Abstract
In the adenosine receptor (AR) subfamily of G protein-coupled receptors (GPCRs), biased agonism has been described for the human A1AR, A2BAR and A3AR. While diverse A3AR agonists have been evaluated for receptor binding and Gi-mediated cAMP signalling, the β-arrestin2 (βarr2) pathway has been left largely unexplored. We screened nineteen diverse adenosine derivatives for βarr2 recruitment using a stable hA3AR-NanoBit®-βarr2 HEK293T cell line. Their activity profiles were compared with a cAMP accumulation assay in stable hA3AR CHO cells. Structural features linked to βarr2 activation were further investigated by the evaluation of an additional ten A3AR ligands. The A3AR-selective reference agonist 2-Cl-IB-MECA, which is a full agonist in terms of cAMP inhibition, only showed partial agonist behaviour in βarr2 recruitment. Highly A3AR-selective (N)-methanocarba 5’-uronamide adenosine derivatives displayed higher potency in both cAMP signalling and βarr2 recruitment than reference agonists NECA and 2-Cl-IB-MECA. Their A3AR-preferred conformation tolerates C2-position substitutions, for increased βarr2 efficacy, better than the flexible scaffolds of ribose derivatives. The different amino functionalities in the adenosine scaffold of these derivatives each seem to be important for signalling as well. In conclusion, we have provided insights into ligand features that can help to guide the future therapeutic development of biased A3AR ligands with respect to G-protein and βarr2 signalling.
Keywords: G protein-coupled receptor, A3 adenosine receptor, β-arrestin2, structure-activity relationship, biased signaling
Graphical Abstract
1. INTRODUCTION
The family of purinergic adenosine receptors (ARs) is involved in different (patho)physiological processes in the human body via modulation of the nervous system, immune response, vascular function and metabolism. The endogenous agonist adenosine, at resting physiological concentrations of less than 1 µM, may bind the four AR subtypes (A1, A2A, A2B and A3). The elevation of adenosine levels due to acute stress or ischemic conditions can benefit cellular adaptation. Under pathological conditions, sustained levels of excess adenosine can contribute to the development and/or progression of various diseases [1–3]. The human A3AR (hA3AR) is highly expressed in lung, liver and immune cells, with lower expression levels in the heart, brain and eye [4]. In recent years, the A3AR has attracted interest as a therapeutic target due to its role in the pathogenesis of heart and vascular diseases, autoimmune inflammatory disorders, COPD and asthma, along with different types of cancer [5–8]. A3AR expression is upregulated in various tumour cells, modulating tumour growth depending on the tumour type, the other AR subtypes present, the surrounding immune cells and micro-environmental conditions involved [9, 10]. More recently, targeting of the A3AR has been suggested as a promising, safe and effective therapeutic approach for the management of chronic neuropathic pain of various etiologies [11–13].
Given the delicate role of adenosine in tissue homeostasis and the plethora of therapeutic opportunities, significant work has been done in synthetic ligand design to target the human A3AR. Structure-based molecular modeling has led to the rational design of an impressive panel of potent, highly-selective A3AR ligands [8]. For therapeutic application, these synthetic ligands must display good pharmacokinetic properties, as well as excellent A3AR binding affinity, selectivity, efficacy and potency [14, 15]. Currently, functional evaluation of these A3AR ligands relies mainly on the measurement of Gi protein-mediated signalling using cAMP accumulation assays. However, as with all GPCRs, the A3AR couples to other downstream signalling proteins besides the G protein. There has been increased interest in designing and developing GPCR agonists that show biased signalling towards certain signalling pathway(s). In recent years, one of the most studied non-G-protein signalling pathways in GPCR systems has been that involving the adaptor protein β-arrestin2 (βarr2) [16, 17]. βarr2 can induce both downstream signalling and GPCR desensitization, thus influencing the duration of a therapeutic effect and/or tolerance in a physiological setting. The biased activation of Gi-dependent or βarr2-dependent signalling pathways in A3AR agonism has remained insufficiently explored. The application of functional assays to expand the structure-activity relationships of A3AR ligands may help link desired therapeutic profiles to in vivo models in the future. Furthermore, this could allow for the reorientation of the therapeutic profiles of previously synthesized ligands.
Here, we report on the development of a stable hA3AR HEK293T cell assay system, which allows real-time monitoring of βarr2 recruitment by application of NanoBit® technology (Promega). This technology relies on the functional complementation of the NanoLuc luciferase enzyme. The applicability of the system to study hA3AR-βarr2 interaction was previously demonstrated by our research group, using transiently transfected HEK293T cells [18]. After establishing that the stable cell line provided a reproducible and concentration-dependent response with a known A3AR agonist (2-Cl-IB-MECA), we compared the βarr2 activity profiles of nineteen compounds with those for cAMP signalling, which were obtained with a cAMP accumulation assay performed in CHO cells stably transfected with the hA3AR. Structural features linked to βarr2 activity were further investigated via the evaluation of an additional panel of ten A3AR ligands, providing insight into the structure-activity relationship of A3AR agonists with respect to βarr2 recruitment.
2. MATERIALS & METHODS
2.1. Chemicals and reagents
HEK293T cells (passage 20) were kindly provided by Prof. O. De Wever (Laboratory of Experimental Cancer Research, Department of Radiation Oncology and Experimental Cancer Research, Ghent University Hospital, Belgium). The human A3AR construct (NM_000677.2, transcript variant 2 of the ADORA3 gene) and human βarr2 construct (NM_004313) were purchased from Origene Technologies (Rockville, MD, USA). DOTAP Liposomal Transfection Reagent was purchased from Sigma Aldrich (Steinheim, Germany), and the anti-dNGFR antibody was purchased from Chromaprobe (Maryland Heights, MO, USA). Reference agonists 2-Cl-IB-MECA, IB-MECA, CGS21680 and NECA were purchased from Tocris Bioscience (Bio-techne, Abingdon, UK). All other chemicals and reagents used were purchased from the same suppliers as described previously [18] or synthesized at NIDDK, National Institutes of Health as reported [19–33].
2.2. Development of a stable A3AR-NanoBit®-βarr2 HEK293T cell line by retroviral transduction
2.2.1. Production of retrovirus using the PhoenixA packaging cell line
To generate retroviral expression vectors, the coding sequences for the A3AR-LgBit and SmBit-βarr2 fusion proteins (reported in [18]) were transferred to the retroviral vectors pLZRS-IRES-EGFP and pLZRS-pBMN-link-I-dNGFR, respectively, using standard cloning procedures, as described previously [18]. Primers, PCR conditions and restriction enzymes used are given in Table 1. The resulting retroviral expression vectors lead to co-expression of the fusion proteins A3AR-LgBit and SmBit-βarr2 with enhanced green fluorescent protein (EGFP) and truncated human nerve growth factor (dNGFR), respectively, in the cell line of choice (in this case HEK293T cells).
Table 1:
PCR conditions (a-c) and restriction enzymes (re; d) used to construct the retroviral expression vectors*: a: primers containing the specific restriction site (underlined), kozak sequence (bold italic) and coding sequence of interest (italic). A stop codon (bold) to ensure a correct reading frame was added where necessary.
| Coding Sequence | Primers (F: forward – R: reverse)a | Tm (°C)b |
Ext. time (s)c |
REd | Retroviral expression vector* | |
|---|---|---|---|---|---|---|
| A3AR-LgBit | F | ACTCAAGGATCCACCATGCCCAACAACAGC | BamHI | pLZRS-A3AR-LgBit-IRES-EGFP | ||
| R | ACTCAATACGTATTAGCTGTTGATGGTTACTCGG | 69.5 | 40 | - | ||
| SnaBI | ||||||
| SmBit-βarr2 | F | ACTCAAGGATCCACCATGGTGACCGGCTACCG | BamHI | pLZRS-SmBit-βarr2-IRES-dNGFR | ||
| R | ACTCAAGCGGCCGCTCAGCAGAGTTGATCATCATAGTCG | 74 | 38 | - | ||
| NotI | ||||||
The retroviral vectors, as well as the protocol for production of retrovirus, were adapted from the research group of Prof. Bruno Verhasselt (Department of Clinical Chemistry, Microbiology, and Immunology, Ghent University, Belgium). The Phoenix-AMPHO (ΦNX-A) packaging cell line was cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) (Thermo Fisher Scientific, Pittsburgh, PA, USA) supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 100 IU/mL penicillin, 100 µg/mL streptomycin and 0.25 µg/mL amphotericin B (full IMDM) under humidified atmosphere at 37°C, 5% CO2. Twenty-four h before transfection, cells were seeded at a density of 106 cells/6 cm dish in full IMDM. The next day, cells were transiently transfected using the Calcium Phosphate Transfection method (Thermo Fisher Scientific, Pittsburgh, PA, USA) according to the manufacturer’s instructions, using 20 µg of retroviral expression vector DNA. Five min prior to transfection, chloroquine (Sigma Aldrich, Steinheim, Germany) was added to the cells at a final concentration of 25 µM, followed by dropwise addition of the transfection mixture. After overnight incubation, the medium was refreshed. Forty-eight h after transfection, the first viral supernatant was harvested, centrifuged (10 min, 350×g, 4°C), aliquoted on ice without disturbing the pellet, and stored at −80°C. After the first harvest, puromycin selection (2 µg/mL) (Sigma Aldrich, Steinheim, Germany) was carried out for 3 rounds of 48 h, punctuated each time with a 48-h incubation in puromycin-free full IMDM. The day before the last viral harvest, the medium was again refreshed. The second viral supernatant was harvested, centrifuged and stored as was done for the first harvest.
