidine kinase genes are adjacent to the genes that code for response regulators in the genome, suggesting that strain DM2 has a highly efficient two-component signaling system and may respond efficiently to the environmental signals [36]. Diguanylate cyclase catalyzes the formation of cyclic diguanylate monophosphate and acts as the ubiquitous secondary messenger involved in various bacterial metabolic and growth processes [37]. The common Sec-dependent secretion system and twin arginine targeting secretion system in the genome are beneficial to the substance exchange across cell membrane and even remold the environment for its growth [38]. In addition, strain DM2 contains 18 chaperone genes, including RNA chaperone, molecular chaperone, nitrate reductase molybdenum cofactor assembly chaperone, copper chaperone, flagellar biosynthesis chaperone, heat shock proteins, and cold shock proteins. Chaperones stabilize the protein conformations and have been shown to contribute to bacterial growth at low temperatures [39]. Strain DM2 possesses two biotin carboxylase genes that have been reported to be expressed at low temperature [40].
Table 7.
The two-component signaling systems of B. subtilis DM2.
| Gene families | Genes | Description | KOs | Protein ID |
|---|---|---|---|---|
| LytTR | natK | Sensor histidine kinase | K11640 | AXF31619 |
| natR | Response regulator | K11641 | AXF31620 | |
| lytT/lytR | Response regulator | K07705 | AXF34190 | |
| lytS | Sensor histidine kinase | K07704 | AXF34191 | |
|
| ||||
| CitB | dctS | Sensor histidine kinase | K11691 | AXF31786 |
| dctR | Response regulator | K11692 | AXF31787 | |
| citS | Sensor histidine kinase | K11637 | AXF32055 | |
| citT | Response regulator | K11638 | AXF32056 | |
| yufL/malK | Sensor histidine kinase | K11614 | AXF34443 | |
| malR | Response regulator | K11615 | AXF34444 | |
|
| ||||
| NarL | ydfH | Sensor histidine kinase | K11623 | AXF31854 |
| ydfI | Response regulator | K11624 | AXF31855 | |
| desK | Sensor histidine kinase | K07778 | AXF33258 | |
| desR | Response regulator | K07693 | AXF33259 | |
| comA | Two-component response regulator | K07691 | AXF34459 | |
| comP | Sensor histidine kinase | K07680 | AXF34460 | |
| liaR | Response regulator | K11618 | AXF34595 | |
| liaS | Sensor histidine kinase | K11617 | AXF34596 | |
| desR | Response regulator | K07693 | AXF34697 | |
| desK | Sensor histidine kinase | K07778 | AXF35550 | |
| degU | Response regulation | K07692 | AXF34832 | |
| degS | Sensor histidine kinase | K07777 | AXF34833 | |
|
| ||||
| OmpR | resE | Sensor histidine kinase | K07651 | AXF33616 |
| resD | Response regulator | K07775 | AXF33617 | |
| phoR | Phosphate regulon histidine kinase | K07636 | AXF34207 | |
| phoB1/phoP | Alkaline phosphatase synthesis response regulator | K07658 | AXF34208 | |
| bceS | Bacitracin resistance histidine kinase | K11629 | AXF34327 | |
| bceR | Bacitracin resistance response regulator | K11630 | AXF34328 | |
| cssR | Response regulator | K07770 | AXF34586 | |
| cssS | Sensor histidine kinase | K07650 | AXF34587 | |
| yxdK | Sensor histidine kinase | K11633 | AXF35248 | |
| yxdJ | Response regulator | K11634 | AXF35249 | |
| vicK | Sensor histidine kinase | K07652 | AXF35323 | |
| vicR | Response regulator | K07668 | AXF35575 | |
|
| ||||
| Unknown | ycbA/glnK | Sensor histidine kinase | K07717 | AXF35385 |
| ycbB/glnL | Response regulator | K07719 | AXF31592 | |
| yesM | Sensor histidine kinase | K07718 | AXF31995 | |
| yesN | Response regulator | K07720 | AXF31996 | |
3.9.5. ABC Transport Systems
Another prominent feature of the genome of strain DM2 is the powerful membrane transport systems, particularly the ABC transport systems. A total of 310 genes are related to the membrane transport systems. Among them, 138 genes encode the ABC transporters, including ATP-binding protein, substrate-binding proteins, and permeases (Table 8). ABC transporters play important roles in active transmembrane transport, acting as alkanesulfonate transporters, glycine betaine/proline phosphate transporters, amino acid transporters, and osmoprotectants. ABC transporters mediate the transport of glutamine/cystine/D-methionine, maltose/maltodextrin/galactose oligomer, raffinose/stachyose/melibiose, oligopeptide, dipeptide, biotin, and bacitracin, as well as iron complex/iron II, zinc, manganese, and Na+ (Table 6). In addition, the DM2 genome contains several phosphotransferase systems (PTS), which is a major carbohydrate active transport system, indicating that these transporters are responsible for the carbohydrate transport into cells.
