Abstract
Background
A robust circadian clock has been implicated in plant resilience, resource-use efficiency, competitive growth and yield. A huge number of physiological processes are under circadian control in plants including: responses to biotic and abiotic stresses; flowering time; plant metabolism; and mineral uptake. Understanding how the clock functions in crops such as Triticum aestivum (bread wheat) and Brassica napus (oilseed rape) therefore has great agricultural potential. Delayed fluorescence (DF) imaging has been shown to be applicable to a wide range of plant species and requires no genetic transformation. Although DF has been used to measure period length of both mutants and wild ecotypes of Arabidopsis, this assay has never been systematically optimised for crop plants. The physical size of both B. napus and T. aestivum led us to develop a representative sampling strategy which enables high-throughput imaging of these crops.
Results
In this study, we describe the plant-specific optimisation of DF imaging to obtain reliable circadian phenotypes with the robustness and reproducibility to detect diverging periods between cultivars of the same species. We find that the age of plant material, light regime and temperature conditions all significantly effect DF rhythms and describe the optimal conditions for measuring robust rhythms in each species. We also show that sections of leaf can be used to obtain period estimates with improved throughput for larger sample size experiments.
Conclusions
We present an optimized protocol for high-throughput phenotyping of circadian period specific to two economically valuable crop plants. Application of this method revealed significant differences between the periods of several widely grown elite cultivars. This method also identified intriguing differential responses of circadian rhythms in T. aestivum compared to B. napus; specifically the dramatic change to rhythm robustness when plants were imaged under constant light versus constant darkness. This points towards diverging networks underlying circadian control in these two species.
Electronic supplementary material
The online version of this article (10.1186/s13007-019-0436-6) contains supplementary material, which is available to authorized users.
Keywords: Circadian period, Delayed fluorescence, Free-running conditions, Hexaploid wheat, Oilseed rape, Aging, L:L, D:D, Temperature, Rhythm robustness
Background
A circadian clock is an endogenous oscillator entrained by external temporal cues. Circadian control of gene expression is a ubiquitous feature which appears to have arisen independently in bacteria, fungi, plants and animals [1]. Since the discovery of the first Arabidopsis circadian mutant in 1995 [2], the significance of the circadian clock in plants has become increasingly evident. Approximately 30% of genes in Arabidopsis are predicted to be under circadian control, regulating photosynthetic, metabolic and developmental pathways [3, 4]. Moreover, a selective advantage resulting from a clock which is matched to the exogenous day-length has been demonstrated in mammals, insects, bacteria and plants [5–9].
The most recent model for the molecular control of the Arabidopsis clock is comprised of a series of interlocking negative transcriptional feedback loops regulated by key activators which control the oscillation of clock gene expression [10]. To ascertain the underlying nature of circadian rhythms, a clock-controlled output representing the pace of the clock must be measured in constant (free-running) conditions. Previously, this research has been conducted by studying leaf movement rhythms or by following luciferase gene expression under the control of a circadian regulated promoter [11–13]. Delayed fluorescence (DF) imaging provides an alternative to these methods that does not require plant transformation. It has previously been shown to work in a variety of plants for which leaf movement assays are not feasible [14, 15]. Delayed fluorescence occurs when excited electrons in photosystem II (PSII) undergo spin-conversion to a triplet excited state before charge recombination allows them to return to their ground state releasing light energy [16]. Measurements of DF have been correlated with the photosynthetic state of PSII [17] and the amount of DF production is regulated by the circadian clock. DF can be measured with a low-light imaging system identical to that used for luciferase imaging and output rhythms have been shown to oscillate with a comparable period to those estimated from luciferase reporter experiments [14]. The output from a DF experiment is a waveform which has parameters that can be mathematically defined and therefore quantified. These parameters include ‘period’ (the time taken to complete one cycle), ‘phase’ (the time of day at which this peaks) and ‘amplitude’ (the distance between the peak and the baseline of the oscillation). Important to circadian dynamics is also the idea of ‘rhythm robustness’ i.e. whether these parameters change over time. In this paper, rhythm robustness was assessed by: the percentage of samples classified as rhythmic; the relative amplitude error (RAE); the period coefficient of variation (CV) and the average period error threshold (all defined in Additional file 1). Together, these parameters allow the effects of different imaging conditions to be quantified.
As DF measurement is correlated with the oscillations in photosynthetic status of PSII, leaf material is the logical choice for a representative sample. Rhythms have been shown to persist in excised leaves in several species [18–22]. However, previous research has demonstrated that independent clocks run at different periods throughout the plant under constant conditions, coordinated by a degree of intercellular coupling [23–27]. The extent to which the clock is affected by dissecting leaf material into small segments is investigated in this paper.
Alongside these spatial differences, the clock has also been shown to be temporally dynamic and is affected by the life history of the plant. Both the systemic age of the plant and the ‘emergence age’ of the individual leaves on a plant have been reported to effect the clock in Arabidopsis, with increasing age associated with period reduction [28]. Conversely, the timing of leaf senescence has also been shown to be directly regulated by core circadian genes [29].
In addition to this endogenous entrainment, the clock is also responsive to external stimuli; the most well characterized of which are light and temperature cues. Increasing light intensity causes a shortening of period in free-running conditions [30–32] and these rhythms rapidly dampen in amplitude under continuous darkness [33]. Circadian systems are relatively buffered against temperature changes compared to other biochemical reactions but are not completely independent of it [34]. Period shortening of 1.8–4.2 h have been reported following temperature increases from 17 to 27 °C determined by both leaf-movement assays and luciferase reporters under circadian regulation in Arabidopsis [31, 35, 36]. Seedlings grown at 17 °C also have rhythms with lower period variability and RAE values than plants grown at 27 °C [35, 37]. The extent to which rhythms are temperature compensated is described using the inverse of the temperature coefficient Q10; the change in the rate of a process over a temperature change of 10 °C [38].
Here we present an optimized protocol for high-throughput phenotyping of circadian period using two crop plant models; Triticum aestivum (bread wheat) and Brassica napus (oilseed rape). B. napus (AACC) and T. aestivum (AABBDD) are both recent polyploids still undergoing genomic rearrangements. The contribution of each genome to clock function remains to be investigated. B. napus is a dicot recently diverged from Arabidopsis [39] and so is likely to have clock homologs with similar functions. T. aestivum is a monocot with an incompletely understood clock mechanism [40]. These species therefore provide interesting insights into two genetically diverse families. Both T. aestivum and B. napus have been influenced by human domestication, genome duplication events and geographical speciation as the use of these crops became globalized. The specific and combined effects of these factors on the control of the clock is yet to be investigated.
Within this paper we show that both the age of the plant and the developmental age of leaves have significant effects on period with older material displaying shorter rhythms. To make our method high-throughput whilst still providing reliable rhythms, tissues were segmented into various sizes and compared to whole leaf samples. We identify regions of the plant leaves which are the most robustly rhythmic and give the most consistent period estimates. Both the light regime and temperature conditions also had large effects on period estimation and we describe conditions optimal for each species.
Finally, we applied our optimized, high-throughput DF method to investigate differences between elite cultivars in both B. napus and T. aestivum and demonstrate it to be a useful tool for assaying circadian rhythms in these crop species.