2.2.2. Retroviral transduction of HEK293T cells
HEK293T cells were seeded in a 96-well plate at 104 cells/well in full DMEM and incubated overnight. For retroviral transduction, the medium was replaced by retroviral supernatant, pre-incubated with DOTAP Liposomal Transfection Reagent according to the manufacturer’s instructions, consisting of a 1:1 mixture of both A3AR- and βarr2-sequence containing retroviruses. Subsequently, the plate was centrifuged for 90 min at 950×g (32°C) to increase transduction efficiency. After overnight incubation, the medium was refreshed, and cells were further cultured in full DMEM. Forty-eight h after transduction, expression efficiency was evaluated with flow-cytometry by measuring the level of EGFP or dNGFR (by pre-incubation with a Allophycocyanin-linked anti-dNGFR antibody), which are co-expressed with the A3AR- and βarr2-fusion proteins, respectively.
2.2.3. Cell sorting of stably transduced HEK293T cells
After routine culture of the stably transduced HEK293T cells for 3 passages, cell sorting was performed with a BDFACSAria III cell sorter (BD Biosciences). In this stage, co-expression of EGFP and dNGFR was used to select a subpopulation of cells with the desired expression levels of A3AR- and βarr2 fusion proteins, respectively. Cells were maintained in full DMEM medium. Stability of the cell line was monitored every 3 to 5 passages by flow cytometry, as described above.
2.3. Screening of synthetic A3AR ligands
2.3.1. A3AR NanoBit® βarr2 assay
In total, a panel of twenty-nine synthetic ligands was subjected to the stable A3AR-NanoBit®-βarr2 HEK293T cell line, using a 2-day assay protocol. On the first day, cells were seeded on PDL-pre-coated, white 96-well plates at 5×104 cells/well and incubated overnight (37°C, 5% CO2). On day 2, the assay read-out was performed at room temperature, as described previously [18]. Briefly, following 2 washing steps, 90 μL of Opti-MEM I reduced serum medium was added to the cells, followed by addition of 25 µL of freshly prepared detection reagent and monitoring of luminescence equilibration. Once a stable signal was observed, 20 µL of 6.75× concentrated agonist solution in Opti-MEM I was added, and luminescence monitored for at least 90 min. All stock solutions of synthetic ligands were DMSO-based. The final in-well concentrations were (100 pM) - (500 pM) - 1 nM – (2 nM) – (5 nM) – 10 nM – 50 nM – 100 nM – 1 µM – (2 µM) - 5 µM – (10 µM) - (25 µM) agonist (maximum 0.5% DMSO). A solvent control (blank sample) of 0.001–0.5% DMSO in Opti-MEM I was included each time.
2.3.2. cAMP assay
The levels of intracellular 3′,5′- cyclic AMP (cAMP) were measured by modification of the originally described competitive protein binding method [34, 35], which is widely used. Briefly, CHO cells stably expressing the human A3AR were treated with the synthetic ligands in the presence of rolipram (10 µM) and adenosine deaminase (3 units/ml). After 30 min, forskolin (10 µM) was added to the medium, and incubation was continued for an additional 15 min. The reaction was terminated by removing the supernatant, and cells were lysed by addition of 0.1 M ice-cold HCl. The cell lysate was resuspended and stored at −20°C until analysis by enzyme immunoassay.
2.3.3. Binding assay
Binding experiments were performed as described previously [26, 28, 36], using [125I]-N6-(4-amino-3-iodobenzyl)adenosine-5’-N-methyluronamide ([125I]I-AB-MECA) as a radioligand. Briefly, membranes were prepared from CHO cells stably expressing the human A3AR. Cells were detached and resuspended in 50 mM Tris HCl buffer (pH 8.0) containing 10 mM MgCl2, and 1 mM EDTA prior to homogenization with an electric homogenizer (10s), and re-centrifugation at 20,000g for 20 min (4 °C). The membrane pellets were resuspended in buffer in the presence of adenosine deaminase (3 U/mL), and suspensions were stored at −80°C until the binding assay. For competition experiments, each tube contained 100 μL of membrane suspension (±20 μg protein), 50 μL of [125I]I-AB-MECA (concentration 0.2 – 1 nM), and 50 μL of increasing concentrations of compounds in Tris–HCl buffer (50 mM, pH 8.0) containing 10 mM MgCl2, 1 mM EDTA. Non-specific binding was determined using 10 μM 2-Cl-IB-MECA. The mixtures were incubated at 25°C for 60 min. Binding reactions were terminated by filtration through Whatman GF/B filters under reduced pressure using an MT-24 cell harvester (Brandell). Filters were washed three times with ice-cold buffer. Radioactivity was determined in a Beckman 5500B γ-counter. IC50 values were obtained from competition curves and converted to Ki values using the Cheng-Prusoff equation [37].
2.4. Data analysis
Concentration-responses (area under the curve; AUCs) were calculated, where the absolute signals were corrected for solvent control, and for inter-well variability. For calculation of logEC50 values, a sigmoidal curve was fitted to the normalized responses by analysing dose-response data using GraphPad Prism software (San Diego, CA, USA). Data points for the highest agonist concentrations were excluded when the signal height showed more than 20% reduction compared to the maximum signal of the closest lower concentration. A non-linear regression model (Hill Slope 1) was fitted to the normalized responses.
3. RESULTS
3.1. Development of a stable A3AR reporter assay for real-time monitoring of βarr2 recruitment
The originally-described reporter system for evaluation of βarr2 coupling to A3AR [18] was characterized by a transient assay set-up with a 4-day protocol, in which the transfection efficiency varied between different assays, complicating inter-day comparisons. In order to obtain a system that could serve high-throughput screening and would more easily allow comparative analysis of a panel of compounds, a stable A3AR-βarr2 reporter cell line was developed by retroviral transduction, reducing the assay protocol to 2 days. Moreover, a stable expression level of both fusion proteins can be ensured by monitoring the level of co-expressed markers (EGFP and dNGFR) using flow cytometry. If needed, an additional advantage is that cells can be sorted into subpopulations with certain expression levels of both constructs. A subpool was selected with the highest expression levels of both A3AR and βarr2 fusion proteins. A clear concentration-dependent response to the reference agonist 2-Cl-IB-MECA, starting at ~2 nM and reaching a maximum at 1 µM, was obtained with this cell line (Figure 1).
Figure 1:
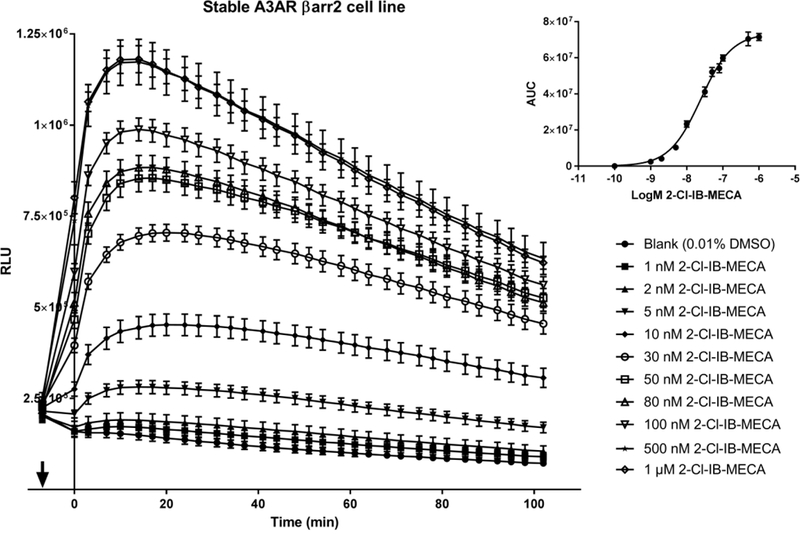
Concentration-dependent effect of reference A3AR agonist 2-Cl-IB-MECA on βarr2 recruitment in a stable hA3AR-NanoBit®-βarr2 HEK293T cell line. 1, 2, 5, 10, 30, 50, 80, 100, 500 nM and 1 µM 2-Cl-IB-MECA was added at the time point indicated by the arrow, and luminescence was measured for >90 min. A solvent control of 0.01% DMSO was included. Insert: Concentration-response (AUCs). AUCs (± SEM) of quadruplicate wells are shown for a representative experiment (n=3).