Table 8.
The ABC transport systems in the genome of B. subtilis DM2.
| Genes | Description | KOs | Protein ID |
|---|---|---|---|
| ssuA | Sulfonate transport system substrate-binding protein | K15553 | AXF32171 |
| ssuC | Sulfonate transport system permease protein | K15554 | AXF32172 |
| ssuB | Sulfonate transport system ATP-binding protein | K15555 | AXF32170 |
| proX | Glycine betaine/proline transport substrate-binding protein | K02002 | AXF31645 |
| proW | Glycine betaine/proline transport permease protein | K02001 | AXF31644 |
| proV | Glycine betaine/proline transport ATP-binding protein | K02000 | AXF31643 |
| msmE | Raffinose/stachyose/melibiose substrate-binding protein | K10117 | AXF34316/AXF34543 |
| msmF | Raffinose/stachyose/melibiose transport permease protein | K10118 | AXF34317/AXF34542 |
| msmG | Raffinose/stachyose/melibiose transport permease protein | K10119 | AXF34318/AXF34541 |
| msmK | Multiple sugar transport system ATP-binding protein | K10112 | AXF34539/AXF35170 |
| rbsB | Ribose transport system substrate-binding protein | K10439 | AXF34880 |
| rbsC | Ribose transport system permease protein | K10440 | AXF34879 |
| rbsD | D-Ribose pyranase | K06726 | AXF34877 |
| rbsA | Ribose transport system ATP-binding protein | K10441 | AXF34878 |
| pstS | Phosphate transport system substrate-binding protein | K02040 | AXF33794 |
| pstC | Phosphate transport system permease protein | K02037 | AXF33793 |
| pstA | Phosphate transport system permease protein | K02038 | AXF33792 |
| pstB | Phosphate transport system ATP-binding protein | K02036 | AXF33791 |
| yxeM | Amino acid transport system substrate-binding protein | K16961 | AXF35234 |
| yxeN | Amino acid transport system permease protein | K16962 | AXF35233 |
| yxeO | Amino acid transport system ATP-binding protein | K16963 | AXF35232 |
| fhuD | Iron complex transport system substrate-binding protein | K02016 | AXF31727/AXF32049/AXF34604/AXF34618 |
| fhuB | Iron complex transport system permease protein | K02015 | AXF31724/AXF31725/AXF32047/AXF32048/AXF34603/AXF34617 |
| fhuC | Iron complex transport system ATP-binding protein | K02013 | AXF31726/AXF32046/AXF35542/AXF34616 |
| znuA | Zinc transport system substrate-binding protein | K09815 | AXF31630/AXF33983 |
| znuB | Zinc transport system permease protein | K09816 | AXF31632 |
| znuC | Zinc transport system ATP-binding protein | K09817 | AXF31631 |
| troA | Manganese/zinc/iron transport substrate-binding protein | K11707 | AXF34365 |
| troC | Manganese/zinc/iron transport system permease protein | K11708 | AXF34363 |
| troD | Manganese/zinc/iron transport system permease protein | K11709 | AXF34362 |
| troB | Manganese/zinc/iron transport system ATP-binding protein | K11710 | AXF34364 |
| bioY | Biotin transport system substrate-specific component | K03523 | AXF32324/AXF34491 |
| ecfT | Energy-coupling factor transport system permease protein | K16785 | AXF31508/AXF32671 |