Results
Circadian variability due to leaf development and age of plant
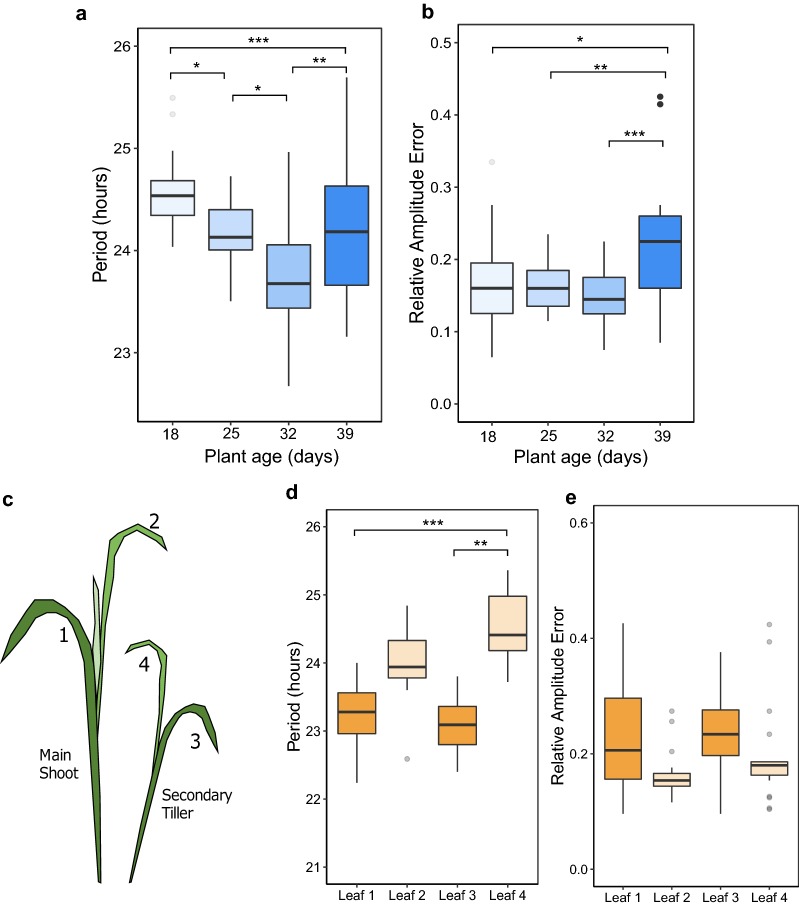
We tested the effect of both plant and leaf aging on period estimates from Brassica and wheat seedlings. Previous studies in Arabidopsis have reported that the pace of the clock increases as the plant ages and that earlier emerged leaves have a shorter period than those which emerge later within the same individual [28]. Our results mirror these findings for young wheat and Brassica plants, however this association was lost for older material (Fig. 1a). For wheat we calculated period estimates from the second leaf of plants at 18, 25, 32 and 39 days after sowing and show that between 18 and 32 days period decreases linearly at a rate of approximately half an hour per week while maintaining a near constant relative amplitude error (RAE) (Fig. 1a, b). However, in leaves from 39 day old plants there was an increase in both average period and relative amplitude error, potentially due to metabolic changes as a leaf changes from a source to a sink tissue or due to the onset of senescence in these samples. A one-way analysis of variance yielded a significant effect of wheat age on both period and RAE (F(3,90) = 12.13, p < 0.001) and (F(3,90) = 7.018, p < 0.001) respectively. Based on our investigation, we recommend using plants between 25 and 32 days after sowing. At 25 days 100% of samples were classified as rhythmic and period CV was 1.52 h. 32 day old samples were also robust, having the lowest RAE (0.15) and period error (0.43) averages (see Additional file 1: Supplementary material S1).
Fig. 1.
DF rhythms in wheat change with the age of the plant and between leaves on the same plant. The wheat plant age experiment (a, b) used ‘leaf 2’ from plants grown for 18, 25, 32 or 39 days. Blue boxplots show differences in period (a) and RAE (b) at each plant age. The wheat leaf variation experiment used 4 leaves sampled from 25 day old plants following the leaf numbering system described in c. Orange boxplots show differences in period (d) and RAE (e) at each leaf age. Colour scales reflect an ageing gradient with lighter colours representing younger material. Data represents results from two imaging cabinets run in parallel as technical replicates and normalised for the between-cabinet effects. Period estimates were calculated using FFT-NLLS (BAMP de-trended data, 24–120 h cut-off). N values reflect the number of samples for which period was estimated out of the total number of individuals sampled. Age 18 (N = 26/26), age 25 (N = 24/24), age 32 (N = 25/26), age 39 (N = 19/23). Leaf 1 (N = 22/22), leaf 2 (N = 22/22), leaf 3 (N = 22/22), leaf 4 (N = 21/22). Significance codes: ***p < 0.001, **p < 0.01 *p < 0.05
In a separate experiment, we analyzed 4 leaves from 25 day old wheat plants as is shown in Fig. 1c, where leaves 1 and 3 were the oldest leaves and leaves 2 and 4 the second oldest leaves from the main and secondary tiller, respectively. There was a statistically significant difference between the mean periods at each leaf age (one-way ANOVA (F(3,83) = 7.434, p < 0.001). Within each tiller pair, the older leaf had a shorter period than the younger leaf and had higher RAE averages (Fig. 1d, e). The mean period for leaf 4 (24.50 h) was found to be significantly longer than both leaf 1 (23.15 h) and leaf 3 (23.32 h) (Tukey HSD). We recommend using leaf 2 as it had the best overall circadian robustness with regards to the % samples returned (100%), RAE (0.18) and period error (0.50) (Additional file 1: Supplementary material S2).
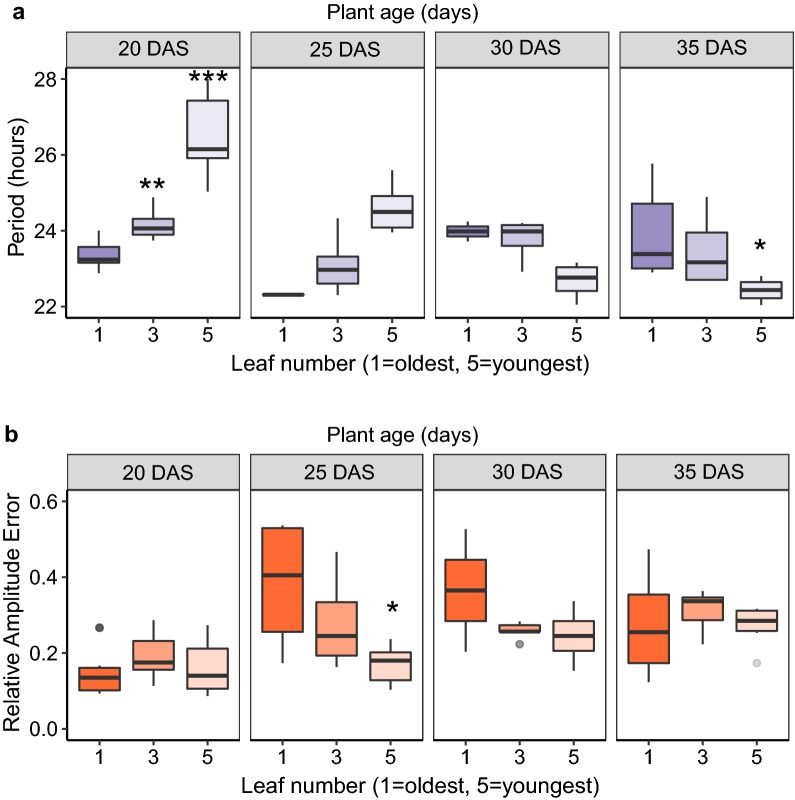
For Brassica seedlings, plants were grown to 4 different ages: 20, 25, 30 and 35 days after sowing and leaf 1, 3 and 5 were sampled in the same experiment with leaf 1 being the earliest emerged leaf and leaf 5 the most recently emerged leaf (Fig. 2). We conducted a nested ANOVA to test the effects of both plant age and within-plant leaf-age on period. We found that variation in plant-age had a significant effect on period, with increasing age causing a shortening of period (F(3,53) = 8.48, p < 0.001). The nested effect of leaf age within each plant age-group was also found to be significant (F(8,53) = 5.45, p < 0.001). The largest difference between leaves in each age-group was seen for 20 day old plants where a difference of 3.14 h was observed between leaf 1 and 5 (p < 0.001, Tukey HSD) (Fig. 2a). Brassica plant-age was also found to have a significant effect on RAE averages with younger plants having a lower mean RAE (F(3,53) = 5.953, p < 0.01) (Fig. 2b). Additional file 1: Supplementary material S3 shows robustness statistics for all plant ages and leaf-ages tested. We recommend using leaf 1 from 20 day old plants as they had the lowest RAE (0.15) and period error threshold (0.47).