3.2. Screening of A3AR ligands for βarr2 recruitment and cAMP signalling
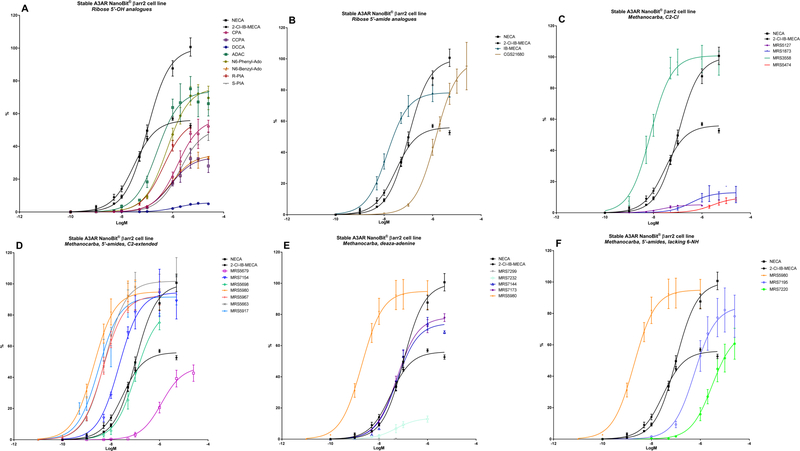
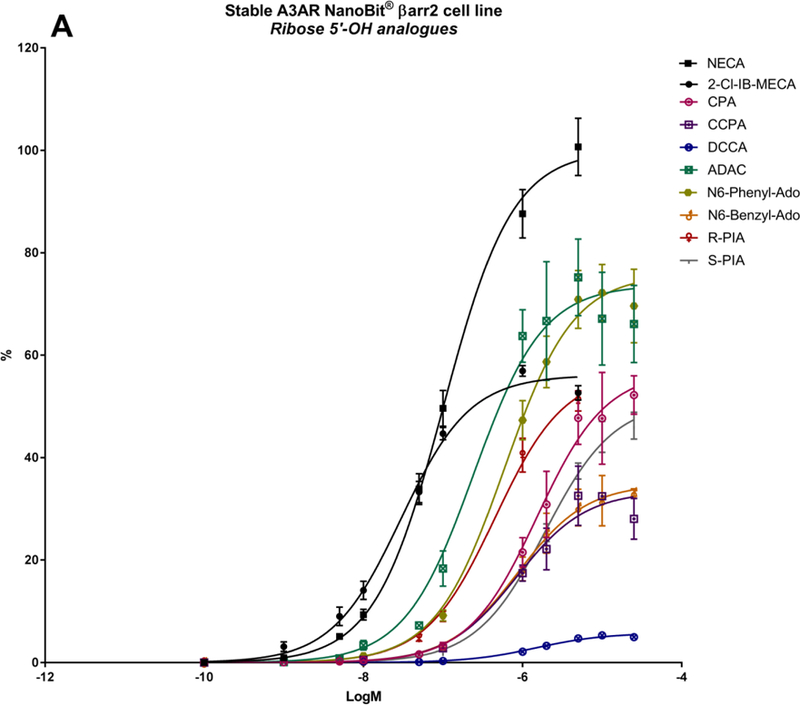
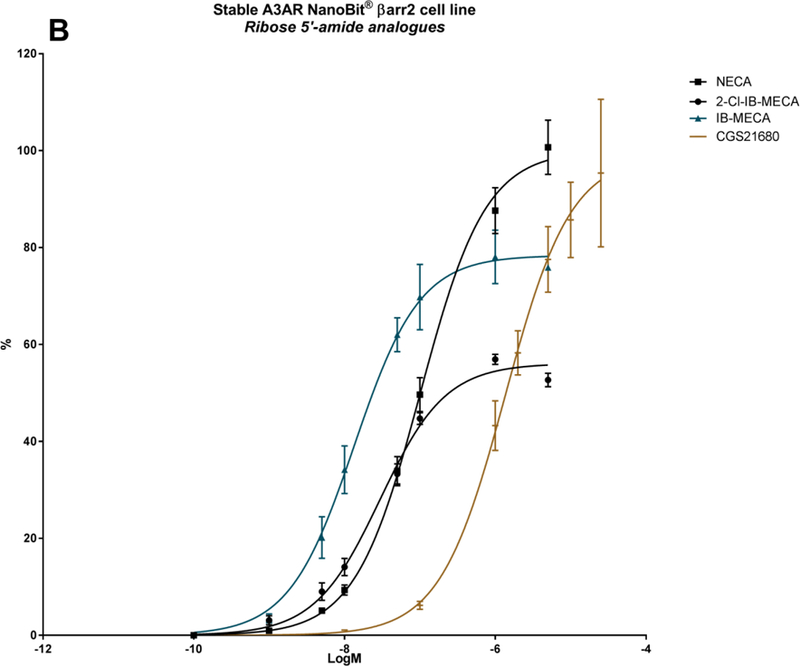
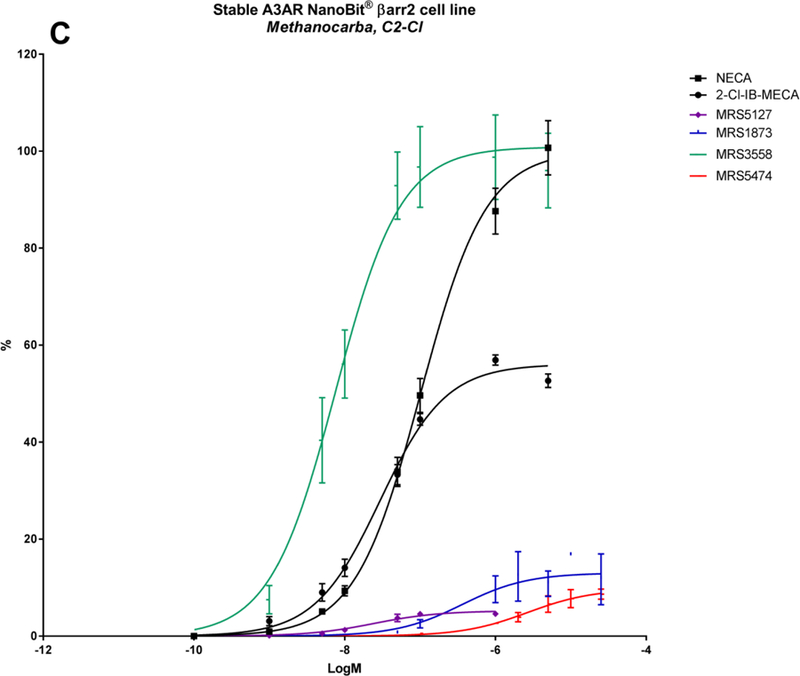
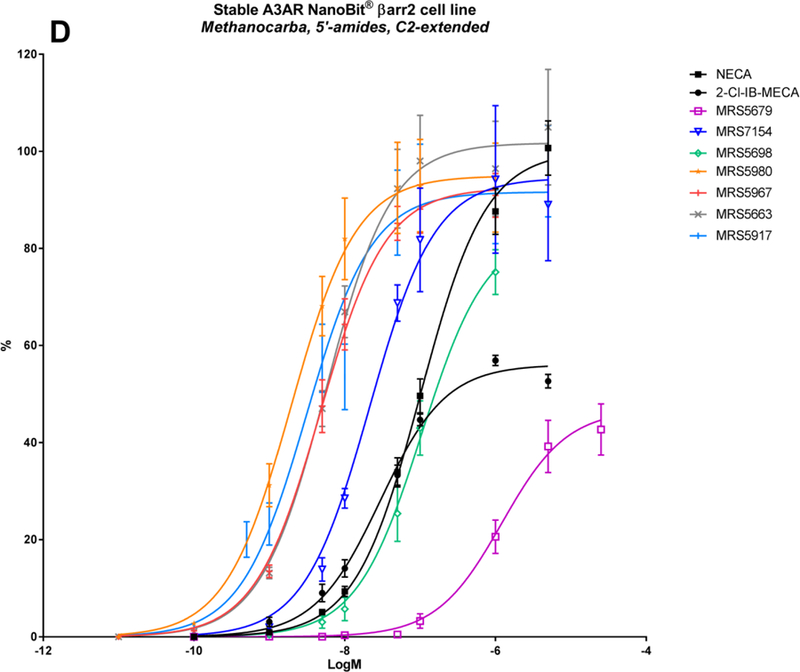
A panel of nineteen synthetic A3AR ligands was evaluated for βarr2 recruitment, using the stable hA3AR-NanoBit®-βarr2 HEK293T cell line, and G-protein dependent cAMP signalling, using a cAMP accumulation assay in stable hA3AR CHO cells. In Table 2, the maximal efficacy (Emax) and potency (logEC50) of these compounds is shown for both signalling pathways, as well as their A3AR binding Ki values and structures. To gain more insight into their βarr2-related activity profiles, an additional ten structural analogues were tested with the hA3AR-βarr2 cell line (Table 3). The sigmoidal dose-response curves depicted in Figure 2 show different extents of maximal βarr2 recruitment for all compounds tested, relative to reference agonists NECA and 2-Cl-IB-MECA (black curves).