| ecfA1 | Energy-coupling factor transport ATP-binding protein | K16786 | AXF31506 |
| ecfA2 | Energy-coupling factor transport ATP-binding protein | K16787 | AXF31507/AXF32672 |
| natB | Sodium transport system permease protein | K09696 | AXF31622 |
| natA | Sodium transport system ATP-binding protein | K09697 | AXF31621 |
| bceB | Bacitracin transport system permease protein | K11632 | AXF34325 |
| bceA | Bacitracin transport system ATP-binding protein | K11631 | AXF34326 |
4. Conclusion
The B. subtilis strain DM2 isolated from petroleum-contaminated soil on the Tibetan Plateau displays a great capacity to degrade petroleum at a low temperature. The complete genome sequencing and genomic analysis of strain DM2 help us to unravel its biological features that enable it to successfully utilize hydrocarbons as carbon source and potentially withstand other environmental challenges. Strain DM2 is clustered as a separate and a higher evolutionary clade in the phylogenetic tree based on 24 housekeeping protein sequences, implying its unique position with respect to other B. subtilis strains. Strain DM2 possesses the largest genome and the most protein-coding genes relative to the other compared B. subtilis strains. DDH values show that strain DM2 belongs to B. subtilis subsp. subtilis, but significant variations in the genome occurred with respect to the other strains or subspecies. Comparative genomic analysis identified the core proteome common to strain DM2, model strain B. subtilis subsp. subtilis 168, and other B. subtilis strains. Strain DM2 possesses almost the same strain-specific proteins as strain MJ01, which is another oil-degrading B. subtilis strain, unlike strain B. subtilis subsp. subtilis 168. Furthermore, many strain DM2-specific proteins were also identified, such as carboxymuconolactone decarboxylase family protein, gfo/Idh/MocA family oxidoreductases, GlsB/YeaQ/YmgE family stress response membrane protein, HlyC/CorC family transporters, LLM class flavin-dependent oxidoreductase, and LPXTG cell wall anchor domain-containing protein. Most of these strain-specific proteins have been shown to be involved in the pathways related to stress response, signaling, and hydrocarbon degradation, suggesting that the main feature of the DM2 genome is the evolutionary occurrence of many genes related to environmental adaptation and carbon utilization. The genomic information provided by the present study might help us to further reveal the genetic and genomic characters of Bacillus subtilis, which is a ubiquitous and important bacterial species.
Acknowledgments
This work was supported by the National Natural Science Funds of China (Grant Nos. 31760110 and 31560121).
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Supplementary Materials
Supplementary Table S1: comparative analysis of COG categories between strain DM2 and strain 168. Supplementary Table S2: list of the strain DM2-specific proteins versus strains PY97, 168, and MJ01.