Fig. 2.
DF rhythms in Brassica change with the age of the plant and between different leaf ages. Brassica seedlings were grown to 4 different ages: 20, 25, 30 and 35 days after sowing. Leaves 1, 3 and 5 were sampled from each plant in the experiment with leaf 1 being the earliest emerged leaf and leaf 5 the most recently emerged leaf. Boxplots show differences in period (a) and RAE (b) for each leaf age within each plant age. Colour scales reflect an ageing gradient with lighter colours representing younger material. Periods and RAE estimates were calculated using FFT NLLS (BAMP dtr, 24–120 h cut-off). Data represents results from two imaging cabinets run in parallel as technical replicates and normalised for the between-cabinet effects. Age 20: leaf 1 (N = 6/6), leaf 3 (N = 6/6), leaf 5 (N = 6/6). Age 25: leaf 1 (N = 4/6), leaf 3 (N = 6/6), leaf 5 (N = 6/6). Age 30: leaf 1 (N = 2/6), leaf 3 (N = 5/6), leaf 5 (N = 6/6). Age 35: leaf 1 (N = 6/6), leaf 3 (N = 6/6), leaf 5 (N = 6/6). Significance codes: ***p < 0.001, **p < 0.01 *p < 0.05, all significance markers are relative to leaf 1 at each age
To approximate the period shortening due to plant aging in Brassica we followed the changes in average period in leaf 5 across plant ages from 20 days after sowing to 30 days after sowing. Our analysis revealed that period shortened by approximately 3 h per week from a mean of 26.50 h (SD 1.17) to 22.38 h (SD 0.62).
Finding an optimal size of leaf sample
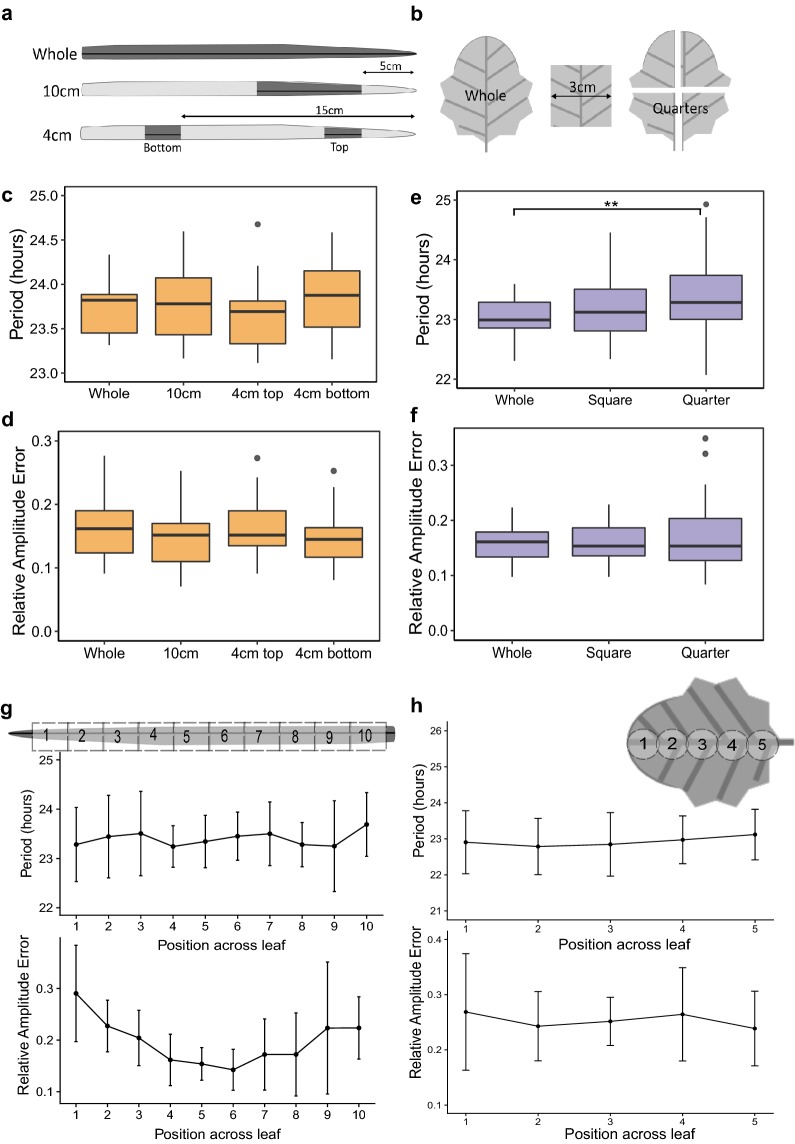
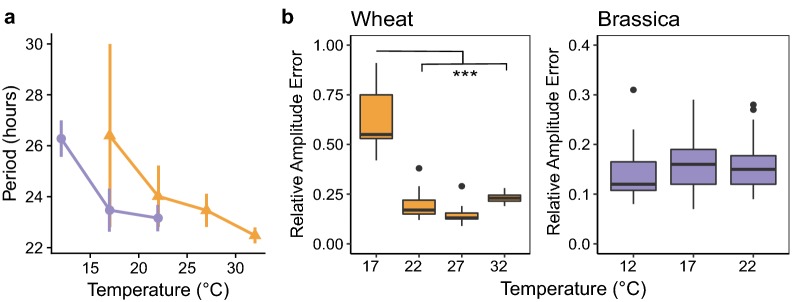
We needed to identify representative leaf sections which allowed a sufficient number of samples to be analyzed on one plate without compromising the robustness of rhythms for period estimation. For wheat, we selected leaf 2 from 25 day old plants and analyzed the periods and circadian robustness given by whole leaves compared to leaves cut into 10 cm sections and leaves cut into 4 cm sections as shown in Fig. 3a. By taking 4 cm samples from 2 regions on the same leaf (5 or 15 cm down from the tip) we could investigate changes in period across the length of the leaf. For Brassica seedlings we selected leaf 1 from 21 day old plants and then kept them whole, took 3 cm square samples from the centre or quartered them (Fig. 3b). This helped inform whether any changes in circadian characteristics were a result of size reduction or from sub-sectioning regions of the leaf. Our data showed that period and RAE averages were not significantly affected by cutting samples in wheat (Fig. 3c, d) (F(3,75) = 2.066, p > 0.1, one-way ANOVA). However, cutting Brassica leaves did significantly affect period estimates; quartered segments had a slightly longer period than whole samples (F(2,113) = 5.46, p < 0.01, one-way ANOVA, Tukey HSD (Whole-Quarter p < 0.01) but RAE means were similar (Fig. 3e, f) (Additional file 1: Supplementary material S4). From this data we recommend using 10 cm segments for wheat and 3 cm square sections for Brassica imaging as these gave similar results to whole leaves and increased throughput by 44% for Brassica and 100% for wheat.
Fig. 3.