Table 2:
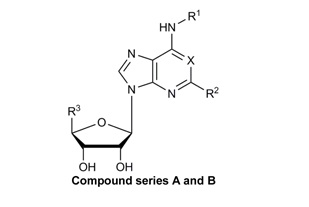
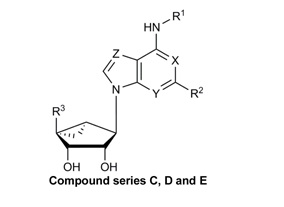
Structures, efficacies (Emax), potencies (logEC50), and hA3AR binding affinities (Ki) of nineteen known A3AR ligands. Data for βarr2 recruitment were obtained with the stable hA3AR-NanoBit®-βarr2 HEK293T cell line, and those for cAMP inhibition were obtained with a stable hA3AR CHO cell line.
 |
 |
 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | X | Y | Z | Stable
A3AR NanoBit® HEK293T cell lineA |
Stable
A3AR CHO cell lineB |
Ki (nM) ± SEM | ||
| Emax (%) | LogEC50 ± SEM | Emax (%) | LogEC50 | ||||||||
| A. Ribose 5’-OH analogues | |||||||||||
| CPA | cyclopentyl | H | CH2OH | N | - | - | 56.9 ± 3.7 | −5.82 ± 0.11 | 97 ± 4 | −6.91 | 72 ± 12 |
| CCPA | cyclopentyl | Cl | CH2OH | N | - | - | 33.5 ± 2.7 | −6.08 ± 0.15 | 0 | NA | 38 ± 6 |
| R-PIA | H | CH2OH | N | - | - | 56.6 ± 1.9 | −6.35 ± 0.06 | 102 ± 6 | −7.57 | 8.7 ± 0.9 | |
| S-PIA | H | CH2OH | N | - | - | 51.0 ± 1.4 | −5.72 ± 0.04 | 97 ± 3 | −6.68 | 68 ± 12 | |
| ADAC | H | CH2OH | N | - | - | 73.7 ± 3.1 | −6.61 ± 0.12 | 103 ± 4 | −7.00 | 13.3 ± 3.0 | |
| B. Ribose 5’-amide analogues | |||||||||||
| NECA | H | H | CONHC2H5 | N | - | - | 100 ± 2.5 | −6.99 ± 0.05 | 103 | −7.42 | 35 ± 12 |
| CGS21680 | H | CONHC2H5 | N | - | - | 99.0 ± 5.5 | −5.87 ± 0.10 | 98 ± 5 | −6.62 | 114 ± 16 | |
| IB-MECA | 3-I-benzyl | H | CONHCH3 | N | - | - | 78.4 ± 2.5 | −7.87 ± 0.07 | 100 | −8.44 | 1.8 ± 0.7 |
| 2-Cl-IB-MECA | 3-I-benzyl | Cl | CONHCH3 | N | - | - | 56.1 ± 1.1 | −7.53 ± 0.04 | 100 | −8.55 | 1.4 ± 0.3 |
| C. Methanocarba, C2-Cl | |||||||||||
| MRS3558 | 3-Cl-benzyl | Cl | CONHCH3 | N | N | N | 100.9 ± 3.7 | −8.12 ± 0.08 | 103 ± 7 | −9.42 | 0.29 ± 0.04 |
| MRS1873 | H | Cl | CH2OH | N | N | N | 13.0 ± 1.6 | −6.44 ± 0. 31 | 41 ± 5 | −6.1 | 353 ± 54 |
| MRS5127 | 3-I-benzyl | Cl | H | N | N | N | 5.2 ± 0.4 | −7.61 ± 0.13 | 44 ± 6 | −7.9 | 1.44 ± 0.6 |
| MRS5474 | dicyclopropylmethyl | Cl | H | N | N | N | 9.8 ± 1.2 | −5.57 ± 0.17 | 56.0 ± 4.8 | −5.86 ± 0.15 | 470 ± 15 |
| D. Methanocarba, 5’-amides, C2-extended | |||||||||||
| MRS5698 | 3-Cl-benzyl | (3,4-F2-phenyl)ethynyl | CONHCH3 | N | N | N | 83.5 ± 4.6 | −6.98 ± 0.07 | 95.7 ± 6.4 | −8.3 | 3.49 ± 1.84 |
| MRS5917 | CH3 | (thien-2-yl)ethynyl | CONHCH3 | N | N | N | 91.6 ± 3.4 | −8.51 ± 0.10 | 97.3 ± 2.9 | −9.38 | 0.57 ± 0.10 |
| E. Methanocarba, deaza-adenine | |||||||||||
| MRS7144 | CH2CH3 | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | CH | N | N | 74.4 ± 2.3 | −7.27 ± 0.06 | 95.5±2.3 | −8.82 | 1.7 ± 0.4 |
| MRS7299 | CH3 | (5-Cl-thien-2-yl)ethynyl | CO2C2H5 | N | N | CH | NA | NA | 1±7 | NA | 448 ± 13 |
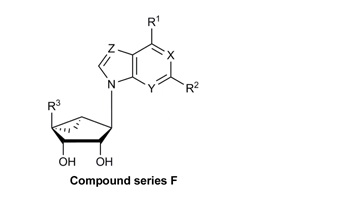
| F. Methanocarba, 5’-amides, lacking 6-NH | |||||||||||
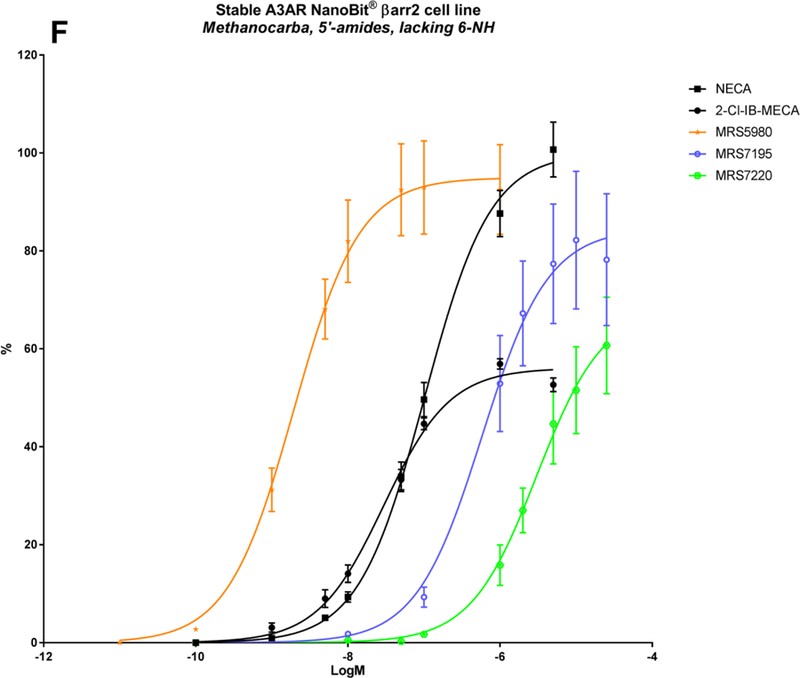
| MRS7195 | CH3 | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | N | N | N | 84.5 ± 6.4 | −6.24 ± 0.19 | 107 ± 26 | −7.9 | 42.2 ± 17.3 |
| MRS7220 | H | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | N | N | N | 68.4 ± 5.3 | −5.52 ± 0.11 | 97.8 ± 0.1 | −7.57 | 60 ± 19 |
Emax is relative to that of reference agonist NECA
Emax is relative to that of reference agonist NECA at 10 µM
NA, not significantly active at 10 µM
Table 3:
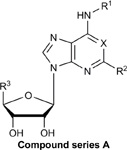
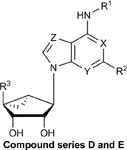
Structures, efficacies (Emax), potencies (logEC50), and hA3AR binding affinities (Ki) of ten additional structural analogs tested in the stable hA3AR-NanoBit®-βarr2 HEK293T cell line.