References
- 1.Yang Y., Wang J., Liao J., Xie S., Huang Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Applied Microbiology and Biotechnology. 2015;99(4):1935–1946. doi: 10.1007/s00253-014-6074-z. [DOI] [PubMed] [Google Scholar]
- 2.Alcaraz L., Moreno-Hagelsieb G., Eguiarte L. E., Souza V., Herrera-Estrella L., Olmedo G. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics. 2010;11(1):p. 332. doi: 10.1186/1471-2164-11-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Earl A. M., Losick R., Kolter R. Ecology and genomics of Bacillus subtilis . Trends in Microbiology. 2008;16(6):269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeo I. C., Lee N. K., Hahm Y. T. Genome sequencing of Bacillus subtilis SC-8, antagonistic to the Bacillus cereus group, isolated from traditional Korean fermented-soybean food. Journal of Bacteriology. 2012;194(2):536–537. doi: 10.1128/JB.06442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng W., Chen G., Tang Z., Wu H., Shu L., Liang Z. Draft genome sequence of Bacillus subtilis GXA-28, a thermophilic strain with high productivity of poly-γ-glutamic acid. Genome Announcements. 2014;2(6, article e01259-14) doi: 10.1128/genomea.01259-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schyns G., Serra C. R., Lapointe T., et al. Genome of a gut strain of Bacillus subtilis . Genome Announcements. 2013;1(1, article e00184-12) doi: 10.1128/genomea.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishnoi U., Polson S. W., Sherrier D. J., Bais H. P. Draft genome sequence of a natural root isolate, Bacillus subtilis UD1022, a potential plant growth-promoting biocontrol agent. Genome Announcements. 2015;3(4, article e00696-15) doi: 10.1128/genomea.00696-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gond S. K., Bergen M. S., Torres M. S., White Jr J. F. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiological Research. 2015;172:79–87. doi: 10.1016/j.micres.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Jeong H., Sim Y. M., Park S. H., Choi S. K. Complete genome sequence of Bacillus subtilis strain ATCC 6051a, a potential host for high-level secretion of industrial enzymes. Genome Announcements. 2015;3(3, article e00532-15) doi: 10.1128/genomea.00532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasc C., Richard J. Y., Peyret P. Genome sequence of Bacillus subtilis strain HUK15, isolated from hexachlorocyclohexane-contaminated soil. Genome Announcements. 2016;4(2, article e00273-16) doi: 10.1128/genomea.00273-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Q., Li S., Lu X., Zhang X., Wang P., Ma P. Complete genome sequence of Bacillus subtilis BAB-1, a biocontrol agent for suppression of tomato gray mold. Genome Announcements. 2014;2(4, article e00744-14) doi: 10.1128/genomea.00744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahimi T., Niazi A., Deihimi T., Taghavi S. M., Ayatollahi S., Ebrahimie E. Genome annotation and comparative genomic analysis of Bacillus subtilis MJ01, a new bio-degradation strain isolated from oil-contaminated soil. Functional & Integrative Genomics. 2018;18(5):533–543. doi: 10.1007/s10142-018-0604-1. [DOI] [PubMed] [Google Scholar]
- 13.Khatri I., Sharma S., Ramya T. N. C., Subramanian S. Complete genomes of Bacillus coagulans S-lac and Bacillus subtilis TO-A JPC, two phylogenetically distinct probiotics. PLoS ONE. 2016;11(6, article e0156745) doi: 10.1371/journal.pone.0156745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borriss R., Danchin A., Harwood C. R., et al. Bacillus subtilis, the model Gram-positive bacterium: 20 years of annotation refinement. Microbial Biotechnology. 2018;11(1):3–17. doi: 10.1111/1751-7915.13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earl A. M., Losick R., Kolter R. Bacillus subtilis genome diversity. Journal of Bacteriology. 2007;189(3):1163–1170. doi: 10.1128/JB.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G., Wang X. C., Tian W. H., et al. Genomic diversity and evolution of Bacillus subtilis . Biomedical and Environmental Science. 2015;28(8):620–625. doi: 10.3967/bes2015.087. [DOI] [PubMed] [Google Scholar]
- 17.Zeigler D. R. The genome sequence of Bacillus subtilis subsp. spizizenii W23: insights into speciation within the B. subtilis complex and into the history of B. subtilis genetics. Microbiology. 2011;157(7):2033–2041. doi: 10.1099/mic.0.048520-0. [DOI] [PubMed] [Google Scholar]
- 18.Parthipan P., Preetham E., Machuca L. L., Rahman P. K. S. M., Murugan K., Rajasekar A. Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Frontiers in Microbiology. 2017;8(193) doi: 10.3389/fmicb.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kureel M. K., Geed S. R., Giri B. S., Rai B. N., Singh R. S. Biodegradation and kinetic study of benzene in bioreactor packed with PUF and alginate beads and immobilized with Bacillus sp. M3. Bioresource Technology. 2017;242:92–100. doi: 10.1016/j.biortech.2017.03.167. [DOI] [PubMed] [Google Scholar]