Cut sections of leaf material can be used to accurately make period estimates. The second leaf from 25 day old wheat seedlings was either left whole or sectioned into 10 cm or 4 cm fragments cut either 5 cm or 15 cm from the tip (shown as dark grey sections in a). The first leaf from 21 day old Brassica plants was either left whole, sectioned into a 3 cm square or quartered (b). Orange boxplots show differences in period (c) and RAE (d) for wheat sections. Purple boxplots show differences in period (e) and RAE (f) for brassica sections. Period and RAE were estimated using FFT NLLS, BAMP dtr, 24–120 h cut-off. Whole leaves were digitally sectioned along the axis of the leaf post image-acquisition. Wheat period and RAE means for each section are shown in g. Brassica period and RAE means are plotted corresponding to the sectioning shown in h. Error bars show standard deviation. Data represents results from two experiments normalised for the between-experiment effects. Wheat: Whole (N = 15/15), 10 cm (N = 23/23), 4 cm top (N = 20/21), 4 cm bottom (N = 22/22). Brassica: Whole (N = 20/20), Square (N = 21/21), Quarter (N = 75/76). Significance codes: **p < 0.01
We next wanted to investigate whether period estimates changed across the axis of the leaf. We selected only the whole leaf images and digitally sectioned them into 10 or 5 regions of interest for wheat and Brassica leaves respectively (Fig. 3g, h). Using this approach we observed an average within-leaf variance of 0.45 h in wheat and 0.42 h in Brassica leaves. This variation was larger than the leaf-to-leaf variation determined for wheat (0.04 h) and Brassica (0.3 h) leaves. The mean period and RAE for each section across these leaves was calculated and plotted (Fig. 3g, h). No significant difference was observed between the period of wheat or Brassica segments; however the RAE was significantly different across wheat leaves (F(9,139) = 6.077, p < 0.001, One-way ANOVA). The middle segments (4, 5, 6, 7 and 8) had significantly lower RAE averages compared to the tip (segment 1) suggesting that this middle region may give the most robust DF rhythms.
Constant free-running conditions: dark versus light
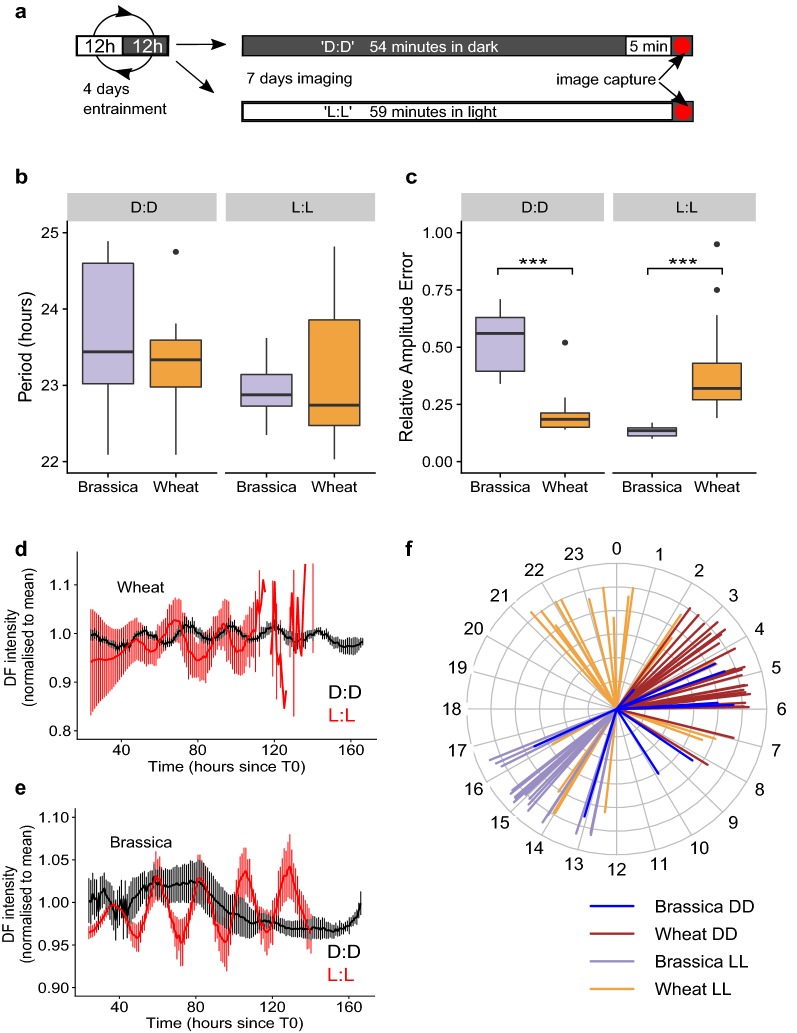
Two light regimes were tested which allowed free-running DF rhythms to be recorded. We entrained plants for 4 days in 12:12 light:dark (L:D) cycles at 22 °C before sampling and imaging every hour under constant conditions, as described in Fig. 4a. In both D:D and L:L conditions the exposure time was kept at 1 min. Figure 4b shows the striking differences in period estimate accuracy obtained from wheat and Brassica under the two light regimes. For wheat, periods from leaves under a D:D regime had much lower variance than those under the L:L regime (D:D mean 23.29 h, SD 0.53; L:L mean 23.54 h, SD 3.19). For Brassica the opposite was observed; rhythms were more accurate under L:L (D:D mean 24.89 h, SD 2.91; L:L mean 22.92 h, SD 0.31). A shortening of period was observed for both Brassica and wheat under L:L compared to D:D based on median values, however the increased variance observed within wheat-L:L and Brassica-D:D resulted in these differences having low significance (wheat t(23.16) = −0.38, p > 0.5; Brassica t(10.14) = 2.24, p = 0.048 Welch’s t test).
Fig. 4.
Effect of either L:L or D:D free-running light conditions on DF rhythms. Wheat and Brassica seedlings were entrained for 4 days in L:D at 22 °C before sections were cut, plated and imaged. A D:D free run consisted of a loop of 54 min of darkness followed by 5 min of light exposure and then image capture. A L:L free-run consisted of 59 min of light exposure before image capture (a). Boxplots of period (b) and RAE (c) are shown for Brassica and wheat in D:D and L:L conditions where Brassica data is displayed in purple and wheat data in orange. Period and RAE were estimated using FFT NLLS, BAMP dtr, 24–120 h cut-off. Examples of oscillation traces are shown in D:D (black lines) and L:L (red lines) for wheat (d) and Brassica (e). Thick lines represent the mean trace of 6 mean-normalised individuals with error bars representing standard deviations. Estimated individual circadian phases are shown in the clock plot in f where the length of the line reflects the inverse circadian phase error (longer lines imply more confidence in the phase prediction). All phase estimates are relative to 0 where 0 represents entrainment dawn and 12 represents dusk. D:D Brassica (N = 12/18); D:D Wheat (N = 24/24); L:L Brassica (N = 18/18); L:L Wheat (N = 23/24). Data is consistent with additional preliminary experiments which can be seen in Additional file 1: Supplementary materials S11 and S12. Significance codes: ***p < 0.001
RAE ratios reflected the accuracy seen in period estimation between the regimes (Fig. 4c). RAE averages were smaller in D:D for wheat (D:D mean 0.20, SD 0.08; L:L mean 0.38, SD 0.18) and in L:L for Brassica (D:D mean 0.53, SD 0.13; L:L mean 0.13, SD 0.02). RAE differences were significant between regimes for both species (Wheat t(29.23) = −4.38, p < 0.001; Brassica t(10.39) = 9.92, p < 0.001 Welch’s t test). Figure 4d, e show mean oscillation traces which demonstrate how DF rhythms were sustained in wheat and Brassica under the different light conditions. Interestingly, DF rhythms also had a dawn-phased peak in wheat and a dusk-phased peak in Brassica which became shifted as different light conditions were applied (Fig. 4f).
Wheat samples under the D:D conditions returned 100% of samples from period estimation (L:L = 95.83%), an average RAE ratio of 0.20 (L:L = 0.38), a period CV of 2.25% (L:L = 13.53%) and a period error threshold of 0.55 (L:L = 1.17). For Brassica, rhythms under the L:L regime returned 100% from period estimation (D:D = 66.67%), a RAE average of 0.13 (D:D = 0.53), a period CV of 1.34% (D:D = 11.80) and a period error threshold of 0.41 (D:D = 1.62). See Additional file 1: Supplementary material S5. We would therefore recommend running wheat DF experiments under D:D conditions and Brassica DF experiments under L:L.