 |
 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | X | Y | Z | Stable
A3AR NanoBit® HEK293T cell lineA |
Ki (nM) ± SEM | |
| Emax (%) | LogEC50 ± SEM | ||||||||
| A. Ribose 5’-OH analogues | |||||||||
| DCCA | cyclopentyl | Cl | CH2OH | CH | - | - | 5.8 ± 0.2 | −5.83 ± 0.07 | 244± 37 |
| N6-benzyl-adenosine | benzyl | H | CH2OH | N | - | - | 34.9 ± 2.3 | −6.07 ± 0.13 | 41.7 ± 5.3 |
| N6-phenyl-adenosine | phenyl | H | CH2OH | N | - | - | 75.8 ± 3.0 | −6.23 ± 0.0 | 14.9 ± 3.1 |
| D. Methanocarba, 5’-amides, C2-extended | |||||||||
| MRS5679 | 3-Cl-benzyl | (4-phenyl-phenyl)ethynyl | CONHCH3 | N | N | N | 46.8 ± 3.3 | −5.92 ± 0.11 | 3.06 ± 1.35 |
| MRS5967 | CH3 | (2-CH3O-phenyl)ethynyl | CONHCH3 | N | N | N | 92.5 ± 2.4 | −8.33 ± 0.06 | 0.77 ± 0.17 |
| MRS5663 | CH3 | (2-Cl-phenyl)ethynyl | CONHCH3 | N | N | N | 101.7 ± 3.6 | −8.25 ± 0.08 | 0.58 ± 0.04 |
| MRS5980 | CH3 | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | N | N | N | 95.0 ± 4.0 | −8.71 ± 0.11 | 0.7 ± 0.11 |
| MRS7154 | (CH2)2CH3 | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | N | N | N | 94.5 ± 4.8 | −7.67 ± 0.11 | 1.1 ± 0.3 |
| E. Methanocarba, deaza-adenine | |||||||||
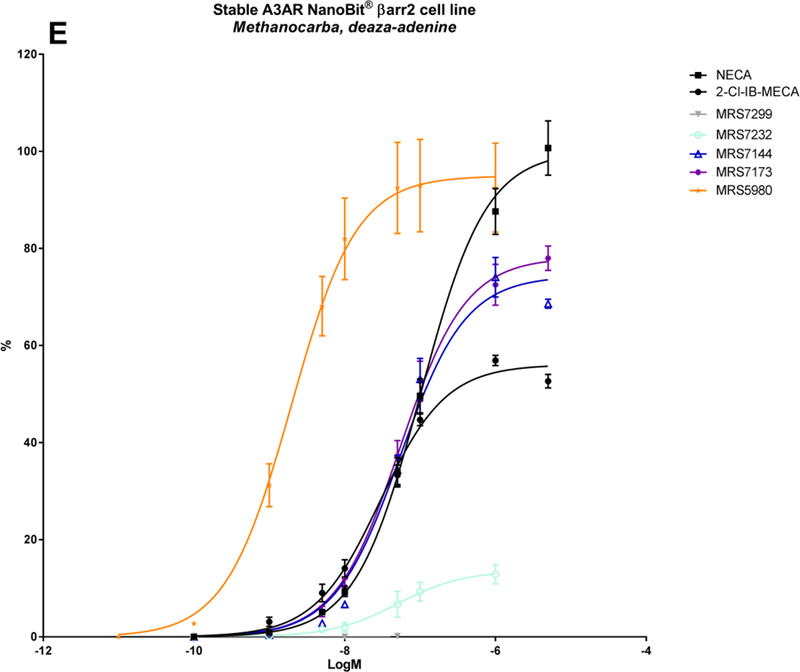
| MRS7173 | CH3 | (5-Cl-thien-2-yl)ethynyl | CONHCH3 | N | CH | N | 78.2 ± 1.8 | −7.26 ± 0.04 | 1.56 ± 0.2 |
| MRS7232 | CH3 | (5-Cl-thien-2-yl)ethynyl | CO2C2H5 | N | N | N | 13.6 ± 1.6 | −7.30 ± 0.17 | 5.38 ± 0.03 |
Emax is relative to that of reference agonist NECA
Figure 2,
A-F: Non-linear regression analysis of the sigmoidal dose-response curves for all twenty-nine synthetic A3AR ligands tested in the stable hA3AR-NanoBit®-βarr2 cell line. The curves were normalized to that of reference agonist NECA. AUCs (± SEM) were calculated based on at least 3 experiments performed in duplicate or triplicate.
4. DISCUSSION
The human A3 adenosine receptor is gaining interest in the field of drug discovery as a therapeutic target for inflammatory diseases and cancer [4]. Two A3AR agonists, IB-MECA (CF101, Piclidenoson) and 2-Cl-IB-MECA (CF102, Namodenoson) have shown safety and efficacy in Phase I and II clinical trials. The former is entering Phase III for treatment of rheumatoid arthritis and psoriasis [38], while its 2-chloro analogue is currently in Phase II for treatment of nonalcoholic steatohepatitis (NASH) and hepatocellular carcinoma [39, 40]. Given the A3AR’s therapeutic potential, highly selective and potent ligands have been synthesized, and their structure-activity relationship (SAR) has been extensively explored and reviewed [8, 19, 41–43]. This SAR is based on an evaluation of the ligands’ binding affinities, as well as their efficacies and potencies in activating downstream A3AR signalling pathways. Radioligand binding assays are originally carried out with membranes from AR-overexpressing mammalian cells, from which the affinities at different AR species homologues can be compared [36]. The availability of an A3AR homology model, which uses an agonist-bound A2AAR X-ray structure [44], but not the recently solved antagonist-bound A1AR [45], as a template, as well as opsin and the β2AR receptor, has enabled additional prediction of nucleoside binding interactions at the A3AR [46–48]. However, how this binding is translated into differential signalling has remained elusive.
Although the A3AR generally couples to the Gi protein, inhibiting adenylate cyclase and leading to a decrease in cAMP levels, there is substantial coupling to Gβγ and possibly Gq, leading to activation of phospholipase C (PLC) and a downstream rise in intracellular calcium [4, 8]. Furthermore, other signalling mediators have been described, such as the nuclear factor kappa B (NF-κB) and the Wnt signalling pathways. For multiple other GPCRs, biased ligands have been developed that preferentially activate a certain subset of signalling pathway(s) in order to obtain a desired therapeutic profile. One of the most explored crossroads for signalling bias in GPCR drug development is the one between the G protein and βarr2 [49–55].
In the AR field, the first steps towards a favoured profile of biased agonism have recently been set for the A1AR, the A2BAR, and A3AR [56–60]. However, so far, evaluation of the βarr2 pathway and a possible bias versus the G-protein pathway, has largely remained unexplored for the human A3AR. First reports of the interaction of the human A3AR with βarr2 were based on experiments using the PathHunter system (DiscoverX), which records activity at a single time point after 2 h of agonist incubation [61, 62]. We recently reported on the development of a live-cell assay system for real-time monitoring of βarr2 recruitment, based on the functional complementation of the Nanoluc luciferase enzyme [18]. Here, we report on the development of a stable hA3AR-NanoBit®-βarr2 HEK293T cell line that was used to screen a panel of nineteen synthetic ligands for βarr2 recruitment to the hA3AR. The resulting βarr2-recruitment activity profiles and those obtained for Gi protein coupling (Table 2), using the cAMP accumulation assay in stable A3AR CHO cells, were compared (Figure 3). Ten additional structural analogues of this parent compound panel were evaluated for the confirmation or more in-depth analysis of SAR features related to the βarr2 pathway (Table 3, Figure 2). The efficacies reported are expressed relative to the non-selective AR reference agonist NECA, for consistency with respect to previous results reported for A3AR signalling. NECA is considered to be a full agonist in multiple A3AR signalling pathways — including the cAMP- and βarr2-pathways, as well as A3AR mediated membrane hyperpolarization and intracellular Ca2+ mobilization [62]. The A3AR-selective reference agonist 2-Cl-IB-MECA has also been reported as a full agonist in the cAMP- and βarr2-pathways based on the aforementioned PathHunter system, but as a partial agonist in membrane hyperpolarization and Ca2+ mobilization (58% and 55% relative to NECA) [62]. Interestingly, in this study, 2-Cl-IB-MECA displayed a partial agonist Emax of only 56% relative to NECA in the βarr2-recruitment assay. The difference in Emax for βarr2 recruitment, when comparing these results with previously published data, might arise from several factors. First, previous data relate to a single time-point readout (PathHunter system), while this study implements a more comprehensive kinetic readout using AUCs (NanoBit® system). Second, we cannot dismiss the possibility that A3AR expression levels in the different assays may have varied and influenced the signal measured. However, with the NanoBit® system, we also evaluated βarr2 recruitment for 2-Cl-IB-MECA in CHO-K1 cells; potencies did not differ substantially between HEK293T cells (logEC50 of −7.53 ± 0.04) and transiently transfected (logEC50 of −7.26 ± 0.08) as well as stably-transduced (logEC50 of −7.24 ± 0.05) CHO-K1 cells. The Emax of 2-Cl-IB-MECA in CHO-K1 cells was 61% relative to NECA. Whilst the similar results obtained in different cell lines for 2-Cl-IB-MECA do indicate that the data obtained for our βarr2 recruitment assay are not confined to a single cell system, it is difficult to predict to what extent this can be extrapolated to the complete panel of compounds. The βarr2-recruitment activity profiles of the ligands tested here were generally similar to the structure-activity relationship for cAMP activity, which has been fine-tuned over years by step-wise structural modification, with differences as described below. In general, we observed a consistent difference of one log unit between logEC50s for cAMP signalling and βarr2 recruitment (Figure 3B).