- 20.Tuo B., Yan J., Fan B., Yang Z., Liu J. Biodegradation characteristics and bioaugmentation potential of a novel quinoline-degrading strain of Bacillus sp. isolated from petroleum-contaminated soil. Bioresource Technology. 2012;107:55–60. doi: 10.1016/j.biortech.2011.12.114. [DOI] [PubMed] [Google Scholar]
- 21.Kesel S., Moormann F., Gümperlein I., et al. Draft genome sequence of the biofilm-producing Bacillus subtilis strain B-1, isolated from an oil field. Genome Announcements. 2014;2(6, article e01163-14) doi: 10.1128/genomea.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latha R., Kalaivani R. Bacterial degradation of crude oil by gravimetric analysis. Advance in Applied Science Research. 2012;3:2789–2795. [Google Scholar]
- 23.Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder J. W., Simmons L. A. Complete genome sequence of Bacillus subtilis strain PY79. Genome Announcements. 2013;1(6, article e01085-13) doi: 10.1128/genomea.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinidis K. T., Tiedje J. M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proceedings of the National Academy of Sciences. 2004;101(9):3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantinidis K. T., Braff J., Karl D. M., Delong E. F. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Applied and Environmental Microbiology. 2009;75(16):5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busse H.-J., Tindall B. J., Ludwig W., Rosselló-Móra R., Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. International Journal of Systematic and Evolutionary Microbiology. 2010;60(1):249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14(1):p. 60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lateef A., Adelere I. A., Gueguim-Kana E. B. Bacillus safensisLAU 13: a new source of keratinase and its multi-functional biocatalytic applications. Biotechnology & Biotechnological Equipment. 2015;29(1):54–63. doi: 10.1080/13102818.2014.986360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padaria J. C., Sarkar K., Lone S. A., Srivastava S. Molecular characterization of cellulose-degrading Bacillus pumilus from the soil of tea garden, Darjeeling hills India. Journal of Environmental Biology. 2014;35(3):555–561. [PubMed] [Google Scholar]
- 31.Yumoto I., Yamaga S., Sogabe Y., et al. Bacillus krulwichiae sp. nov., a halotolerant obligate alkaliphile that utilizes benzoate and m-hydroxybenzoate. International Journal of Systematic and Evolutionary Microbiology. 2003;53(5):1531–1536. doi: 10.1099/ijs.0.02596-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Li P., Luo L., Simpson D. J., Gänzle M. G. Daqu fermentation selects for heat-resistant enterobacteriaceae and bacilli. Applied and Environmental Microbiology. 2018;84(21, article e01483-18) doi: 10.1128/aem.01483-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langille M. G. I., Hsiao W. W. L., Brinkman F. S. L. Detecting genomic islands using bioinformatics approaches. Nature Reviews Microbiology. 2010;8(5):373–382. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 34.Bowman J. S., Deming J. W. Alkane hydroxylase genes in psychrophile genomes and the potential for cold active catalysis. BMC Genomics. 2014;15(1):p. 1120. doi: 10.1186/1471-2164-15-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skerker J. M., Prasol M. S., Perchuk B. S., Biondi E. G., Laub M. T. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PloS Biology. 2005;3(10, article e334) doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierman W. C., Feldblyum T. V., Laub M. T., et al. Complete genome sequence of Caulobacter crescentus . Proceedings of the National Academy of Sciences. 2001;98(7):4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romling U., Gomelsky M., Galperin M. Y. C-di-GMP: the dawning of a novel bacterial signalling system. Molecular Microbiology. 2005;57(3):629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 38.Wickner W., Schekman R. Protein translocation across biological membranes. Science. 2005;310(5753):1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 39.Ferrer M., Chernikova T. N., Yakimov M. M., Golyshin P. N., Timmis K. N. Chaperonins govern growth of Escherichia coli at low temperatures. Nature Biotechnology. 2003;21(11):1266–1267. doi: 10.1038/nbt1103-1266. [DOI] [PubMed] [Google Scholar]
- 40.Abat J. K., Deswal R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics. 2009;9(18):4368–4380. doi: 10.1002/pmic.200800985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: comparative analysis of COG categories between strain DM2 and strain 168. Supplementary Table S2: list of the strain DM2-specific proteins versus strains PY97, 168, and MJ01.
Data Availability Statement
The data used to support the findings of this study are included within the article.