Finding an optimum free-running temperature
To investigate the effect of temperature on period and rhythm robustness we tested Brassica and wheat seedlings at a range of constant temperatures. We used the optimal conditions from the variables so far tested and entrained each batch of plants at the imaging temperature for 4 days prior to imaging (see Methods). Both Brassica and wheat experienced an acceleration of the clock at higher temperatures, with the rate increasing most dramatically at lower temperatures (Fig. 5a). Periods decreased from 26.40 h (SD 3.60) at 17 °C to 22.48 h (SD 0.31) at 32 °C in wheat. Periods decreased from 26.28 h (SD 0.72) at 12 °C to 23.16 h (SD 0.52) at 22 °C in Brassica. The temperature coefficient Q10 was calculated as an average across all temperatures (Additional file 1: Supplementary material S7). Q10 was found to be 1.12 for wheat and 1.14 for Brassica indicating a degree of thermal compensation, but to a lesser extent than has been previously reported in Arabidopsis [36]. We next looked at which temperatures gave the best rhythmicity in each crop. Rhythms were most robust in wheat grown at 27 °C: 100% of period estimates were returned, the average RAE ratio was 0.15, period CV was 2.48% and period error threshold was 0.48 (Additional file 1: Supplementary material S6). There was a clear negative trend in period CV as the temperature increased in wheat from 13.63% at 17 °C to 1.38% at 32 °C. Mean RAE at each temperature can be seen in Fig. 5b.
Fig. 5.
Increasing temperature causes a shortening of period and effects rhythm robustness. Wheat and Brassica seedlings were entrained for 4 days in L:D at the temperature being assessed before imaging. Each temperature point represents a separate imaging experiment. Period means decrease with increasing temperatures as is shown in a for Brassica (purple circles) or wheat (orange triangles). Error bars represent standard deviation. Box plots show the effect of temperature on RAE for wheat (orange) or Brassica (purple) (b). Period and RAE were estimated by FFT NLLS, BAMP dtr, 24–120 h cut-off. Wheat: 17 °C (N = 17/23); 22 °C (N = 32/32); 27 °C (N = 11/11); 32 °C (N = 15/15). Brassica: 12 °C (N = 24/24); 17 °C (N = 35/35); 22 °C (N = 30/30). An additional preliminary experiment consistent with these observations can be seen in Additional file 1: Supplementary material S13. Significance codes: ***p < 0.001
Across the temperatures tested Brassica rhythm robustness remained consistent; all samples were returned from FFT-NLLS and RAE, period CV and Period error were similar (Fig. 5b, Additional file 1: Supplementary material S6). We recommend 22 °C for DF using Brassica as it had the lowest period CV of 2.24%.
An optimized DF method can be used in circadian analysis for crops
To see whether the optimized method could be used to investigate circadian differences between cultivars of the same species, we looked at circadian rhythms from seven T. aestivum cultivars and three B. napus cultivars. For the B. napus lines, seeds were obtained from 3 different harvest years to see whether period was constant between batches. The optimized imaging parameters we used for these elite cultivars is outlined in Table 1.
Table 1.
Optimised DF method for circadian phenotyping of Brassica and wheat leaves
| Species | Plant age (days after sowing) | Leaf age (1 = oldest leaf) | Cut sample | Light regime | Temperature (°C) | Throughput (N/imaging cabinet) | % return (from period estimation algorithms) |
|---|---|---|---|---|---|---|---|
| Brassica | 20 | 1 | 3 cm square | L:L | 22 | 36 | 96.3 |
| Wheat | 25 | 2 | 10 cm section | D:D | 27 | 44–48 | 98.8 |
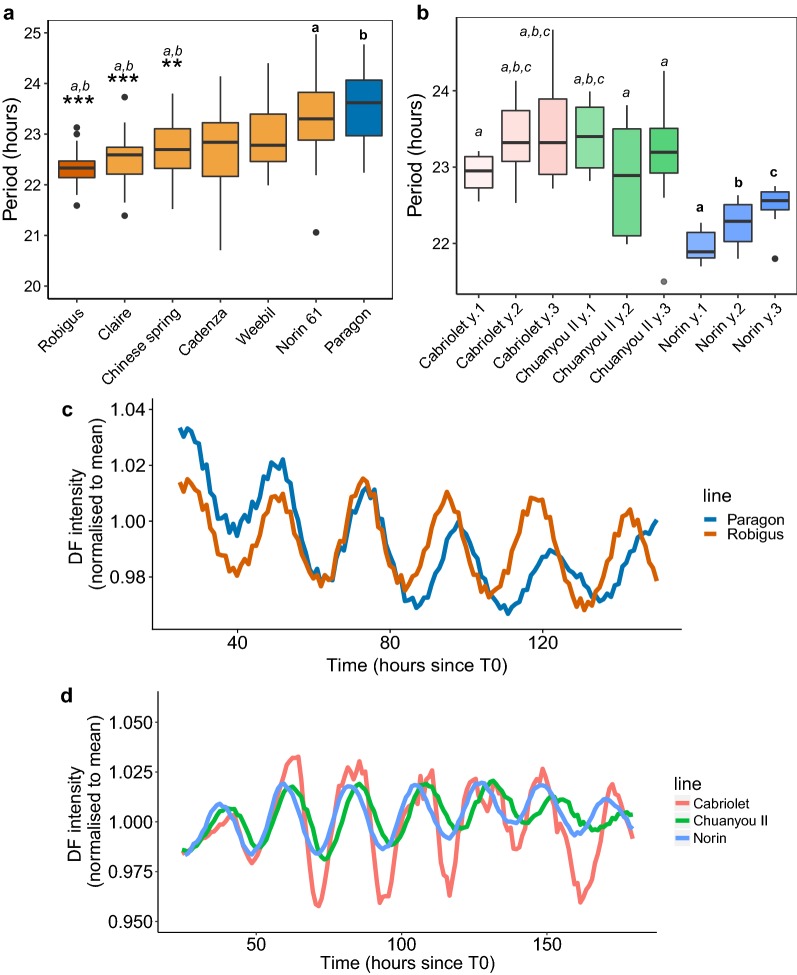
There was significant variation in the periods of the wheat lines tested as shown in Fig. 6a (F(6,152) = 9.81, p < 0.001, one-way ANOVA). A Tukey HSD test showed that Paragon (mean 23.48 h, SD 0.54) and Norin 61 (mean 23.50 h, SD 1.30) both have longer periods than Chinese Spring (mean 22.70 h, SD 0.40), Claire (mean 22.48, SD 0.54) and Robigus (mean 22.32 h, SD 0.34) α = 0.01).
Fig. 6.
DF can be used to measure period differences between elite cultivars in Brassica and wheat. 10 cm sections from the second leaf of 25 day old wheat seedlings were imaged under D:D at 27 °C. 3 cm square sections from the first leaf of 21 day old Brassica seedlings were imaged under L:L at 22 °C. Period boxplots based on the DF oscillations from different cultivars are shown for wheat (a) and Brassica (b). The three replicates in the Brassica data represent different seed batches. Period values were estimated using FFT NLLS, BAMP dtr, 24–120 h cut-off window. BAMP de-trended DF data was normalised to the mean DF intensity across all cultivars and plotted against time in hours after dawn (c, d). Brassica data represents results from two cabinets normalised for between-cabinet effects. Wheat data represents results from two identical experiments with two imaging cabinets run in parallel as technical replicates and normalised for the between-cabinet and experimental-run effects. Wheat: Robigus (N = 25/25), Cadenza (N = 12/12), Chinese Spring (N = 24/25), Claire (N = 27/27), Weebil (N = 22/24), Paragon (N = 24/4), Norin-61 (N = 25/25). Brassica: for each seed batch of Cabriolet, Chuanyou II, Norin (N = 8/8). Brassica data is consistent with previous experiments shown in Additional file 1: Supplementary material S14. Significance codes: for wheat cultivars (a), ***p < 0.001, **p < 0.01 relative to either a = Norin 61 or b = Paragon. For brassica cultivars (b), significantly different periods (p < 0.05) are labelled relative to a = Norin Y1, b = Norin Y2 or c = Norin Y3
Figure 6b shows the variation in period across three Brassica lines taken from three seed batches. We conducted a two-way analysis of variance to compare cultivar ID and seed batch effects as well as the interaction between the two factors. The cultivar ID was found to have a significant effect on period (F(2,60) = 25.47, p < 0.001) but batch year did not significantly account for any variation in period either as a main effect (F(2,60) = 1.73, p > 0.1) or as an interaction with the cultivar ID (F(4,60) = 2.27, p = 0.72). This suggests that the observed differences in period are due to heritable genetic differences. The Brassica cultivar Norin had the shortest overall period of 22.29 h (SD 0.34); shorter than either Cabriolet (23.32 h, SD 0.57) or Chuanyou II (23.18 h, SD 0.72) (p < 0.001, Tukey HSD).