Figure 3:
Comparison of the Emax relative to that of NECA (arbitrarily set at 100%) (A) and negative logEC50 (B) of nineteen synthetic A3AR ligands tested with the stable hA3AR-NanoBit®-βarr2 HEK293T cell line (Barr2) versus the stable hA3AR CHO cell line (cAMP).
Several N6 substituted adenosine derivatives were tested, as they are known to be quite potent A3AR ligands from G-protein-dependent signalling experiments [61, 63]. N6-cyclopentyladenosine (CPA), an A1AR-selective agonist, and the N6-phenylisopropyl R- and S-diastereomers R-PIA and S-PIA, which have demonstrated stereoselectivity in binding at the A1AR, A2AAR and A3AR [19], show only partial agonist efficacy in βarr2 recruitment, in contrast to full agonism in cAMP signaling (Table 2; Figure 2A; Figure 3A). These compounds also have a low potency in βarr2 recruitment (logEC50s of around −5.8), although R-PIA is more potent than S-PIA (logEC50 of −6.35), as is also the case in cAMP signalling (Table 2; Figure 3B). The 2-chloro analogue of N6-cyclopentyladenosine (CCPA) is a full antagonist in the cAMP assay, but a partial agonist in βarr2 recruitment (Emax of 34%; Table 2). The 1-deaza analogue of CCPA, DCCA, showed a further reduced 6% efficacy in βarr2 recruitment (Table 3), suggesting an importance of the 1-position ring nitrogen in βarr2-pathway activation. Furthermore, substitution of N6 with a benzyl or phenyl group instead of a cyclopropyl group (as in CPA) reduces or stimulates βarr2 recruitment, respectively (Table 3). The N6-chain elongated adenosine congener N6-[4-[[[4-[[[(2-aminoethyl)amino]carbonyl]methyl]anilino]-carbonyl]methyl]phenyl]adenosine (ADAC), an agonist with selectivity for human A1AR and A3AR, has been studied for enhanced A3AR-selectivity upon conjugation to a nanocarrier [64]. Amongst the ribose-5’-OH analogues, this compound performs substantially well in βarr2 recruitment (Table 2; Figure 3A). Overall, the N6-substituted adenine 9-riboside (4’-CH2OH) derivatives mentioned here all have intermediate potency for βarr2 recruitment (logEC50s of around −6) compared to reference agonists NECA or 2-Cl-IB-MECA (logEC50s of around −7).
A series of 5’-alkyluronamide adenosine derivatives was developed as selective A3AR agonists, and substitutions at the C2- and/or N6- positions were explored [20]. Focusing on the structure of NECA, these substitutions preserved efficacy and influenced potency of cAMP signalling. The A2AAR-selective agonist CGS21680 [61] contains an extended C2-substituent, decreasing the potency in cAMP signalling, by roughly one log unit (logEC50 from −7.42 to −6.62), as well as βarr2 recruitment (logEC50 from −6.99 to −5.87) (Table 2; Figure 2B). Introduction of a shorter methyluronamide group was reported to favour A3AR binding compared to larger alkyl groups. An N6-halobenzyl group maintains affinity at the A3AR while reducing A1AR and A2AAR affinity, to increase A3AR selectivity. Examples include the prototypical agonists IB-MECA [8] and 2-Cl-IB-MECA (Figure 2B; Figure 3, black curves) [21]. These agonists display high potency in both cAMP signalling and βarr2 recruitment (logEC50s of around −8.5 and −7.5, respectively) (Table 2). However, the combination of the N6-halobenzyl group in IB-MECA with a C2-substituent, as in 2-Cl-IB-MECA, reduces the Emax of βarr2 recruitment (Table 2; Figure 2B). Thus, for (5’-uronamide) adenosine derivatives, the Emax of βarr2 recruitment can be fine-tuned by substitution of the N6- and C2-positions.
Numerous highly A3AR-selective (N)-methanocarba nucleoside ligands were developed that include a rigid bicyclo[3.1.0]hexane ring system in place of the flexible ribose, stabilizing the favoured adenosine receptor-bound isomer conformation [22]. Combining this (N)-methanocarba modification with a 5’-uronamide group has been reported to yield a good efficacy and potency in A3AR activation, which are otherwise reduced when utilizing selectivity-enhancing N6- and/or C2-substituents [23, 24]. This was observed for MRS3558, which shows high efficacy and potency in both cAMP signalling and βarr2 recruitment (logEC50s of −9.42 and −8.12, respectively) (Table 2). Exchanging the 5’-uronamide group, as in the adenosine-like 4’-CH2OH derivative MRS1873 [19] and the truncated 4’-H MRS5127, drastically lowered the Emax and potency towards both signalling pathways (Table 2; Figure 2C). MRS5127 has well-suited antagonist-like properties for radioligand binding assays but partial agonist activity in some functional assays [25, 26, 65]. The N6-iodobenzyl group of MRS5127 tends to preserve potency more than, for example, the N6-dicyclopropylmethyl group of MRS5474 [27] in βarr2 recruitment as well as in cAMP signalling (Table 2; Figure 2C; Figure 3, blue curves).
In SAR studies with (N)-methanocarba 5’-uronamide derivatives, the A3AR binding site appeared to be very flexible in its ability to accommodate extended C2-substituents, such as C2-ethynyl and arylethynyl groups that further increased A3AR selectivity [8, 26, 28]. This is demonstrated with MRS5698, which contains a 3,4-difluorophenylethynyl group at the C2 position [28, 29], notwithstanding a somewhat reduced signalling potency compared to MRS3558 (Table 2; Figure 2D; Figure 3, red curves). To further evaluate C2-extension, the βarr2 activity profile of analogue MRS5679, containing a biphenyl substituent at the C2-position, was tested. Interestingly, this compound showed much lower potency (logEC50 of −5.92), and only half the Emax of MRS5698 (Table 3; Figure 2D). This might be explained by the characteristics of transmembrane helix 2 (TM2). When relying solely on the A2AAR structure as a template for the generation of an A3AR homology model, there are three cysteine bridges present in the extracellular regions, which restrict the flexibility of the TM helices. However, there is only one cysteine bridge present in the A3AR and the extracellular part of TM2 is expected to be more flexible. Sensibly, a hybrid model basing the TM2 conformation on the activated β2-adrenergic receptor and rhodopsin, better accommodated the ligands with extended C2-substituents [28, 30, 47]. The proposed outer displacement of TM2 logically requires overcoming a greater energy barrier, which might be reflected here in the lower potency of MRS5679. Once this has occurred, a maximal effect can still be obtained, at least comparable to that of 2-Cl-IB-MECA (47%). Known to favour human A3AR association [28, 30], the N6-methyl substitution in derivatives MRS5967 and MRS5663 also promoted βarr2 recruitment (Table 3; Figure 2D). Both compounds, only differing slightly at the ortho-position of the C2- phenylethynyl group, display equally good efficacy and potency as the highly potent MRS3558 (logEC50s of around −8.3). They have more pronounced activities than compounds bearing N6-benzyl substitutions, a modification that is mostly made to obtain species-independent A3AR selectivity. Thus, in contrast to ribose analogs, (N)-methanocarba 5’-uronamide adenosine derivatives seem to occupy the A3AR binding site in a manner that maintains efficacy for βarr2 recruitment when certain C2 modifications are present.
(N)-Methanocarba 5′-methyluronamide adenosine derivatives containing 2-arylethynyl groups have been screened in an in vivo mouse pain model, surpassing the effect of MRS5698 [30]. Compound MRS5917 showed very high potency in both cAMP signalling and βarr2 recruitment (logEC50 of −9.38 and −8.5, respectively) (Table 2; Figure 2D). By testing two additional 5-chlorothien-2-yl analogues, MRS5980 and MRS7154 [31], it was confirmed that a small methyl group at the N6-position favours βarr2 signalling (Table 3). MRS5917 and its arylethynyl congeners are highly promising orally active A3AR agonists for the treatment of chronic pain, displaying a prolonged in vivo effect [30], independent of endogenous opioid or endocannabinoid pathways [12]. In cell systems, the A3AR is subject to desensitization and downregulation [66], which is known to be correlated with βarr2 recruitment. However, protection in animal models of pain results from prolonged A3AR agonist action, which can be prevented by coadministration of an A3AR antagonist [12]. Despite potential A3AR downregulation, animal models have already revealed the persistent anti-inflammatory and anticancer effects of A3AR agonists. Thus, the roles of G-protein versus βarr2- recruitment signalling pathways and receptor desensitization in the downstream inhibition of key regulatory proteins involved in inflammation/tumour growth and in pain, i.e. the targeted mechanism of action, warrant further investigation [13].