DF oscillations in both Brassica and wheat remained rhythmic throughout the experiment allowing confident period estimation over 4 days (24–120 h following T0). The average DF oscillations for the two most divergent wheat lines is shown in Fig. 6c; the other lines have been omitted for clarity. DF expression from all 3 years was averaged to make the oscillation plots for the Brassica lines as shown in Fig. 6d. The percentage of DF rhythms returned from period estimation was high for both Brassica (96.3%) and wheat (98.8%) proving that the method is both efficient and reliable.
The overall throughput of this assay is dependent on the expected exclusion rate from period analysis and on the number of imaging cabinets available. Designs with 3 replicates per plate allow 10 independent lines to be assayed per cabinet over one experiment allowing for an expected 5% loss of samples. If rhythms are expected to be less robust, for example in a mutant screen, we suggest using a larger number of replicates. The scaling-up of this imaging assay to multiple cabinets is also becoming increasingly affordable as the CCD camera technology progresses.
Discussion
Manipulating the circadian clock has potential for influencing crop productivity, efficiency and resilience; however research has been hindered by the lack of high-throughput circadian protocols which can be reliably applied to crop plants. Transcriptional assays, luciferase constructs and fluorescent markers have been used to investigate circadian rhythms in tobacco [13], tomato [41], potato [42, 43], Brassica rapa [44], rice [45–47], barley [48] and wheat [49]. However, these approaches are either manually intensive, technologically expensive or require genetic modification to systematically investigate each component and so are low throughput. We have optimized a delayed fluorescence imaging method for reliable circadian phenotyping of either Brassica or wheat seedlings. Several differences between the function of these clocks have been exposed through the factors examined in this paper. The opposing robustness of clocks under D:D or L:L and the fact that DF rhythms peak with distinct phases under each condition is indicative of diverging networks underlying circadian control of each species. Lower temperatures (17 °C) also seem to have a detrimental effect on the robustness of the clock in T. aestivum but not B. napus, suggesting that temperature may be a stronger zeitgeber for wheat than for Brassica within this temperature range. The DF rhythms in both T. aestivum and B. napus have reduced temperature compensation compared to those reported for leaf movement in Arabidopsis [36, 37]. However, it is important to recognize that our rhythms were measured in dissected sections of leaves and may not be truly analogous to rhythms from whole Arabidopsis individuals. The difference between intact and excised leaves has been previously reported in Hall et al. [22]. Our analysis of the homogeneity of periods across a single leaf also revealed variability of period robustness across the axis of wheat leaves but relatively little variation across Brassica leaves.
In this study, we have shown that in both Brassica and wheat there is a strong interaction between circadian period and age due to both systemic aging and leaf-specific developmental aging. Previous research in Arabidopsis has asked whether the onset of senescence is a result of a faster running clock or vice versa [28, 50]. Our results suggest that the acceleration of the clock occurs in very young plants before senescence phase, raising the possibility that the clock could be artificially manipulated to moderate senescence and control timing of peak productivity in crops.
Natural variation of circadian phenotypes has been previously demonstrated in wild Arabidopsis accessions [35, 36, 51] revealing a selection pressure for circadian traits specific to different ecological settings. The extent to which circadian fitness has been selected-for in modern crop plants has not yet been investigated. Application of our optimized protocol in this study demonstrates that diverging rhythms are present within elite cultivars of the same species. This variation in circadian period suggests that some level of circadian diversity exists, but the question remains as to whether each cultivar is currently optimized to enhance individual plant fitness. Crop plants with ‘optimized circadian clocks’ may have the capacity to improve yield, efficiency and resilience potentially overlooked by traditional plant breeding methods.
Conclusions
In this study, we investigated several important factors influencing circadian rhythms in Brassica napus and Triticum aestivum and reveal intriguing differences between the two crops. We provide an optimized DF methodology which can be reliably used for high-throughput measurement of circadian rhythms. This research highlights the considerable plasticity of the circadian clock under free-running conditions. It is our hope that these results may inform future research by showing the extent to which controllable variables can affect period estimation and how these may differ depending on the model species being studied.
Method
Plant material and growth conditions
Brassica seedlings used were from the winter varieties Cabriolet and Norin and the semi-winter variety Chuanyou II from the OREGIN Brassica napus Diversity Fixed Foundation Set (BnDFFS) [52]. Wheat seedlings used were all hexaploid elite cultivars ordered from the Genome Resource Unit (John Innes Centre) (Additional file 1: Supplementary material S8).
Brassica plants were grown in Levington’s F2 mix in FP11 pots, spaced 5 plants to a pot. They were grown in controlled greenhouse conditions, (16:8 h L:D at 22:20 °C). After 17 days, plants were transferred to a plant growth chamber set at 12:12 L:D cycle at 22 °C under approximately 200 µmol m−2 s−1 white light for 4 days entrainment (light spectra can be seen in Additional file 1: Supplementary material S9).
Wheat plants were imbibed at 4 °C for 6 days before being planted in Petersfield cereal mix in FP9 pots, spaced two to a pot. They were then grown in controlled greenhouse conditions (16:8 h L:D 17:12 °C). After 21 days plants were transferred to a plant growth chamber set at the cabinet conditions above. For temperature experiments, plants were entrained at the temperatures in which they would be imaged.
Image acquisition-standard conditions
Leaves were removed just after entrainment dawn and placed face up onto 24 cm square petri dishes (Stratlab LTD, cat no. 163-PB-007) containing 0.5% water agar (Sigma-Aldrich, SKU A1296). Unless otherwise stated, 3 cm squares were cut from the second true leaf of 21 day old B. napus seedlings. A segment of 10 cm was taken from the second leaf of the main tiller of 25 day old T. aestivum seedlings, beginning 5 cm down from the tip. A small strip of agar was placed over the ends of wheat sections to prevent leaf curling during the experiment. Plates were secured with masking tape around the periphery.
The imaging set-up is adapted from that described by Southern et al. [53]. A set-up schematic can be seen in Additional file 1: Supplementary material S10. We use Lumo Reteiga CCD cameras (QImaging, Canada), which we have found to have comparable image quality to the Orca II (Hamamatsu Photonics, Japan) without the need to run a water-cooling pump. Cameras were fitted with a Xenon 0.95/25 mm lens (Schneider-Kreuznach, Germany).
A custom built 25 × 25 red/blue LED rig (approx. 60 µmol m−2 s−1) was controlled by μManager software (v1.4.19, Open Imaging) through an Arduino Uno microcontroller board [54]. LED spectra for cabinets can be viewed in Additional file 1: Supplementary material S9. μManager was used to configure both the supplied camera driver software (PVCam v3.7.1.0) and program the Arduino after installing the firmware source code available online [55]).
Both camera and LEDs were housed in a temperature controlled growth cabinet (Sanyo MIR-553) in a dark room. The temperature was set to 22 °C unless otherwise specified [changed for the temperature experiments (Fig. 5) and for the wheat cultivar experiment (Fig. 6)]. Camera properties were kept the same in each experiment (Binning = 4, Gain = 1, Readout-Rate = 0.650195 MHz 16 bit) and camera exposure was initiated 500 ms after the lights were turned off. A ‘L:L’ script refers to a regime of 59 min of light followed by a 1 min exposure in the dark. A ‘D:D’ script refers to 54 min of darkness followed by 5 min light and then the 1 min exposure. BeanShell scripts run by μManager have been adapted from scripts used previously [56] and are available to view as Additional files 2 and 3. Wheat imaging used the D:D script and Brassica imaging used the L:L script with the exception of experiments in Fig. 4.
Processing in FIJI and BioDare2 parameters
Image stacks were imported into FIJI [57] and regions of interest were selected. Measurements for integrated density were taken for these regions across the stack using the Multi-measure plugin. Each region was then labeled in Excel and an offset time series added. The ‘offset time’ is the difference between the time of the first image (T1) and entrainment dawn (ZT) in decimal hours. Data can then be uploaded to BioDare2 as described online [58, 59]. BioDare2 is an open-access web tool for analyzing timeseries data and predicting circadian parameters. For our data we found that Baseline and amplitude (BAMP) de-trending was most appropriate but recommend visual inspection of the detrending methods available to find the least intrusive method which removes any baseline trends. Period estimation was done using the Fast Fourier Transform Non-Linear Least Squares (FFT-NLLS) algorithm [60] on a data window of 24–120 h with expected periods set to 18–34 h. Manual inspection of resulting periods ensured that all arrhythmic traces were excluded from further analysis.
Rhythm Robustness analysis
We summarized rhythm robustness metrics based on several BioDare2 outputs. ‘% returned’ is the number of samples for which periods could be estimated out of the number of samples originally imaged. The RAE (relative amplitude error) is the ratio of amplitude error to amplitude and represents amplitude robustness. A RAE of 1 indicates the most irregular waveform which can still be classified as rythmic whereas a RAE of 0 indicates a perfect sine wave with no amplitude error. The period coefficient of variation (CV) is the standard deviation of period estimates adjusted for the mean period and represents between sample variation [58, 61]. Period error is the extent to which the period estimate could vary and still give a good fit to the model. Error scores close to 0 indicate a tight fit of the model to the observed data and a high within sample period robustness. See Additional file 1: Supplementary materials S1–S6 for statistic tables and further descriptions.
Normalization for experimental effects
After circadian parameters were estimated in BioDare2, data was normalized to account for the following random experimental effects. For the wheat plant age and leaf age experiments (Fig. 1), Brassica plant-leaf age experiments (Fig. 2) and the Brassica cultivar experiments (Fig. 6b) samples were split between two imaging cabinets run in parallel in a single experiment. The predicted parameters (e.g. period) for each sample from the two cabinets were adjusted so that the cabinet means were then equivalent. This was achieved by dividing the cabinet means by the overall mean to get an adjustment factor for each cabinet and then dividing each individual value by that factor to get a cabinet-normalized value. For the cutting data (Fig. 3), the experiments were replicated in two separate imaging weeks and then adjusted for the between-experiment effects. For the wheat cultivar experiments (Fig. 6a) data was obtained from two cabinets over two separate experiments and was normalized for both effects in a similar way. The light regime (Fig. 4) and the temperature experiments (Fig. 5) measured each variable in one cabinet at a time and therefore did not require any normalization. The conclusions from these experiments is consistent with preliminary experiments presented in Additional file 1: Supplementary materials S11–S14.
Statistical analysis was carried out in RStudio v1.1.423 using aov and t.test functions fit with an appropriate linear model in the format specified in the Results. Datasets used to produce each figure are available in the online version of this article as Additional files 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17.
Additional files
Additional file 1. Supplementary materials S1–S14.
Additional file 4. Wheat plant ages (Fig. 1).
Additional file 5. Wheat leaf ages (Fig. 1).
Additional file 6. Brassica Leaf and age (Fig. 2).
Additional file 7. Brassica crossection (Fig. 3).
Additional file 8. Brassica cutting (Fig. 3).
Additional file 9.Wheat crossection (1–10) (Fig. 3).
Additional file 10. Wheat cutting (Fig. 3).
Additional file 11. DDLL oscillation data (Fig. 4).
Additional file 13. Temperature Brassica + Wheat (Fig. 5).
Additional file 14. Brassica cultivars (Fig. 6).
Additional file 15. Brassica oscillation (Fig. 6).
Additional file 16. Wheat cultivars (Fig. 6).
Additional file 17. Wheat oscillation (Fig. 6).
Acknowledgements
Not applicable.
Abbreviations
- DF
delayed fluorescence
- FFT-NLLS
fast Fourier transform non-linear least squares
- RAE
relative amplitude error
- PSII
photosystem II
- L:L
constant light
- L:D
light–dark cycles
- D:D
constant dark
- BnDFFS
Brassica napus diversity fixed foundation set
- ZT
zeitgeber time
- BAMP
baseline and amplitude
- CV
coefficient of variation
Authors’ contributions
This project was conceptualized by HR and AH. HR designed and conducted experiments, carried out data processing and analysis and wrote the manuscript with contributions from AH. All authors read and approved the final manuscript.
Funding
This project was supported by the BBSRC via the Earlham institute CSP (BB/P016774/1, AH SD, HR) and BBSRC Design Future Wheat (BB/P016855/1, AH).
Availability of data and materials
The datasets generated during the current study are available as additional files 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 in the online version of this article. Raw image files are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hannah Rees, Email: hannah.rees@earlham.ac.uk.
Susan Duncan, Email: susan.duncan@earlham.ac.uk.
Peter Gould, Email: peter.gould@liverpool.ac.uk.
Rachel Wells, Email: rachel.wells@jic.ac.uk.
Mark Greenwood, Email: mark.greenwood@slcu.cam.ac.uk.
Thomas Brabbs, Email: thomas.brabbs@earlham.ac.uk.
Anthony Hall, Email: anthony.hall@earlham.ac.uk.