From the structures of the above-mentioned compounds, some final SAR conclusions can be drawn. The 1-deaza analogue MRS7144 shows that the N1-group is more dispensable for βarr2 recruitment, although it is characterized by a somewhat reduced but still substantially good logEC50 of approximately −7.3 (Table 2; Figure 2E; Figure 3, purple curve). Thus, the more stabilized A3AR interaction of (N)-methanocarba 5′-methyluronamide adenosine derivatives tends to compensate for the loss of N1 in βarr2 recruitment, as well as cAMP signalling [31]. Also, the additionally tested 3-deaza analogue MRS7173 was fully efficacious in recruiting βarr2 (Table 3). Replacement of the 5′-N-methyluronamide with an ethyl ester group, as in the 7-deaza analogue MRS7299, completely abolishes cAMP signalling as well as βarr2 recruitment (Table 2; Figure 2E; Figure 3, bright blue curve) [33]. When re-introducing the amino group (MRS7232), βarr2 recruitment was partially restored (Table 3; Figure 2E). The necessity of N7 is a well-described feature in literature concerning A3AR binding activation, reflecting its proposed function as a H-bond acceptor with Asn250 (6.55). The same residue accepts a H-bond from the 6-amino group (bidentate ligand coordination). However, it was shown that a suitable combination of stabilizing interactions in these kind of hypermodified A3AR-selective ligands can partially compensate for the lack of an exocyclic amine, an otherwise important contributor to recognition in the A3AR binding site [32]. This is demonstrated by the sustained A3AR activity of MRS7195 and MRS7220 in cAMP signalling as well as in βarr2 recruitment (Table 2; Figure 2F; Figure 3, mustard curves), although MRS7220 has a substantially reduced Emax and logEC50 compared to those of other (N)-methanocarba 5′-methyluronamide derivatives.
In summary, the screening of a panel of synthetic nucleosides as A3AR ligands using a βarr2-recruitment assay, and comparison with the Gi-mediated cAMP-pathway, has provided us insight into ligand features that can be of meaning for future development of biased A3AR ligands. A next step will be to elucidate which of these pathways is key for a certain therapeutic profile that is (mostly) devoid of side effects; in doing so, the role of G-protein versus βarr2 signalling, as well as A3AR desensitization, remains to be determined for the different therapeutic applications.
ACKNOWLEDGMENTS
The authors acknowledge support from the Ghent University (Belgium) Special Research Council (BOF) for granting a Ph.D. fellowship to J. Storme (application number 01D31913), as well as the NIDDK Intramural Research Program (ZIADK31117). Edward Will is acknowledged for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Borea PA, Gessi S, Merighi S, Varani K. Adenosine as a Multi-Signalling Guardian Angel in Human Diseases: When, Where and How Does it Exert its Protective Effects? Trends in Pharmacological Sciences 2016;37:419–34. [DOI] [PubMed] [Google Scholar]
- [2].Liu H, Xia Y. Beneficial and detrimental role of adenosine signaling in diseases and therapy. J Appl Physiol 2015;119:1173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Peleli M, Fredholm BB, Sobrevia L, Carlstrom M. Pharmacological targeting of adenosine receptor signaling. Mol Aspects Med 2017;55:4–8. [DOI] [PubMed] [Google Scholar]
- [4].Borea PA, Varani K, Vincenzi F, Baraldi PG, Tabrizi MA, Merighi S, et al. The A3 adenosine receptor: history and perspectives. Pharmacol Rev 2015;67:74–102. [DOI] [PubMed] [Google Scholar]
- [5].Burnstock G Purinergic Signalling: Therapeutic Developments. Front Pharmacol 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tian YK, Marshall M, French BA, Linden J, Yang ZQ. The infarct-sparing effect of IB-MECA against myocardial ischemia/reperfusion injury in mice is mediated by sequential activation of adenosine A(3) and A(2A) receptors. Basic Res Cardiol 2015;110. [DOI] [PubMed] [Google Scholar]
- [7].Cronstein BN, Sitkovsky M. Adenosine and adenosine receptors in the pathogenesis and treatment of rheumatic diseases. Nat Rev Rheumatol 2017;13:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jacobson KA, Merighi S, Varani K, Borea PA, Baraldi S, Aghazadeh Tabrizi M, et al. A3 Adenosine Receptors as Modulators of Inflammation: From Medicinal Chemistry to Therapy. Med Res Rev 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gessi S, Merighi S, Sacchetto V, Simioni C, Borea PA. Adenosine receptors and cancer. Biochim Biophys Acta 2011;1808:1400–12. [DOI] [PubMed] [Google Scholar]
- [10].Di Virgilio F, Adinolfi E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 2017;36:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen Z, Janes K, Chen C, Doyle T, Bryant L, Tosh DK, et al. Controlling murine and rat chronic pain through A3 adenosine receptor activation. FASEB J 2012;26:1855–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Little JW, Ford A, Symons-Liguori AM, Chen Z, Janes K, Doyle T, et al. Endogenous adenosine A3 receptor activation selectively alleviates persistent pain states. Brain 2015;138:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Janes K, Symons-Liguori AM, Jacobson KA, Salvemini D. Identification of A3 adenosine receptor agonists as novel non-narcotic analgesics. Br J Pharmacol 2016;173:1253–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen JF, Eltzschig HK, Fredholm BB. Adenosine receptors as drug targets--what are the challenges? Nat Rev Drug Discov 2013;12:265–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schmidt J, Ferk P. Safety issues of compounds acting on adenosinergic signalling. J Pharm Pharmacol 2017;69:790–806. [DOI] [PubMed] [Google Scholar]
- [16].Rankovic Z, Brust TF, Bohn LM. Biased agonism: An emerging paradigm in GPCR drug discovery. Bioorg Med Chem Lett 2016;26:241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bologna Z, Teoh JP, Bayoumi AS, Tang Y, Kim IM. Biased G Protein-Coupled Receptor Signaling: New Player in Modulating Physiology and Pathology. Biomol Ther (Seoul) 2017;25:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Storme J, Cannaert A, Van Craenenbroeck K, Stove CP. Molecular dissection of the human A3 adenosine receptor coupling with β-arrestin2. Biochemical pharmacology 2018;148:298–307. [DOI] [PubMed] [Google Scholar]
- [19].Gao ZG, Kim SK, Biadatti T, Chen W, Lee K, Barak D, et al. Structural determinants of A(3) adenosine receptor activation: nucleoside ligands at the agonist/antagonist boundary. J Med Chem 2002;45:4471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gallo-Rodriguez C, Ji XD, Melman N, Siegman BD, Sanders LH, Orlina J, et al. Structure-Activity-Relationships of N6-Benzyladenosine-5’-Uronamides as A(3)-Selective Adenosine Agonists. Journal of Medicinal Chemistry 1994;37:636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim HO, Ji XD, Siddiqi SM, Olah ME, Stiles GL, Jacobson KA. 2-Substitution of N6-Benzyladenosine-5’-Uronamides Enhances Selectivity for A3 Adenosine Receptors. Journal of Medicinal Chemistry 1994;37:3614–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jacobson KA, Ji X, Li AH, Melman N, Siddiqui MA, Shin KJ, et al. Methanocarba analogues of purine nucleosides as potent and selective adenosine receptor agonists. J Med Chem 2000;43:2196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tchilibon S, Joshi BV, Kim SK, Duong HT, Gao ZG, Jacobson KA. (N)-methanocarba 2,N6-disubstituted adenine nucleosides as highly potent and selective A3 adenosine receptor agonists. J Med Chem 2005;48:1745–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Melman A, Gao ZG, Kumar D, Wan TC, Gizewski E, Auchampach JA, et al. Design of (N)-methanocarba adenosine 5’-uronamides as species-independent A3 receptor-selective agonists. Bioorg Med Chem Lett 2008;18:2813–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Melman A, Wang B, Joshi BV, Gao ZG, Castro S, Heller CL, et al. Selective A(3) adenosine receptor antagonists derived from nucleosides containing a bicyclo[3.1.0]hexane ring system. Bioorg Med Chem 2008;16:8546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tosh DK, Chinn M, Ivanov AA, Klutz AM, Gao ZG, Jacobson KA. Functionalized congeners of A3 adenosine receptor-selective nucleosides containing a bicyclo[3.1.0]hexane ring system. J Med Chem 2009;52:7580–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tosh DK, Paoletta S, Deflorian F, Phan K, Moss SM, Gao ZG, et al. Structural sweet spot for A1 adenosine receptor activation by truncated (N)-methanocarba nucleosides: receptor docking and potent anticonvulsant activity. J Med Chem 2012;55:8075–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tosh DK, Deflorian F, Phan K, Gao ZG, Wan TC, Gizewski E, et al. Structure-guided design of A(3) adenosine receptor-selective nucleosides: combination of 2-arylethynyl and bicyclo[3.1.0]hexane substitutions. J Med