References
- 1.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 2.Millar AJ, Straume M, Chory J, Chua NH, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995;267(5201):1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 6.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeCoursey PJ, Walker JK, Smith SA. A circadian pacemaker in free-living chipmunks: essential for survival? J Comp Physiol A. 2000;186(2):169–180. doi: 10.1007/s003590050017. [DOI] [PubMed] [Google Scholar]
- 8.von Saint Paul U, Aschoff J. Longevity among blowflies Phormia terraenovae R.D. kept in non-24-hour light-dark cycles. J Comp Physiol. 1978;127(3):191–195. doi: 10.1007/BF01350109. [DOI] [Google Scholar]
- 9.Pittendrigh CS, Minis DH. Circadian systems: longevity as a function of circadian resonance in Drosophila melanogaster. Proc Natl Acad Sci USA. 1972;69(6):1537–1539. doi: 10.1073/pnas.69.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernando CE, Romanowski A, Yanovsky MJ. Transcriptional and post-transcriptional control of the plant circadian gene regulatory network. Biochim Biophys Acta Gene Regul Mech. 2017;1860:84–94. doi: 10.1016/j.bbagrm.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Edwards KD, Millar AJ. Analysis of circadian leaf movement rhythms in Arabidopsis thaliana. In: Rosato E, editor. Circadian rhythms: methods and protocols. Totowa: Humana Press; 2007. pp. 103–113. [DOI] [PubMed] [Google Scholar]
- 12.Welsh David K., Imaizumi Takato, Kay Steve A. Methods in Enzymology. 2005. Real-Time Reporting of Circadian-Regulated Gene Expression by Luciferase Imaging in Plants and Mammalian Cells; pp. 269–288. [DOI] [PubMed] [Google Scholar]
- 13.Millar AJ, Short SR, Chua N-H, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4(9):1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould PD, Diaz P, Hogben C, Kusakina J, Salem R, Hartwell J, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58(5):893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 15.Gawroński P, Ariyadasa R, Himmelbach A, Poursarebani N, Kilian B, Stein N, et al. A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics. 2014;196(4):1253–1261. doi: 10.1534/genetics.113.158444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goltsev V, Zaharieva I, Chernev P, Strasser RJ. Delayed fluorescence in photosynthesis. Photosynth Res. 2009;101(2–3):217–232. doi: 10.1007/s11120-009-9451-1. [DOI] [PubMed] [Google Scholar]
- 17.Kurzbaum E, Eckert W, Yacobi YZ. Delayed fluorescence as a direct indicator of diurnal variation in quantum and radiant energy utilization efficiencies of phytoplankton. Photosynthetica. 2007;45(4):562–567. doi: 10.1007/s11099-007-0096-z. [DOI] [Google Scholar]
- 18.Gould PD, Diaz P, Hogben C, Kusakina J, Salem R, Hartwell J, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58(5):893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 19.Boxall SF, Dever LV, Kneřová J, Gould PD, Hartwell J. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi. Plant Cell. 2017;29(10):2519–2536. doi: 10.1105/tpc.17.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thain SC, Hall A, Millar AJ. Functional independence of circadian clocks that regulate plant gene expression. Curr Biol. 2000;10(16):951–956. doi: 10.1016/S0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi N, Hirata Y, Aihara K, Mas P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell. 2015;163(1):148–159. doi: 10.1016/j.cell.2015.08.062. [DOI] [PubMed] [Google Scholar]
- 22.Hall A, Kozma-Bognár L, Tóth R, Nagy F, Millar AJ. Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol. 2001;127(4):1808–1818. doi: 10.1104/pp.010294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515(7527):419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002;130(1):102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322(5909):1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 26.Gould PD, Domijan M, Greenwood M, Tokuda IT, Rees H, Kozma-Bognar L, et al. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife. 2018;26(7):e31700. doi: 10.7554/eLife.31700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenden B, Toner DLK, Hodge SK, Grima R, Millar AJ. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc Natl Acad Sci USA. 2012;109(17):6757–6762. doi: 10.1073/pnas.1118814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Kim Y, Yeom M, Lim J, Nam HG. Age-associated circadian period changes in Arabidopsis leaves. J Exp Bot. 2016;67(9):2665–2673. doi: 10.1093/jxb/erw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H, Kim HJ, Vu QT, et al. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci U S A. 2018;115(33):8448–8453. doi: 10.1073/pnas.1722407115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris PJC, Wilkins MB. Light-induced changes in the period of the circadian rhythm of carbon dioxide output in Bryophyllum leaves. Planta. 1976;129(3):253–258. doi: 10.1007/BF00398267. [DOI] [PubMed] [Google Scholar]
- 31.Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125(3):485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 32.Aschoff J. Circadian rhythms: influences of internal and external factors on the period measured in constant conditions 1. Z Tierpsychol. 2010;49(3):225–249. doi: 10.1111/j.1439-0310.1979.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 33.Salomé PA, Xie Q, McClung CR. Circadian timekeeping during early Arabidopsis development. Plant Physiol. 2008;147(3):1110–1125. doi: 10.1104/pp.108.117622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pittendrigh CS. On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sc. 1954;40(10):1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusakina J, Gould PD, Hall A. A fast circadian clock at high temperatures is a conserved feature across Arabidopsis accessions and likely to be important for vegetative yield. Plant Cell Environ. 2014;37(2):327–340. doi: 10.1111/pce.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170(1):387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18(5):1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170(1):387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y-W, Lai K-N, Tai P-Y, Li W-H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between brassica and other angiosperm lineages. J Mol Evol. 1999;48(5):597–604. doi: 10.1007/PL00006502. [DOI] [PubMed] [Google Scholar]
- 40.Campoli C, Shtaya M, Davis SJ, von Korff M. Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012;21(12):97. doi: 10.1186/1471-2229-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Facella P, Lopez L, Carbone F, Galbraith DW, Giuliano G, Perrotta G. Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE. 2008;3(7):e2798. doi: 10.1371/journal.pone.0002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kloosterman B, Abelenda JA, Gomez MDMC, Oortwijn M, de Boer JM, Kowitwanich K, et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495(7440):246–250. doi: 10.1038/nature11912. [DOI] [PubMed] [Google Scholar]
- 43.Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- 44.Xu X, Xie Q, McClung CR. Robust circadian rhythms of gene expression in Brassica rapa tissue culture. Plant Physiol. 2010;153(2):841–850. doi: 10.1104/pp.110.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23(5):1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiyama N, Izawa T, Oikawa T, Shimamoto K. Light regulation of circadian clock-controlled gene expression in rice. Plant J. 2001;26(6):607–615. doi: 10.1046/j.1365-313x.2001.01063.x. [DOI] [PubMed] [Google Scholar]
- 47.Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, et al. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell. 2012;151(6):1358–1369. doi: 10.1016/j.cell.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 48.Ford B, Deng W, Clausen J, Oliver S, Boden S, Hemming M, et al. Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J Exp Bot. 2016;67(18):5517–5528. doi: 10.1093/jxb/erw317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115(5):721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- 50.Kim H, Kim HJ, Vu T, Jung S, Mcclung CR, Hong S, et al. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci. 2018;115(33):8448–8453. doi: 10.1073/pnas.1722407115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302(5647):1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 52.Pink D, Bailey L, McClement S, Hand P, Mathas E, Buchanan-Wollaston V, et al. Double haploids, markers and QTL analysis in vegetable brassicas. Euphytica. 2008;164(2):509–514. doi: 10.1007/s10681-008-9742-1. [DOI] [Google Scholar]
- 53.Southern MM, Brown PE, Hall A. Luciferases as reporter genes. In: Salinas J, Sanchez-Serrano JJ, editors. Arabidopsis protocols. New Jersey: Humana Press; 2006. pp. 293–305. [Google Scholar]
- 54.Arduino. https://www.arduino.cc/. Cited 2018 Nov 14.
- 55.uManager-Arduino. https://micro-manager.org/wiki/Arduino#Firmware. Cited 2018 Nov 15.
- 56.Litthauer S, Battle MW, Lawson T, Jones MA. Phototropins maintain robust circadian oscillation of PSII operating efficiency under blue light. Plant J. 2015;83(6):1034–1045. doi: 10.1111/tpj.12947. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ. Strengths and limitations of period estimation methods for circadian data. PLoS ONE. 2014;9(5):e96462. doi: 10.1371/journal.pone.0096462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.BioDare2—circadian period analysis. https://biodare2.ed.ac.uk/welcome. Cited 2018 Nov 14.
- 60.Straume M, Frasier-Cadoret SG, Johnson ML. Least-squares analysis of fluorescence data. In: Lakowicz JR, editor. Topics in fluorescence spectroscopy. Boston: Kluwer Academic Publishers; 1991. pp. 177–240. [Google Scholar]
- 61.Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127(4):1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary materials S1–S14.
Additional file 4. Wheat plant ages (Fig. 1).
Additional file 5. Wheat leaf ages (Fig. 1).
Additional file 6. Brassica Leaf and age (Fig. 2).
Additional file 7. Brassica crossection (Fig. 3).
Additional file 8. Brassica cutting (Fig. 3).
Additional file 9.Wheat crossection (1–10) (Fig. 3).
Additional file 10. Wheat cutting (Fig. 3).
Additional file 11. DDLL oscillation data (Fig. 4).
Additional file 13. Temperature Brassica + Wheat (Fig. 5).
Additional file 14. Brassica cultivars (Fig. 6).
Additional file 15. Brassica oscillation (Fig. 6).
Additional file 16. Wheat cultivars (Fig. 6).
Additional file 17. Wheat oscillation (Fig. 6).
Data Availability Statement
The datasets generated during the current study are available as additional files 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 in the online version of this article. Raw image files are available from the corresponding author on reasonable request.