Chem 2012;55:4847–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tosh DK, Padia J, Salvemini D, Jacobson KA. Efficient, large-scale synthesis and preclinical studies of MRS5698, a highly selective A3 adenosine receptor agonist that protects against chronic neuropathic pain. Purinergic signalling 2015;11:371–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tosh DK, Finley A, Paoletta S, Moss SM, Gao ZG, Gizewski ET, et al. In vivo phenotypic screening for treating chronic neuropathic pain: modification of C2-arylethynyl group of conformationally constrained A3 adenosine receptor agonists. J Med Chem 2014;57:9901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tosh DK, Crane S, Chen Z, Paoletta S, Gao ZG, Gizewski E, et al. Rigidified A3 Adenosine Receptor Agonists: 1-Deazaadenine Modification Maintains High in Vivo Efficacy. ACS Med Chem Lett 2015;6:804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tosh DK, Ciancetta A, Warnick E, O’Connor R, Chen ZM, Gizewski E, et al. Purine (N)-Methanocarba Nucleoside Derivatives Lacking an Exocyclic Amine as Selective A(3) Adenosine Receptor Agonists. Journal of Medicinal Chemistry 2016;59:3249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tosh DK, Ciancetta A, Mannes P, Warnick E, Janowsky A, Eshleman AJ; Gizewski E, Brust TF, Bohn LM, Auchampach JA, Gao ZG, Jacobson KA . Repurposing of a nucleoside scaffold from adenosine receptor agonists to opioid receptor antagonists. ACS Omega in press, 10.1021/acsomega.8b01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nordstedt C, Fredholm BB. A modification of a protein-binding method for rapid quantification of cAMP in cell-culture supernatants and body fluid. Anal Biochem 1990;189:231–4. [DOI] [PubMed] [Google Scholar]
- [35].Post SR, Ostrom RS, Insel PA. Biochemical methods for detection and measurement of cyclic AMP and adenylyl cyclase activity. Methods Mol Biol 2000;126:363–74. [DOI] [PubMed] [Google Scholar]
- [36].Olah ME, Gallo-Rodriguez C, Jacobson KA, Stiles GL. 125I-4-Aminobenzyl-5’-N-Methylcarboxamidoadenosine, a High-Affinity Radioligand for the Rat A(3) Adenosine Receptor. Molecular pharmacology 1994;45:978–82. [PMC free article] [PubMed] [Google Scholar]
- [37].Cheng Y, Prusoff WH. Relationship between Inhibition Constant (K1) and Concentration of Inhibitor Which Causes 50 Per Cent Inhibition (I50) of an Enzymatic-Reaction. Biochemical pharmacology 1973;22:3099–108. [DOI] [PubMed] [Google Scholar]
- [38].David M, Gospodinov DK, Gheorghe N, Mateev GS, Rusinova MV, Hristakieva E, et al. Treatment of Plaque-Type Psoriasis With Oral CF101: Data from a Phase II/III Multicenter, Randomized, Controlled Trial. J Drugs Dermatol 2016;15:931–8. [PubMed] [Google Scholar]
- [39].Stemmer SM, Benjaminov O, Medalia G, Ciuraru NB, Silverman MH, Bar-Yehuda S, et al. CF102 for the treatment of hepatocellular carcinoma: a phase I/II, open-label, dose-escalation study. Oncologist 2013;18:25–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shen YC, Lin ZZ, Hsu CH, Hsu C, Shao YY, Cheng AL. Clinical trials in hepatocellular carcinoma: an update. Liver Cancer 2013;2:345–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Muller CE. Medicinal chemistry of adenosine A3 receptor ligands. Curr Top Med Chem 2003;3:445–62. [DOI] [PubMed] [Google Scholar]
- [42].Jacobson KA, Klutz AM, Tosh DK, Ivanov AA, Preti D, Baraldi PG. Medicinal chemistry of the A3 adenosine receptor: agonists, antagonists, and receptor engineering. Handb Exp Pharmacol 2009:123–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Baraldi PG, Preti D, Borea PA, Varani K. Medicinal chemistry of A(3) adenosine receptor modulators: pharmacological activities and therapeutic implications. J Med Chem 2012;55:5676–703. [DOI] [PubMed] [Google Scholar]
- [44].Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 2008;322:1211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Glukhova A, Thal DM, Nguyen AT, Vecchio EA, Jorg M, Scammells PJ, et al. Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity. Cell 2017;168:867–77 e13. [DOI] [PubMed] [Google Scholar]
- [46].Cheong SL, Federico S, Venkatesan G, Mandel AL, Shao YM, Moro S, et al. The A3 adenosine receptor as multifaceted therapeutic target: pharmacology, medicinal chemistry, and in silico approaches. Med Res Rev 2013;33:235–335. [DOI] [PubMed] [Google Scholar]
- [47].Ciancetta A, Jacobson KA. Structural Probing and Molecular Modeling of the A(3) Adenosine Receptor: A Focus on Agonist Binding. Molecules 2017;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ciancetta A, Jacobson KA. Breakthrough in GPCR Crystallography and Its Impact on Computer-Aided Drug Design. Methods Mol Biol 2018;1705:45–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. μ-opioid receptor desensitization by β-arrestin-2 determines morphine tolerance but not dependence. Nature 2000;408:720–3. [DOI] [PubMed] [Google Scholar]
- [50].Groer CE, Tidgewell K, Moyer RA, Harding WW, Rothman RB, Prisinzano TE, et al. An opioid agonist that does not induce μ-opioid receptor--arrestin interactions or receptor internalization. Molecular pharmacology 2007;71:549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Siuda ER, Carr R, 3rd, Rominger DH, Violin JD. Biased μ-opioid receptor ligands: a promising new generation of pain therapeutics. Curr Opin Pharmacol 2017;32:77–84. [DOI] [PubMed] [Google Scholar]
- [52].Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, et al. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci U S A 2003;100:10782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, et al. Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther 2010;335:572–9. [DOI] [PubMed] [Google Scholar]
- [54].Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, et al. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci U S A 2007;104:16657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Urs NM, Peterson SM, Caron MG. New Concepts in Dopamine D2 Receptor Biased Signaling and Implications for Schizophrenia Therapy. Biol Psychiatry 2017;81:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Verzijl D, Ijzerman AP. Functional selectivity of adenosine receptor ligands. Purinergic signalling 2011;7:171–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gao ZG, Balasubramanian R, Kiselev E, Wei Q, Jacobson KA. Probing biased/partial agonism at the G protein-coupled A(2B) adenosine receptor. Biochemical pharmacology 2014;90:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baltos JA, Gregory KJ, White PJ, Sexton PM, Christopoulos A, May LT. Quantification of adenosine A(1) receptor biased agonism: Implications for drug discovery. Biochemical pharmacology 2016;99:101–12. [DOI] [PubMed] [Google Scholar]
- [59].Baltos JA, Paoletta S, Nguyen AT, Gregory KJ, Tosh DK, Christopoulos A, et al. Structure-Activity Analysis of Biased Agonism at the Human Adenosine A3 Receptor. Molecular pharmacology 2016;90:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Baltos JA, Vecchio EA, Harris MA, Qin CX, Ritchie RH, Christopoulos A, et al. Capadenoson, a clinically trialed partial adenosine A1 receptor agonist, can stimulate adenosine A2B receptor biased agonism. Biochemical pharmacology 2017;135:79–89. [DOI] [PubMed] [Google Scholar]
- [61].Gao ZG, Jacobson KA. Translocation of arrestin induced by human A(3) adenosine receptor ligands in an engineered cell line: comparison with G protein-dependent pathways. Pharmacological research 2008;57:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gao ZG, Verzijl D, Zweemer A, Ye K, Goblyos A, Ijzerman AP, et al. Functionally biased modulation of A(3) adenosine receptor agonist efficacy and potency by imidazoquinolinamine allosteric enhancers. Biochemical pharmacology 2011;82:658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gao ZG, Blaustein JB, Gross AS, Melman N, Jacobson KA. N6-Substituted adenosine derivatives: selectivity, efficacy, and species differences at A3 adenosine receptors. Biochemical pharmacology 2003;65:1675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Klutz AM, Gao ZG, Lloyd J, Shainberg A, Jacobson KA. Enhanced A3 adenosine receptor selectivity of multivalent nucleoside-dendrimer conjugates. Journal of nanobiotechnology 2008;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Auchampach JA, Gizewski ET, Wan TC, de Castro S, Brown GG, Jr., Jacobson KA. Synthesis and pharmacological characterization of [(125)I]MRS5127, a high affinity, selective agonist radioligand for the A3 adenosine receptor. Biochemical pharmacology 2010;79:967–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Trincavelli ML, Tuscano D, Marroni M, Falleni A, Gremigni V, Ceruti S, et al. A(3) adenosine receptors in human astrocytoma cells: Agonist-mediated desensitization, internalization, and down-regulation. Molecular pharmacology 2002;62:1373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]